Impact of E484Q and L452R Mutations on Structure and Binding Behavior of SARS-CoV-2 B.1.617.1 Using Deep Learning AlphaFold2, Molecular Docking and Dynamics Simulation
Abstract
1. Introduction
2. Results
2.1. Structure Predictions of S Proteins for the WT and Double Mutant Variant
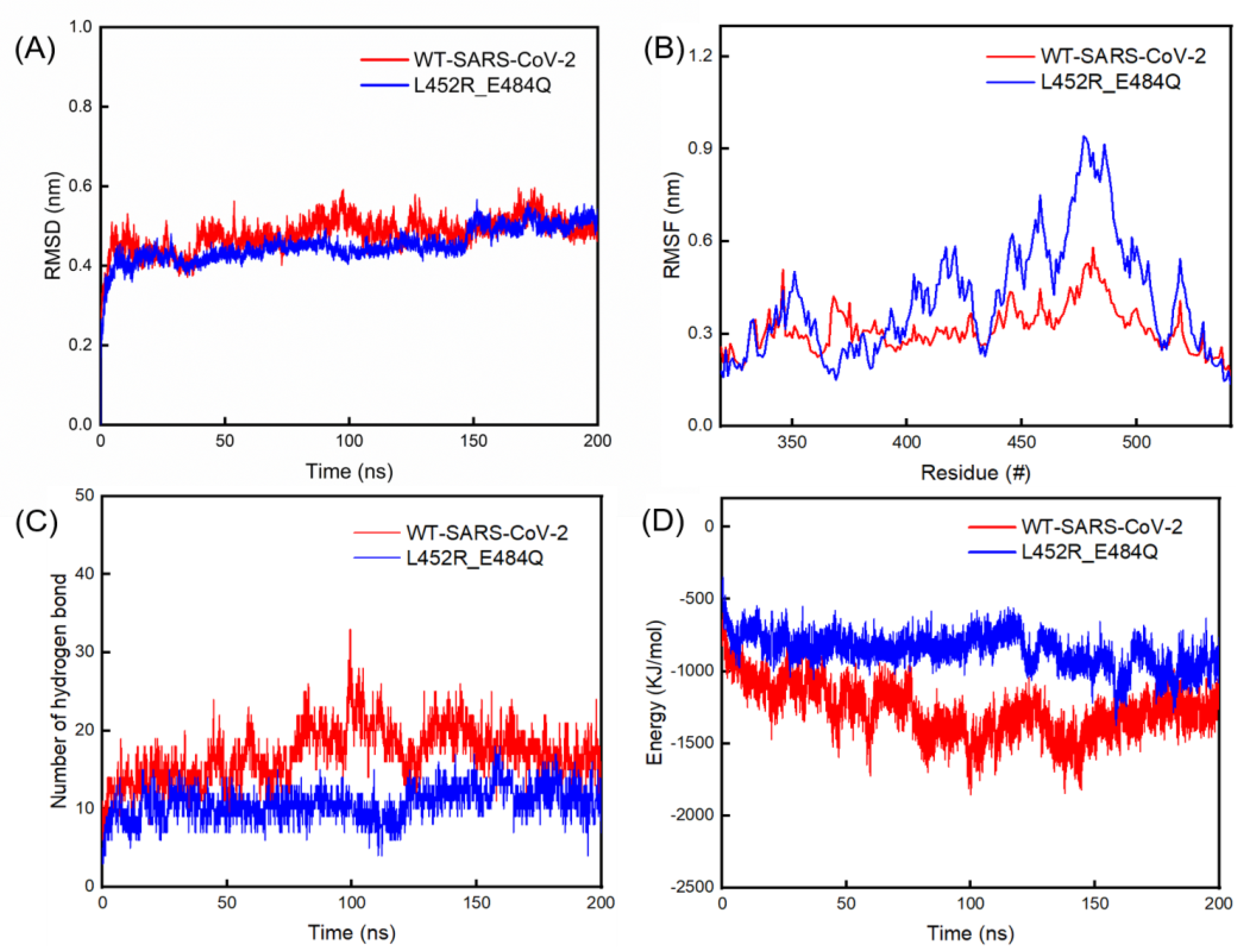
2.2. Results of RMSD and RMSF Values, Hydrogen Bonds and Interaction Energy via MD Simulations
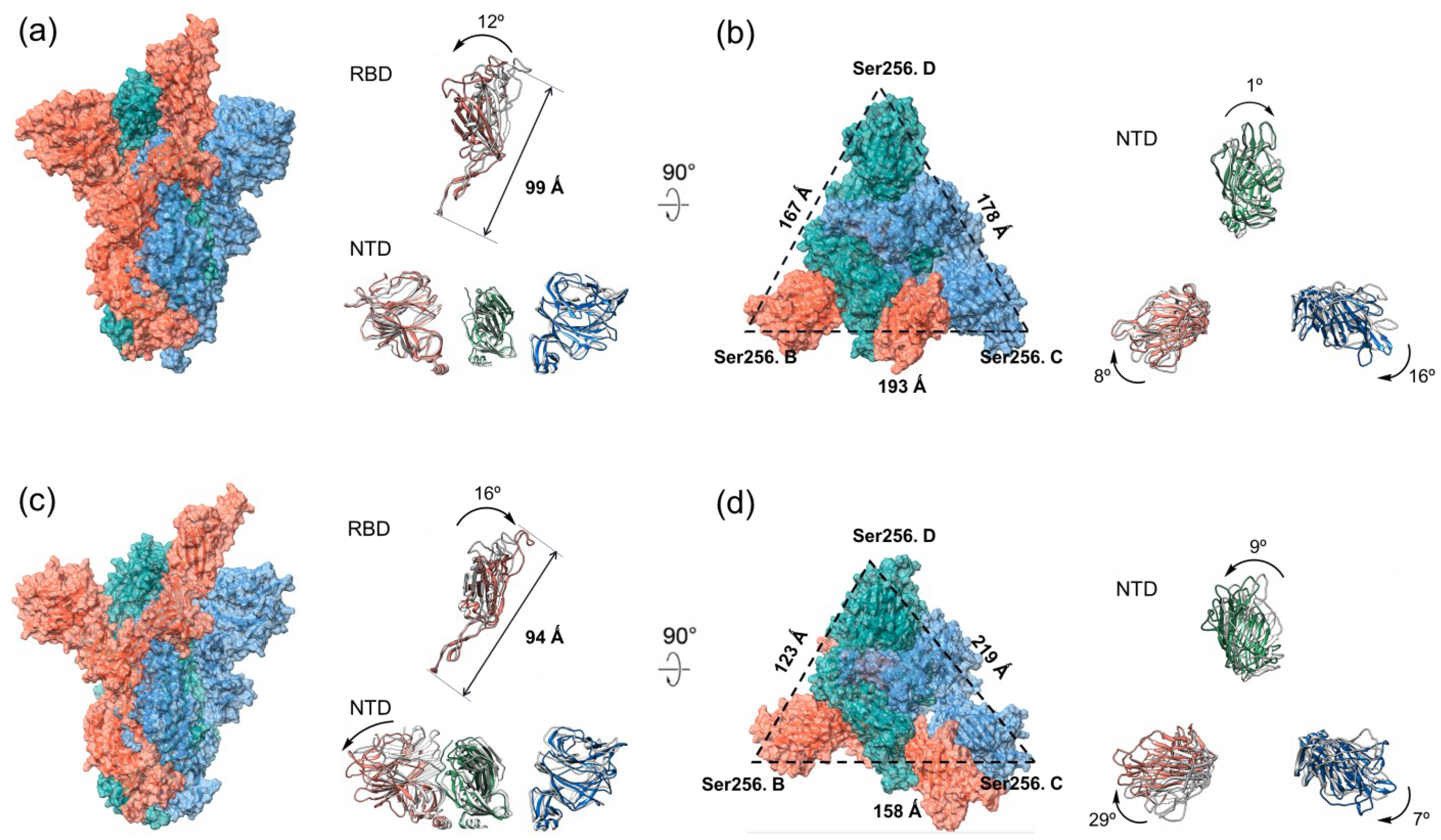
2.3. The Molecular Architectures of S Proteins of WT and Double Mutant Variant after 200 ns MD Simulations
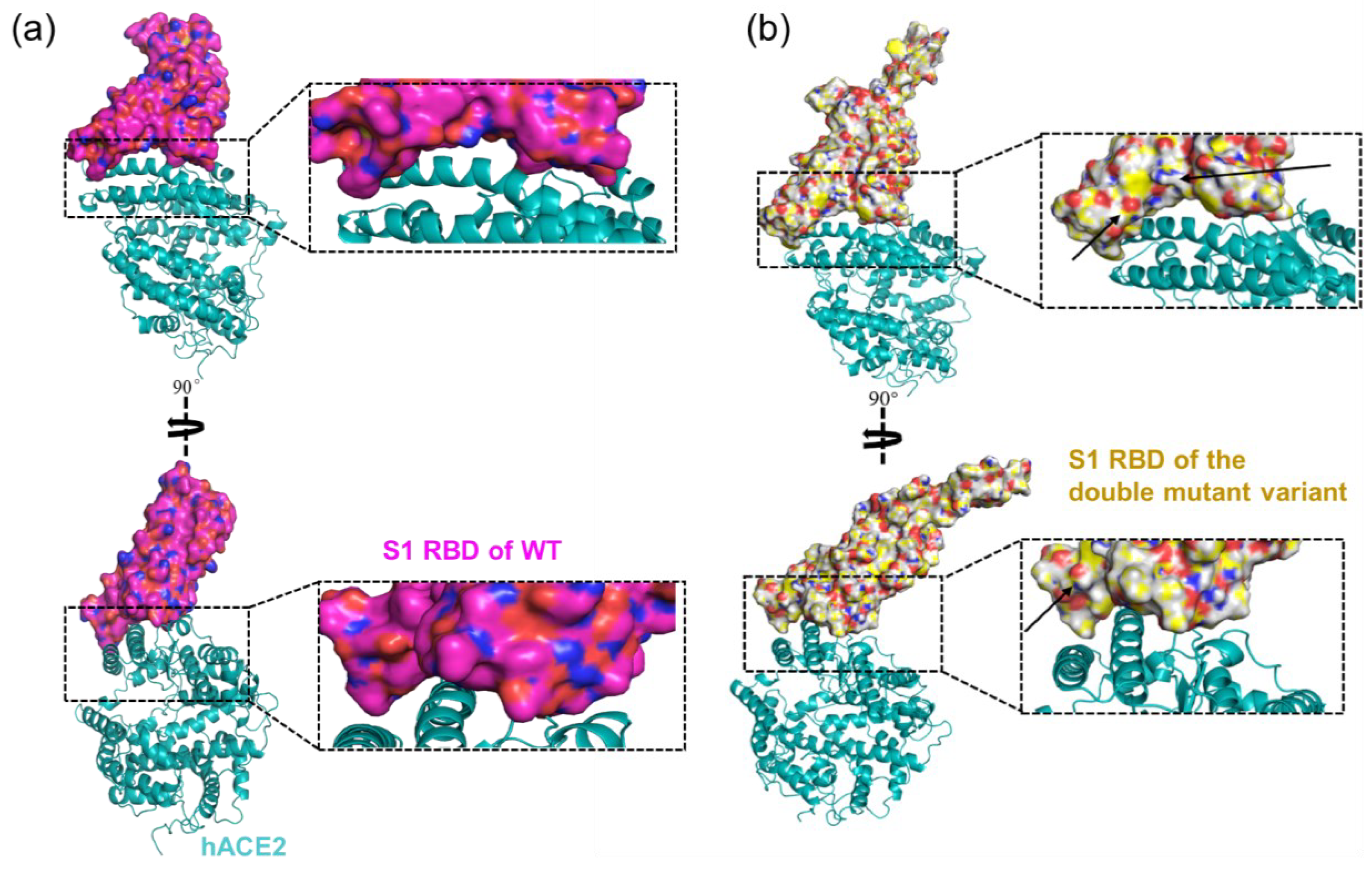
2.4. The Binding of Double Mutant Variant to hACE2 on Host Cells
2.5. The Binding of Double Mutant Variant to Neutralizing mAbs
3. Discussion
4. Materials and Methods
4.1. Structure Modeling with AlphaFold2
4.2. Calculation of the Similarity between Experimental and Predicted Structures
4.3. Molecular Dynamics Simulations
4.4. Molecular Docking Studies of hACE2 with S1 RBDs
4.5. Molecular Docking Studies of mAbs with S1 RBDs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharma, A.; Tiwari, S.; Deb, M.K.; Marty, J.L. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): A global pandemic and treatment strategies. Int. J. Antimicrob. Agents 2020, 56, 106054. [Google Scholar] [CrossRef] [PubMed]
- Dousari, A.S.; Moghadam, M.T.; Satarzadeh, N. COVID-19 (Coronavirus Disease 2019): A New Coronavirus Disease. Infect. Drug Resist. 2020, 13, 2819–2828. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Zheng, Q.; Zhang, H.; Niu, Y.; Lou, Y.; Wang, H. The SARS-CoV-2 Spike Glycoprotein Biosynthesis, Structure, Function, and Antigenicity: Implications for the Design of Spike-Based Vaccine Immunogens. Front. Immunol. 2020, 11, 576622. [Google Scholar] [CrossRef] [PubMed]
- Örd, M.; Faustova, I.; Loog, M. The sequence at Spike S1/S2 site enables cleavage by furin and phospho-regulation in SARS-CoV2 but not in SARS-CoV1 or MERS-CoV. Sci. Rep. 2020, 10, 16944. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell. Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, C.; Xu, X.-F.; Xu, W.; Liu, S.-W. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef]
- Krammer, F. SARS-CoV-2 vaccines in development. Nature 2020, 586, 516–527. [Google Scholar] [CrossRef]
- Excler, J.-L.; Saville, M.; Berkley, S.; Kim, J.H. Vaccine development for emerging infectious diseases. Nat. Med. 2021, 27, 591–600. [Google Scholar] [CrossRef]
- Harvey, W.T.; Carabelli, A.; Jackson, B.; Gupta, R.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Hwang, Y.-C.; Lu, R.-M.; Su, S.-C.; Chiang, P.-Y.; Ko, S.-H.; Ke, F.-Y.; Liang, K.-H.; Hsieh, T.-Y.; Wu, H.-C. Monoclonal antibodies for COVID-19 therapy and SARS-CoV-2 detection. J. Biomed. Sci. 2022, 29, 1. [Google Scholar] [CrossRef]
- Aleem, A.; Akbar Samad, A.B.; Slenker, A.K. Emerging Variants of SARS-CoV-2 And Novel Therapeutics Against Coronavirus (COVID-19). In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2022. [Google Scholar]
- McCallum, M.; Walls, A.C.; Sprouse, K.R.; Bowen, J.E.; Rosen, L.E.; Dang, H.V.; De Marco, A.; Franko, N.; Tilles, S.W.; Logue, J.; et al. Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science 2021, 374, 1621–1626. [Google Scholar] [CrossRef] [PubMed]
- England, P.H. SARS-CoV-2 Variants of Concern and Variants under Investigation in England: Technical Briefing 9. 22 April 2021. Available online: https://www.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variant-variant-of-concern-20201201 (accessed on 13 February 2023).
- Chavda, V.P.; Bezbaruah, R.; Deka, K.; Nongrang, L.; Kalita, T. The Delta and Omicron Variants of SARS-CoV-2: What We Know So Far. Vaccines 2022, 10, 1926. [Google Scholar] [CrossRef] [PubMed]
- Gobeil, S.M.-C.; Janowska, K.; McDowell, S.; Mansouri, K.; Parks, R.; Manne, K.; Stalls, V.; Kopp, M.F.; Henderson, R.; Edwards, R.J.; et al. D614G Mutation Alters SARS-CoV-2 Spike Conformation and Enhances Protease Cleavage at the S1/S2 Junction. Cell Rep. 2020, 34, 108630. [Google Scholar] [CrossRef]
- Liu, C.; Ginn, H.M.; Dejnirattisai, W.; Supasa, P.; Wang, B.; Tuekprakhon, A.; Nutalai, R.; Zhou, D.; Mentzer, A.J.; Zhao, Y.; et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell 2021, 184, 4220–4236.e13. [Google Scholar] [CrossRef] [PubMed]
- Vikkurthi, R.; Ansari, A.; Pai, A.R.; Jha, S.N.; Sachan, S.; Pandit, S.; Nikam, B.; Kalia, A.; Jit, B.P.; Parray, H.A.; et al. Inactivated whole-virion vaccine BBV152/Covaxin elicits robust cellular immune memory to SARS-CoV-2 and variants of concern. Nat. Microbiol. 2022, 7, 974–985. [Google Scholar] [CrossRef]
- Mlcochova, P.; Kemp, S.A.; Dhar, M.S.; Papa, G.; Meng, B.; Ferreira, I.A.T.M.; Datir, R.; Collier, D.A.; Albecka, A.; Singh, S.; et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 2021, 599, 114–119. [Google Scholar] [CrossRef]
- Raman, R.; Patel, K.J.; Ranjan, K. COVID-19: Unmasking Emerging SARS-CoV-2 Variants, Vaccines and Therapeutic Strategies. Biomolecules 2021, 11, 993. [Google Scholar] [CrossRef]
- Gobeil, S.M.-C.; Janowska, K.; McDowell, S.; Mansouri, K.; Parks, R.; Stalls, V.; Kopp, M.F.; Manne, K.; Li, D.; Wiehe, K.; et al. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation, and antigenicity. Science 2021, 373, eabi6226. [Google Scholar] [CrossRef]
- Lauring, A.S.; Hodcroft, E.B. Genetic Variants of SARS-CoV-2—What Do They Mean? JAMA 2021, 325, 529–531. [Google Scholar] [CrossRef]
- Dearlove, B.; Lewitus, E.; Bai, H.; Li, Y.; Reeves, D.B.; Joyce, M.G.; Scott, P.T.; Amare, M.F.; Vasan, S.; Michael, N.L.; et al. A SARS-CoV-2 vaccine candidate would likely match all currently circulating variants. Proc. Natl. Acad. Sci. USA 2020, 117, 23652–23662. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Marks, D.S.; Hopf, T.A.; Sander, C. Protein structure prediction from sequence variation. Nat. Biotechnol. 2012, 30, 1072–1080. [Google Scholar] [CrossRef] [PubMed]
- Zardecki, C.; Dutta, S.; Goodsell, D.S.; Voigt, M.; Burley, S.K. RCSB Protein Data Bank: A Resource for Chemical, Biochemical, and Structural Explorations of Large and Small Biomolecules. J. Chem. Educ. 2016, 93, 569–575. [Google Scholar] [CrossRef]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Žídek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef]
- Stevens, A.O.; He, Y. Benchmarking the Accuracy of AlphaFold 2 in Loop Structure Prediction. Biomolecules 2022, 12, 985. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics 2010, 26, 889–895. [Google Scholar] [CrossRef] [PubMed]
- Siew, N.; Elofsson, A.; Rychlewski, L.; Fischer, D. MaxSub: An automated measure for the assessment of protein structure prediction quality. Bioinformatics 2000, 16, 776–785. [Google Scholar] [CrossRef] [PubMed]
- Olechnovič, K.; Monastyrskyy, B.; Kryshtafovych, A.; Venclovas, Č. Comparative analysis of methods for evaluation of protein models against native structures. Bioinformatics 2019, 35, 937–944. [Google Scholar] [CrossRef]
- Yang, Q.; Jian, X.; Alamdar, A.; Syed, S.; Fahira, A.; Zheng, C.; Zhu, Z.; Wang, K.; Jinmai, Z.; Wen, Y.; et al. Structural Comparison and Drug Screening of Spike Proteins of Ten SARS-CoV-2 Variants. Research 2022, 2022, 9781758. [Google Scholar] [CrossRef]
- Sahihi, M.; Faraudo, J. Molecular Dynamics Simulations of Adsorption of SARS-CoV-2 Spike Protein on Polystyrene Surface. J. Chem. Inf. Model. 2022, 62, 3814–3824. [Google Scholar] [CrossRef]
- Rana, S.K.P.; Sharma, A.; Sharma, S.; Giri, R.; Ghosh, K.S. Identification of Naturally Occurring Antiviral Molecules for SARS-CoV-2 Mitigation. Open COVID J. 2021, 1, 38–46. [Google Scholar] [CrossRef]
- Shrestha, A.; Marahatha, R.; Basnet, S.; Regmi, B.P.; Katuwal, S.; Dahal, S.R.; Sharma, K.R.; Adhikari, A.; Basnyat, R.C.; Parajuli, N. Molecular Docking and Dynamics Simulation of Several Flavonoids Predict Cyanidin as an Effective Drug Candidate against SARS-CoV-2 Spike Protein. Adv. Pharmacol. Pharm. Sci. 2022, 2022, 3742318. [Google Scholar] [CrossRef] [PubMed]
- Shoemark, D.K.; Colenso, C.K.; Toelzer, C.; Gupta, K.; Sessions, R.B.; Davidson, A.D.; Berger, I.; Schaffitzel, C.; Spencer, J.; Mulholland, A.J. Molecular Simulations suggest Vitamins, Retinoids and Steroids as Ligands of the Free Fatty Acid Pocket of the SARS-CoV-2 Spike Protein. Angew. Chem. Int. Ed. 2021, 60, 7098–7110. [Google Scholar] [CrossRef]
- Dominguez, C.; Boelens, R.; Bonvin, A.M.J.J. HADDOCK: A Protein−Protein Docking Approach Based on Biochemical or Biophysical Information. J. Am. Chem. Soc. 2003, 125, 1731–1737. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, D.; Zhou, P.; Li, B.; Huang, S.-Y. HDOCK: A web server for protein–protein and protein–DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017, 45, W365–W373. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Fowler, N.J.; Williamson, M.P. The accuracy of protein structures in solution determined by AlphaFold and NMR. Structure 2022, 30, 925–933.e2. [Google Scholar] [CrossRef]
- Zhang, J.; Xiao, T.; Cai, Y.; Chen, B. Structure of SARS-CoV-2 spike protein. Curr. Opin. Virol. 2021, 50, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, C.; Wang, Y.; Hong, Q.; Zhang, C.; Li, Z.; Xu, S.; Zuo, Q.; Liu, C.; Huang, Z.; et al. Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM. Nat. Commun. 2021, 12, 7345. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Dejnirattisai, W.; Supasa, P.; Liu, C.; Mentzer, A.J.; Ginn, H.M.; Zhao, Y.; Duyvesteyn, H.M.; Tuekprakhon, A.; Nutalai, R.; et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell 2021, 184, 2348–2361.e6. [Google Scholar] [CrossRef]
- Nabel, K.G.; Clark, S.A.; Shankar, S.; Pan, J.; Clark, L.E.; Yang, P.; Coscia, A.; McKay, L.G.A.; Varnum, H.H.; Brusic, V.; et al. Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain. Science 2022, 375, eabl6251. [Google Scholar] [CrossRef]
- Rodriguez-Rivas, J.; Croce, G.; Muscat, M.; Weigt, M. Epistatic models predict mutable sites in SARS-CoV-2 proteins and epitopes. Proc. Natl. Acad. Sci. USA 2022, 119, e2113118119. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Singh, J.; Hasnain, S.E.; Sundar, D. Possible Link between Higher Transmissibility of Alpha, Kappa and Delta Variants of SARS-CoV-2 and Increased Structural Stability of Its Spike Protein and hACE2 Affinity. Int. J. Mol. Sci. 2021, 22, 9131. [Google Scholar] [CrossRef] [PubMed]
- Yi, C.; Sun, X.; Lin, Y.; Gu, C.; Ding, L.; Lu, X.; Yang, Z.; Zhang, Y.; Ma, L.; Gu, W.; et al. Comprehensive mapping of binding hot spots of SARS-CoV-2 RBD-specific neutralizing antibodies for tracking immune escape variants. Genome Med. 2021, 13, 164. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Driesch, L.V.D.; Galiez, C.; Martin, M.-J.; Söding, J.; Steinegger, M. Uniclust databases of clustered and deeply annotated protein sequences and alignments. Nucleic Acids Res. 2017, 45, D170–D176. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wang, Y.; Liu, C.; Zhang, C.; Han, W.; Hong, X.; Wang, Y.; Hong, Q.; Wang, S.; Zhao, Q.; et al. Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM. Sci. Adv. 2021, 7, eabe5575. [Google Scholar] [CrossRef]
- Yuan, S.; Chan, H.S.; Hu, Z. Using PyMOL as a platform for computational drug design. WIREs Comput. Mol. Sci. 2017, 7, e1298. [Google Scholar] [CrossRef]
- Zhang, Y.; Skolnick, J. Scoring function for automated assessment of protein structure template quality. Proteins Struct. Funct. Bioinform. 2004, 57, 702–710. [Google Scholar] [CrossRef]
- Zhang, Y.; Skolnick, J. TM-Align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005, 33, 2302–2309. [Google Scholar] [CrossRef]
- Hegedűs, T.; Geisler, M.; Lukács, G.L.; Farkas, B. Ins and outs of AlphaFold2 transmembrane protein structure predictions. Cell. Mol. Life Sci. 2022, 79, 73. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins Struct. Funct. Bioinform. 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Mark, P.; Nilsson, L. Structure and Dynamics of the TIP3P, SPC, and SPC/E Water Models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]


| WT-hACE2 | Double Mutant Variant-hACE2 | |
|---|---|---|
| HADDOCK score | −116.1 ± 1.6 | −123.7 ± 4.0 |
| RMSD from the overall lowest energy | 6.5 ± 0.2 | 0.8 ± 0.5 |
| Van der Waals energy | −56.4 ± 3.0 | −63.6 ± 4.4 |
| Electrostatic energy | −221.5 ± 11.0 | −248.1 ± 29.2 |
| Buried surface area (Å2) | 1710.8 ± 41.9 | 1834.8 ± 27.4 |
| Z-score | −1.3 | −2.1 |
| HDOCK docking score | −310.19 | −347.29 |
| Ligand RMSD (Å) | 3.27 | 1.94 |
| Antibodies | S Protein | Docking Scores | |
|---|---|---|---|
| HDOCK | HADDOCK | ||
| Bamlanvimab | WT | −125.996 | −275.49 |
| Double mutant variant | −107.493 | −181.94 | |
| Casirivimab | WT | −121.918 | −407.74 |
| Double mutant variant | −94.374 | −94.374 | |
| Etesevimab | WT | −123.423 | −263.07 |
| Double mutant variant | −108.941 | −184.8 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, Y.; Xing, Y.; Sun, Y. Impact of E484Q and L452R Mutations on Structure and Binding Behavior of SARS-CoV-2 B.1.617.1 Using Deep Learning AlphaFold2, Molecular Docking and Dynamics Simulation. Int. J. Mol. Sci. 2023, 24, 11564. https://doi.org/10.3390/ijms241411564
Jiao Y, Xing Y, Sun Y. Impact of E484Q and L452R Mutations on Structure and Binding Behavior of SARS-CoV-2 B.1.617.1 Using Deep Learning AlphaFold2, Molecular Docking and Dynamics Simulation. International Journal of Molecular Sciences. 2023; 24(14):11564. https://doi.org/10.3390/ijms241411564
Chicago/Turabian StyleJiao, Yanqi, Yichen Xing, and Yao Sun. 2023. "Impact of E484Q and L452R Mutations on Structure and Binding Behavior of SARS-CoV-2 B.1.617.1 Using Deep Learning AlphaFold2, Molecular Docking and Dynamics Simulation" International Journal of Molecular Sciences 24, no. 14: 11564. https://doi.org/10.3390/ijms241411564
APA StyleJiao, Y., Xing, Y., & Sun, Y. (2023). Impact of E484Q and L452R Mutations on Structure and Binding Behavior of SARS-CoV-2 B.1.617.1 Using Deep Learning AlphaFold2, Molecular Docking and Dynamics Simulation. International Journal of Molecular Sciences, 24(14), 11564. https://doi.org/10.3390/ijms241411564







