Molecular Characterization of Galectin-3 in Large Yellow Croaker Larimichthys crocea Functioning in Antibacterial Activity
Abstract
1. Introduction
2. Results
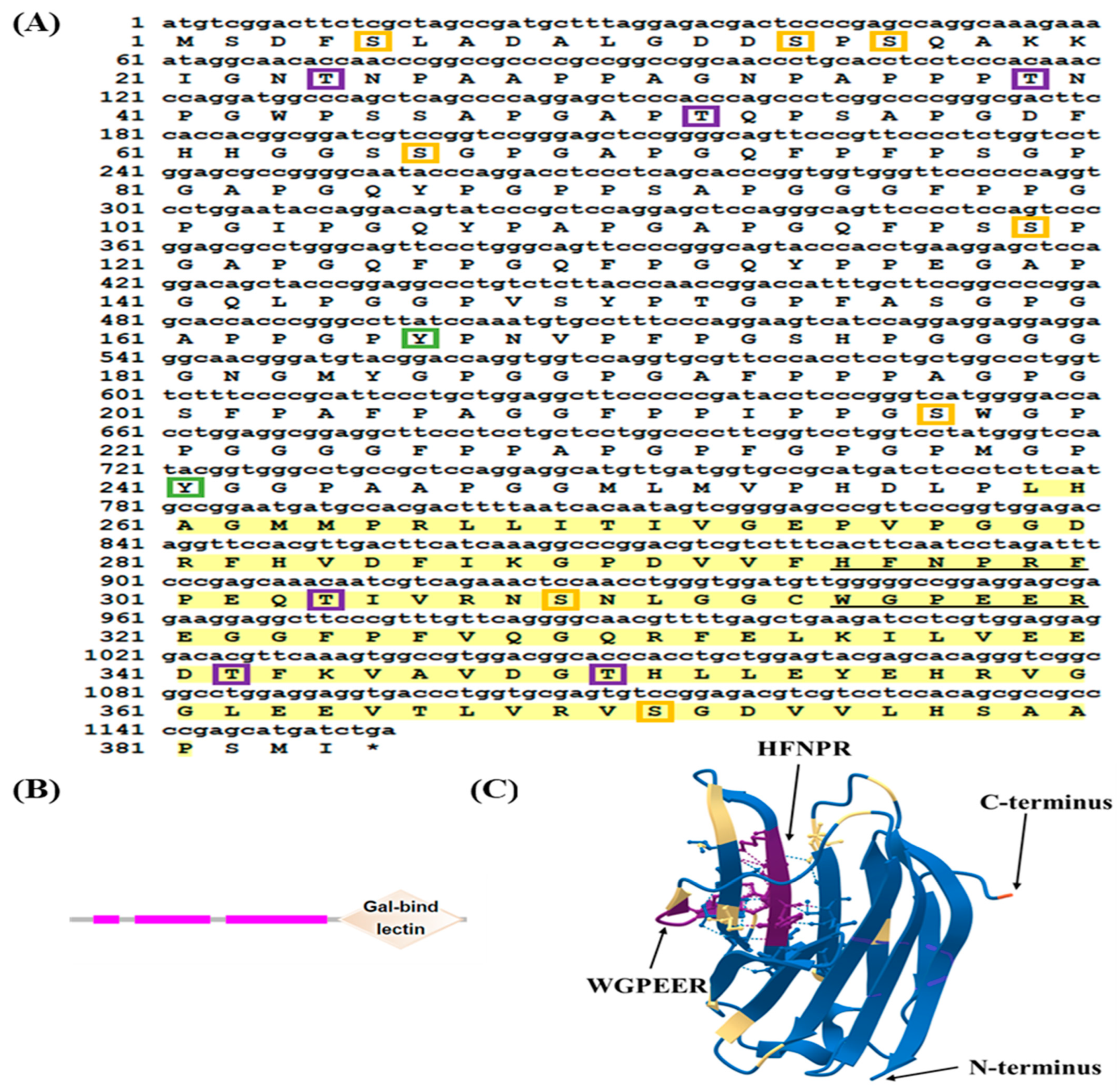
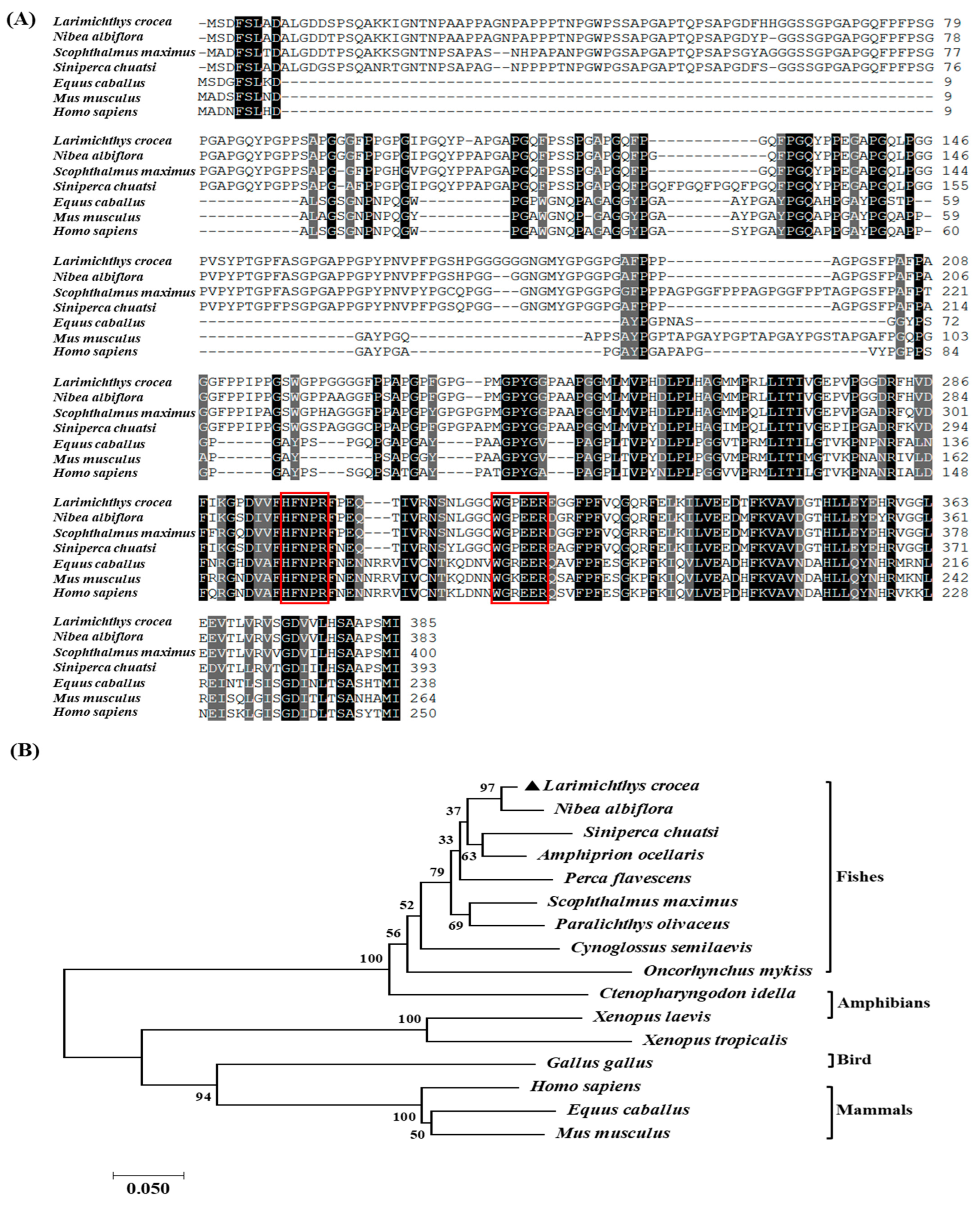
2.1. Sequence Characteristics and Bioinformatics Analysis of LcGal-3
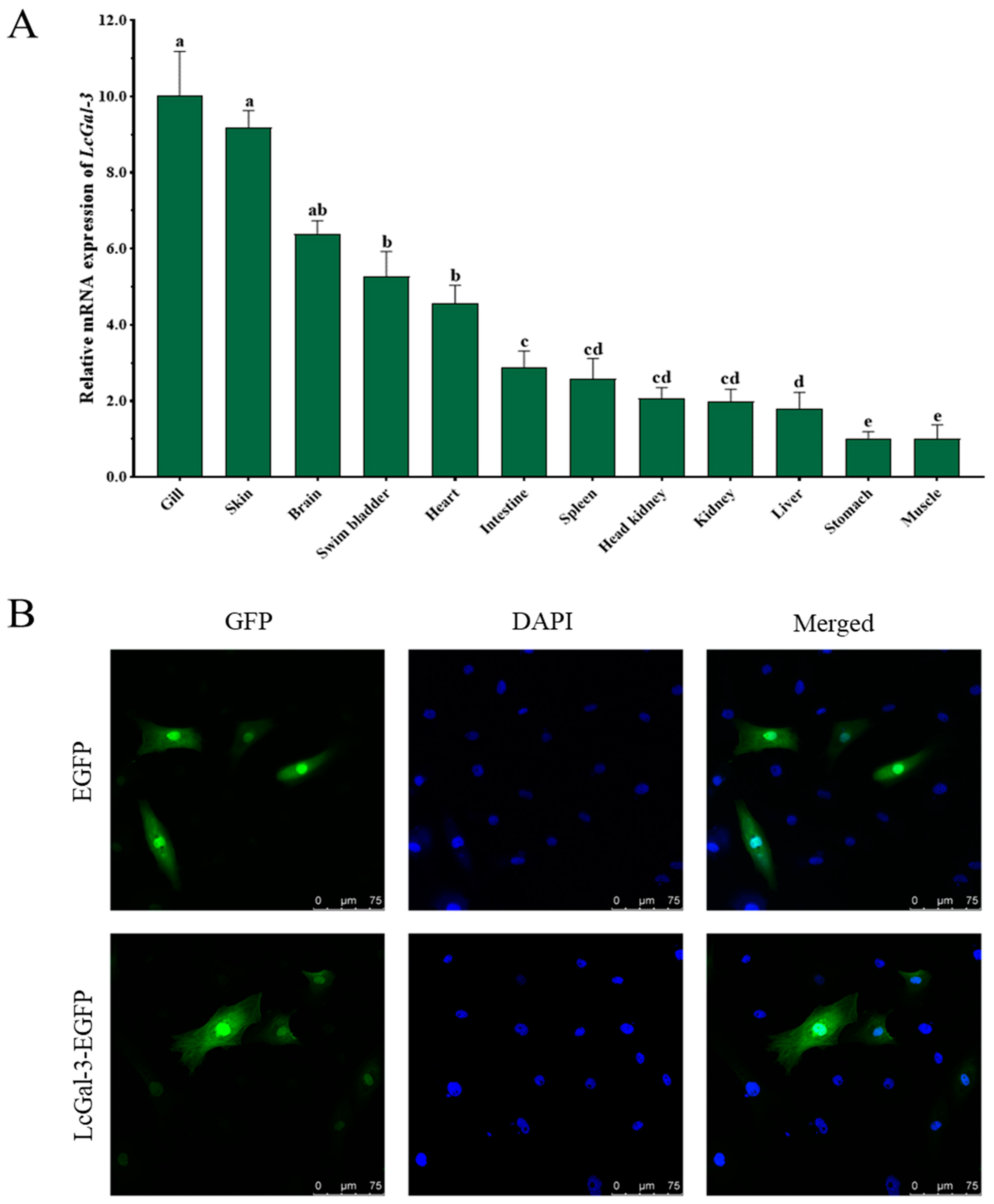
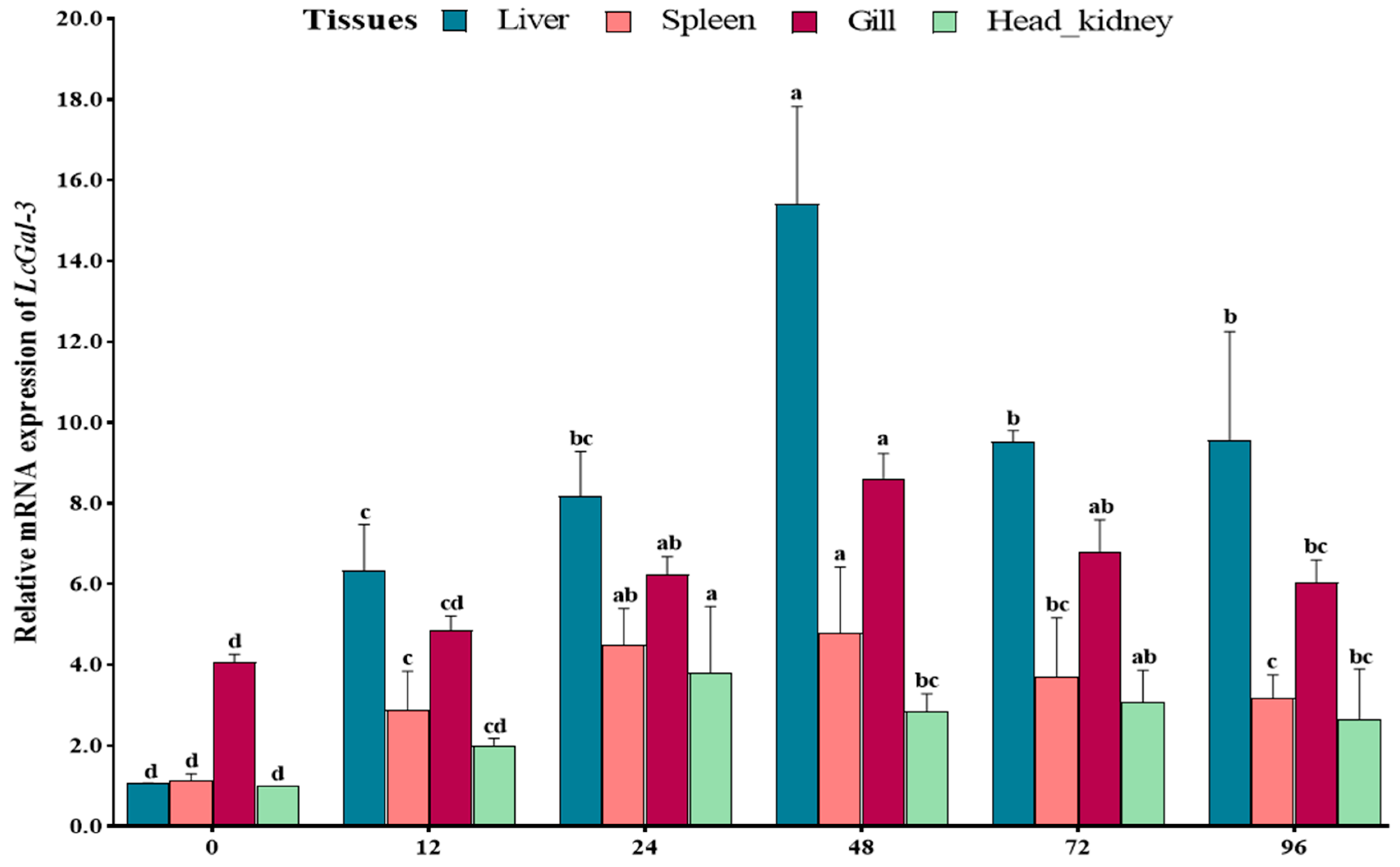
2.2. Tissue Distribution of LcGal-3 and Subcellular Localization of LcGal-3
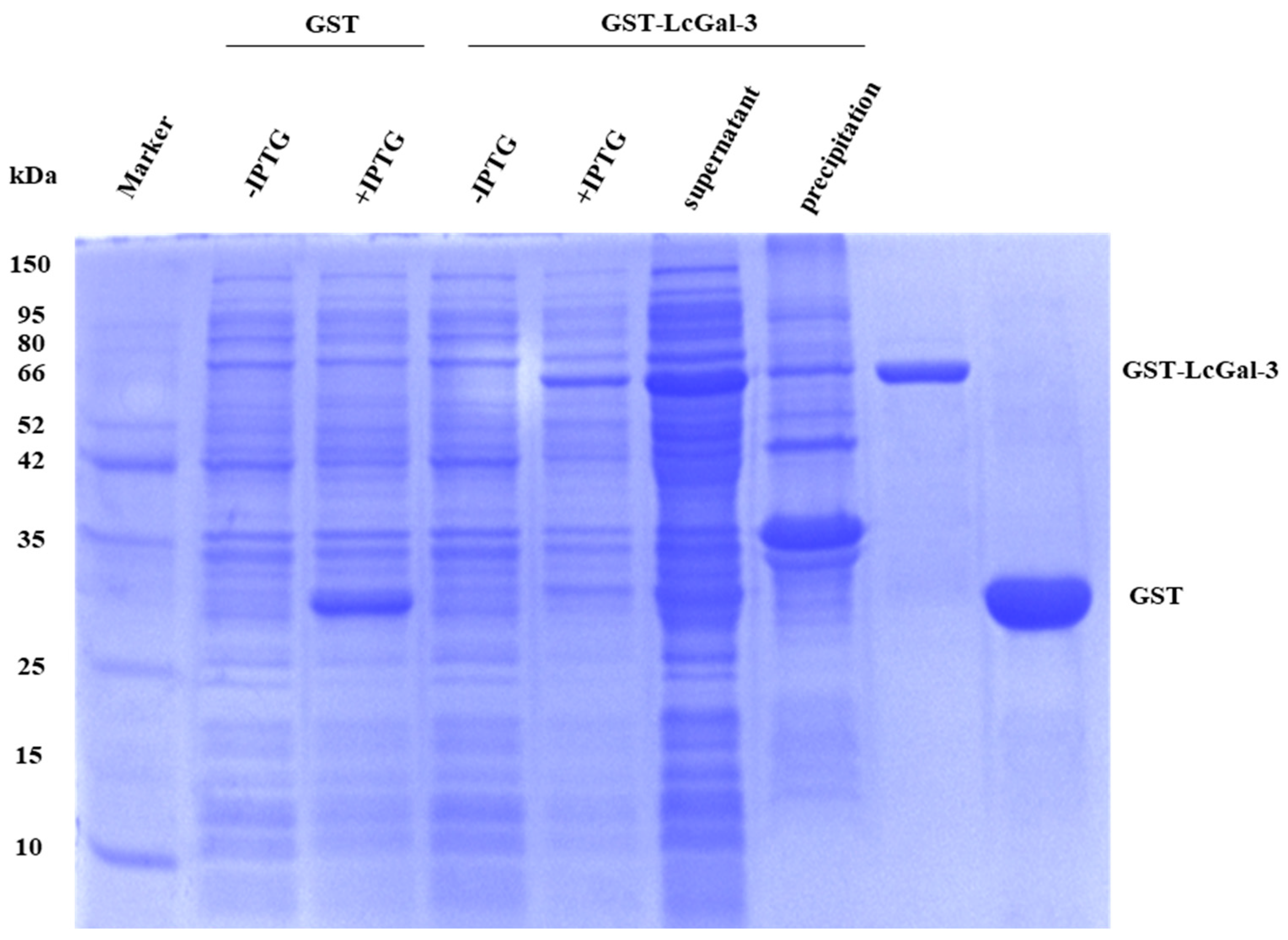
2.3. Prokaryotic Expression and Purification of LcGal-3 Protein
2.4. Hemagglutination and Sugar Inhibition Assays of LcGal-3 Protein
2.5. Defense Response of LcGal-3 against P. plecoglossicida Infection
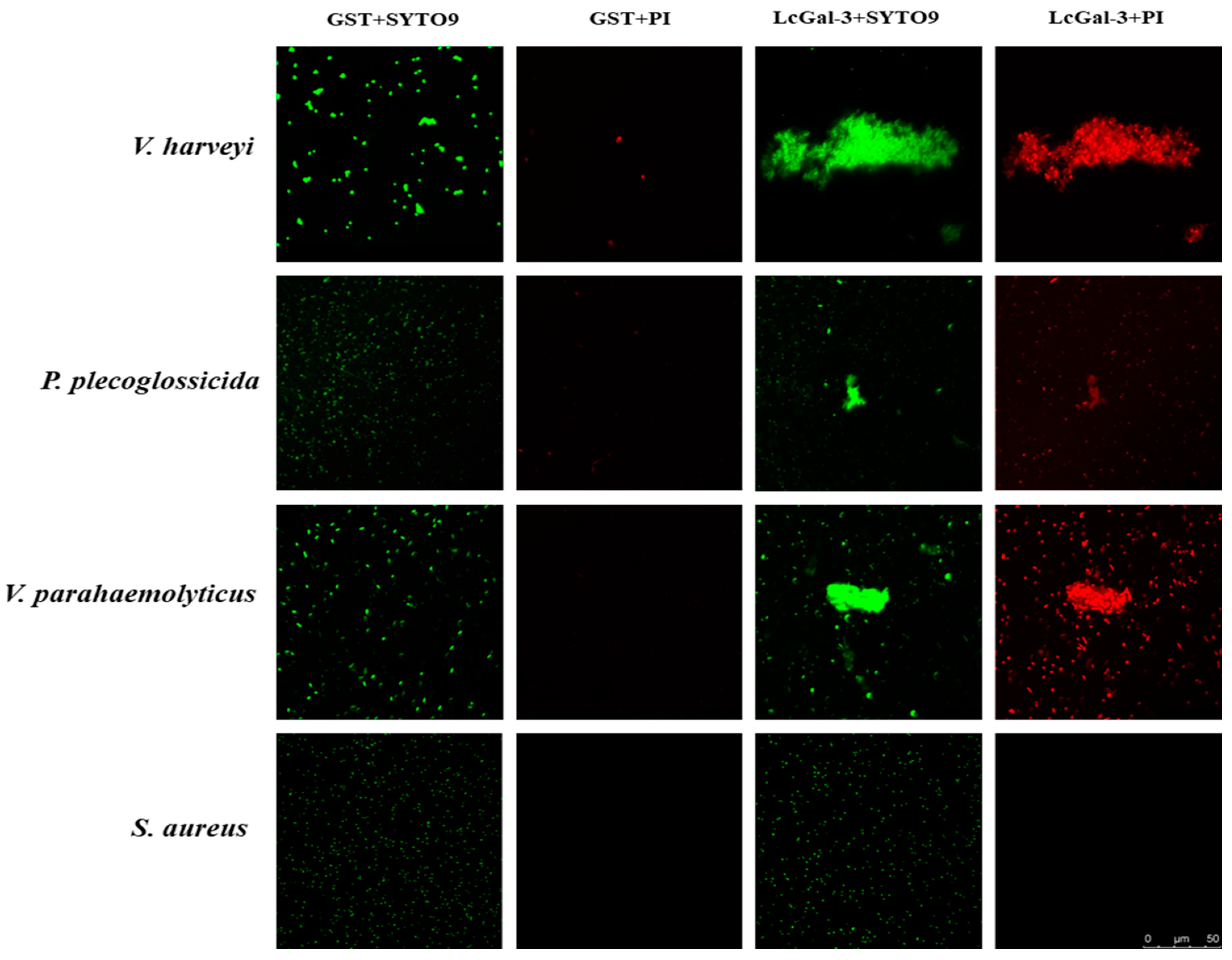
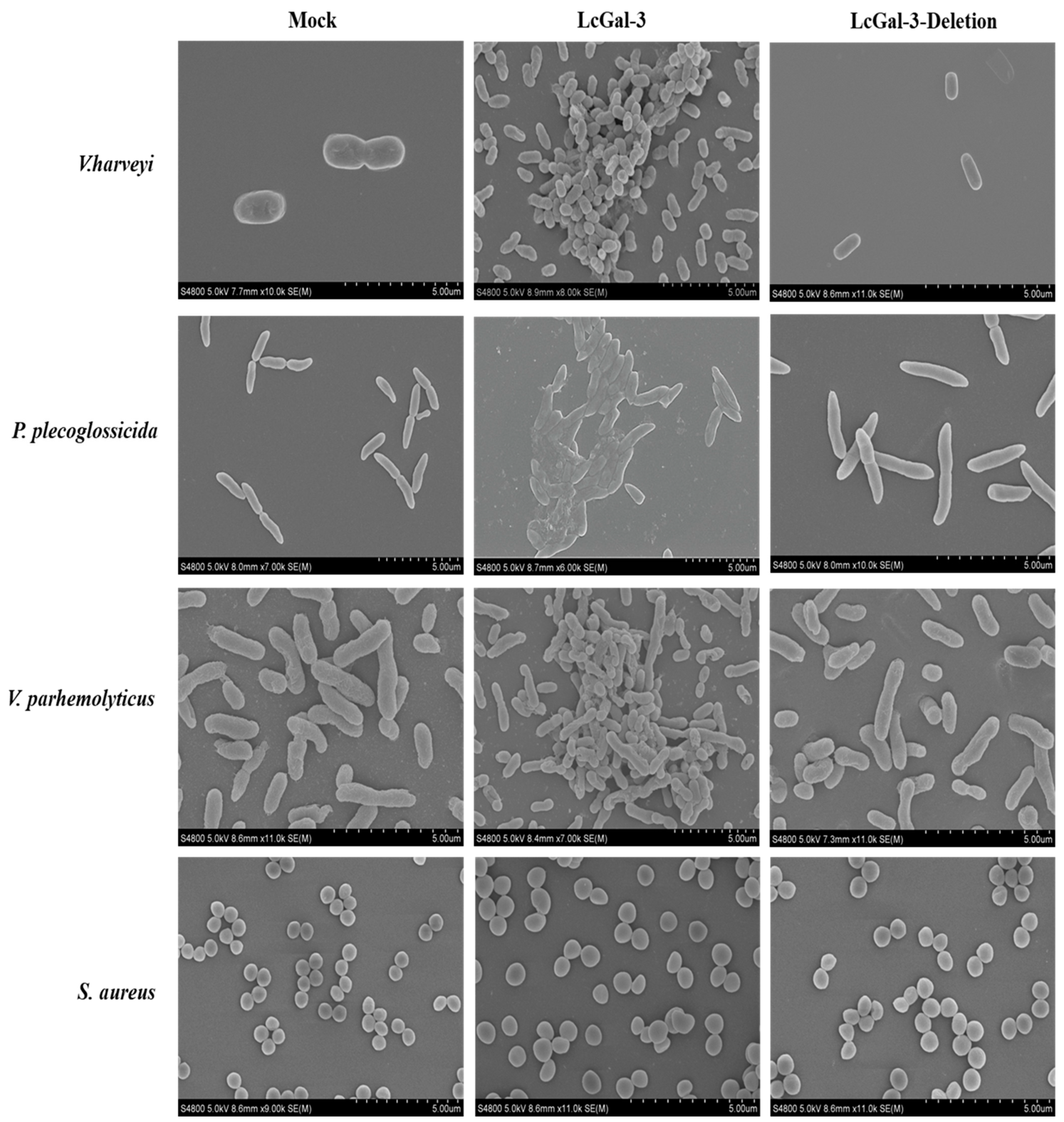
2.6. Agglutination and Bactericidal Effect of LcGal-3 on Gram Negative Bacteria
3. Discussion
4. Materials and Methods
4.1. Experimental Fish and Bacterial Challenge Experiment
4.2. RNA Extraction and cDNA Synthesis
4.3. Primer Design and Clone of LcGal-3 Gene
4.4. Bioinformatics Analysis
4.5. Tissue Distribution of LcGal-3 and Temporal Expression Pattern Post Infection
4.6. Cell Culture and Subcellular Localization
4.7. Prokaryotic Expression and Purification of LcGal-3 Protein and Deletion Mutant LcGal-3a Protein
4.8. Hemagglutination and Glucose Inhibition Assays of LcGal-3 Protein and Deletion Mutant Protein LcGal-3a
4.9. Bacterial Agglutination and Antibacterial Assay
4.10. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Han, D.; Shan, X.; Zhang, W.; Chen, Y.; Wang, Q.; Li, Z.; Zhang, G.; Xu, P.; Li, J.; Xie, S.; et al. A Revisit to Fishmeal Usage and Associated Consequences in Chinese Aquaculture. Rev. Aquac. 2018, 10, 493–507. [Google Scholar] [CrossRef]
- Gong, D.; Cui, X.; Song, M.; Xing, B.; Xu, P.; Tang, Y.; Yin, L. Behavior of Large Yellow Croaker (Larimichthys crocea) in Pen Aquaculture as Measured by Meter-Scale Telemetry. Front. Mar. Sci. 2023, 10, 1177037. [Google Scholar] [CrossRef]
- Bai, Y.; Qu, A.; Liu, Y.; Chen, X.; Wang, J.; Zhao, J.; Ke, Q.; Chen, L.; Chi, H.; Gong, H.; et al. Integrative Analysis of GWAS and Transcriptome Reveals P53 Signaling Pathway Mediates Resistance to Visceral White-Nodules Disease in Large Yellow Croaker. Fish Shellfish Immunol. 2022, 130, 350–358. [Google Scholar] [CrossRef]
- Li, C.; Wang, S.; Ren, Q.; He, T.; Chen, X. An Outbreak of Visceral White Nodules Disease Caused by Pseudomonas Plecoglossicida at a Water Temperature of 12 °C in Cultured Large Yellow Croaker (Larimichthys crocea) in China. J. Fish Dis. 2020, 43, 1353–1361. [Google Scholar] [CrossRef]
- Zhang, X.-Y.; Zhuo, X.; Cheng, J.; Wang, X.; Liang, K.; Chen, X.P. 1 Regulates Cathepsin S Expression in Large Yellow Croaker (Larimichthys crocea) Macrophages. Front. Immunol. 2021, 12, 819029. [Google Scholar] [CrossRef]
- Li, Z.; Fang, M.; Tang, X.; Zhang, D.; Wang, Z. Disentangling Genetic Variation for Endurance and Resistance to Visceral White-Nodules Disease in Large Yellow Croaker (Larimichthys crocea) Using Genome Information. Aquaculture 2023, 564, 739045. [Google Scholar] [CrossRef]
- Thurston, T.L.M.; Wandel, M.P.; von Muhlinen, N.; Foeglein, A.; Randow, F. Galectin 8 Targets Damaged Vesicles for Autophagy to Defend Cells against Bacterial Invasion. Nature 2012, 482, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Chiariotti, L.; Salvatore, P.; Benvenuto, G.; Bruni, C.B. Control of Galectin Gene Expression. Biochimie 1999, 81, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Saccon, F.; Gatto, M.; Ghirardello, A.; Iaccarino, L.; Punzi, L.; Doria, A. Role of Galectin-3 in Autoimmune and Non-Autoimmune Nephropathies. Autoimmun. Rev. 2017, 16, 34–47. [Google Scholar] [CrossRef]
- Elumalai, P.; Rubeena, A.S.; Arockiaraj, J.; Wongpanya, R.; Cammarata, M.; Ringø, E.; Vaseeharan, B. The Role of Lectins in Finfish: A Review. Rev. Fish. Sci. Aquac. 2019, 27, 152–169. [Google Scholar] [CrossRef]
- Wu, B.; Song, Q.; Li, W.; Xie, Y.; Luo, S.; Tian, Q.; Zhao, R.; Liu, T.; Wang, Z.; Han, F. Characterization and Functional Study of a Chimera Galectin from Yellow Drum Nibea Albiflora. Int. J. Biol. Macromol. 2021, 187, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Yang, N.; Zhang, L.; Fu, Q.; Tan, F.; Li, C. Expression Profiling and Functional Characterization of Galectin-3 of Turbot (Scophthalmus maximus L.) in Host Mucosal Immunity. Fish Shellfish Immunol. 2019, 84, 333–340. [Google Scholar] [CrossRef]
- Ji, X.; Liu, X.; Li, X.; Zhou, S.; Xiu, Y. Characterization and Functional Study of Galectin3 from Japanese Flounder (Paralichthys olivaceus). Fish Shellfish Immunol. 2020, 102, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.M.; Kitani, Y.; Korsnes, K.; Iversen, M.H.; Brinchmann, M.F. A Truncated Galectin-3 Isolated from Skin Mucus of Atlantic Salmon Salmo Salar Binds to and Modulates the Proteome of the Gram-Negative Bacteria Moritella Viscosa. Mar. Drugs 2020, 18, 102. [Google Scholar] [CrossRef]
- Zhu, D.; Huang, R.; Chu, P.; Chen, L.; Li, Y.; He, L.; Li, Y.; Liao, L.; Zhu, Z.; Wang, Y. Characterization and Expression of Galectin-3 in Grass Carp (Ctenopharyngodon idella). Dev. Comp. Immunol. 2020, 104, 103567. [Google Scholar] [CrossRef]
- Zhu, J.; Wei, M.; Wang, Q.; Ao, Q.; Tan, Y.; Luo, Y.; Wang, H.; Jiang, H.; Hu, Q. Characterization and Expression of Galectin-3 after Streptococcus Agalactiae and Aeromonas Hydrophila Challenge in GIFT Strain Nile Tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 2019, 86, 974–980. [Google Scholar] [CrossRef]
- Niu, J.; Huang, Y.; Liu, X.; Luo, G.; Tang, J.; Wang, B.; Lu, Y.; Cai, J.; Jian, J. Functional Characterization of Galectin-3 from Nile Tilapia (Oreochromis niloticus) and Its Regulatory Role on Monocytes/Macrophages. Fish Shellfish Immunol. 2019, 95, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Nita-Lazar, M.; Mancini, J.; Feng, C.; González-Montalbán, N.; Ravindran, C.; Jackson, S.; de Las Heras-Sánchez, A.; Giomarelli, B.; Ahmed, H.; Haslam, S.M.; et al. The Zebrafish Galectins Drgal1-L2 and Drgal3-L1 Bind in Vitro to the Infectious Hematopoietic Necrosis Virus (IHNV) Glycoprotein and Reduce Viral Adhesion to Fish Epithelial Cells. Dev. Comp. Immunol. 2016, 55, 241–252. [Google Scholar] [CrossRef]
- Peng, J.; Li, W.; Wang, B.; Zhang, S.; Xiao, Y.; Han, F.; Wang, Z. UBE2G1 Is a Critical Component of Immune Response to the Infection of Pseudomonas plecoglossicida in Large Yellow Croaker (Larimichthys crocea). Int. J. Mol. Sci. 2022, 23, 8298. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Liu, X.; Zhou, J.; Wang, X.; Liu, R.; Peng, H.; Li, B.; Cai, Z.; Jiang, C. Molecular Characterization and Expression Analysis of Galectins in Japanese Pufferfish (Takifugu rubripes) in Response to Vibrio Harveyi Infection. Fish Shellfish Immunol. 2019, 86, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.-T.; Stowell, S.R. The Role of Galectins in Immunity and Infection. Nat. Rev. Immunol. 2023, 1–16. [Google Scholar] [CrossRef]
- Porębska, N.; Poźniak, M.; Matynia, A.; Żukowska, D.; Zakrzewska, M.; Otlewski, J.; Opaliński, Ł. Galectins as Modulators of Receptor Tyrosine Kinases Signaling in Health and Disease. Cytokine Growth Factor. Rev. 2021, 60, 89–106. [Google Scholar] [CrossRef]
- Tian, Q.; Li, W.; Li, J.; Xiao, Y.; Wu, B.; Wang, Z.; Han, F. Towards Understanding PRPS1 as a Molecular Player in Immune Response in Yellow Drum (Nibea Albiflora). Int. J. Mol. Sci. 2022, 23, 6475. [Google Scholar] [CrossRef]
- Li, N.; Hua, J. Immune Cells in Liver Regeneration. Oncotarget 2017, 8, 3628–3639. [Google Scholar] [CrossRef]
- Wang, M.; Wang, L.; Huang, M.; Yi, Q.; Guo, Y.; Gai, Y.; Wang, H.; Zhang, H.; Song, L. A Galectin from Eriocheir sinensis Functions as Pattern Recognition Receptor Enhancing Microbe Agglutination and Haemocytes Encapsulation. Fish Shellfish Immunol. 2016, 55, 10–20. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, W.; Mu, C.; Li, R.; Song, W.; Ye, Y.; Shi, C.; Liu, L.; Wang, H.; Wang, C. Identification and Characterization of a Novel Galectin from the Mud Crab Scylla paramamosain. Fish Shellfish Immunol. 2020, 98, 699–709. [Google Scholar] [CrossRef]
- Clairfeuille, T.; Buchholz, K.R.; Li, Q.; Verschueren, E.; Liu, P.; Sangaraju, D.; Park, S.; Noland, C.L.; Storek, K.M.; Nickerson, N.N.; et al. Structure of the Essential Inner Membrane Lipopolysaccharide-PbgA Complex. Nature 2020, 584, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Yong-Ping, X.U.; Ting-Ting, W.; Yong-Xin, S.; Wen-Jia, L.; Jian-Song, Y.; Li-Ji, J. Current Research on Function and Application of Aquatic Animal Lysozyme. Fish. Sci. 2011, 9, e109666. [Google Scholar] [CrossRef]
- Guest, R.L.; Rutherford, S.T.; Silhavy, T.J. Border Control: Regulating LPS Biogenesis. Trends Microbiol. 2021, 29, 334–345. [Google Scholar] [CrossRef]
- Huang, M.; Zhou, T.; Wu, Y.; Fei, H.; Wang, G.; Li, Z.; Lei, Y.; Liu, Q.; Sun, C.; Lv, Z.; et al. Characterisation and Functional Comparison of Single-CRD and Multidomain Containing Galectins CgGal-2 and CgGal-3 from Oyster Crassostrea gigas. Fish Shellfish Immunol. 2018, 78, 238–247. [Google Scholar] [CrossRef] [PubMed]
- Vasta, G.R. Roles of Galectins in Infection. Nat. Rev. Microbiol. 2009, 7, 424–438. [Google Scholar] [CrossRef]
- Vasta, G.R.; Ahmed, H.; Nita-Lazar, M.; Banerjee, A.; Pasek, M.; Shridhar, S.; Guha, P.; Fernández-Robledo, J.A. Galectins as Self/Non-Self Recognition Receptors in Innate and Adaptive Immunity: An Unresolved Paradox. Front. Immunol. 2012, 3, 199. [Google Scholar] [CrossRef]
- Li, M.; Huang, X.; Wen, J.; Chen, S.; Wu, X.; Ma, W.; Cui, S.W.; Xie, M.; Nie, S. Innate Immune Receptors Co-Recognition of Polysaccharides Initiates Multi-Pathway Synergistic Immune Response. Carbohydr. Polym. 2023, 305, 120533. [Google Scholar] [CrossRef] [PubMed]
- Wolf, A.J.; Underhill, D.M. Peptidoglycan Recognition by the Innate Immune System. Nat. Rev. Immunol. 2018, 18, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Li, F.-Y.; Wang, S.-F.; Bernardes, E.S.; Liu, F.-T. Galectins in Host Defense Against Microbial Infections. Adv. Exp. Med. Biol. 2020, 1204, 141–167. [Google Scholar] [CrossRef] [PubMed]
- Kohatsu, L.; Hsu, D.K.; Jegalian, A.G.; Liu, F.-T.; Baum, L.G. Galectin-3 Induces Death of Candida Species Expressing Specific Beta-1,2-Linked Mannans. J. Immunol. 2006, 177, 4718–4726. [Google Scholar] [CrossRef]
- Almeida, F.; Wolf, J.M.; da Silva, T.A.; DeLeon-Rodriguez, C.M.; Rezende, C.P.; Pessoni, A.M.; Fernandes, F.F.; Silva-Rocha, R.; Martinez, R.; Rodrigues, M.L.; et al. Galectin-3 Impacts Cryptococcus Neoformans Infection through Direct Antifungal Effects. Nat. Commun. 2017, 8, 1968. [Google Scholar] [CrossRef]
- Fowler, M.; Thomas, R.J.; Atherton, J.; Roberts, I.S.; High, N.J. Galectin-3 Binds to Helicobacter Pylori O-Antigen: It Is Upregulated and Rapidly Secreted by Gastric Epithelial Cells in Response to H. pylori Adhesion. Cell Microbiol. 2006, 8, 44–54. [Google Scholar] [CrossRef]
- Hatanaka, O.; Rezende, C.P.; Moreno, P.; Freitas Fernandes, F.; Oliveira Brito, P.K.M.; Martinez, R.; Coelho, C.; Roque-Barreira, M.C.; Casadevall, A.; Almeida, F. Galectin-3 Inhibits Paracoccidioides Brasiliensis Growth and Impacts Paracoccidioidomycosis through Multiple Mechanisms. mSphere 2019, 4, e00209-19. [Google Scholar] [CrossRef]
- Breitenbach Barroso Coelho, L.C.; Marcelino Dos Santos Silva, P.; Felix de Oliveira, W.; de Moura, M.C.; Viana Pontual, E.; Soares Gomes, F.; Guedes Paiva, P.M.; Napoleão, T.H.; Dos Santos Correia, M.T. Lectins as Antimicrobial Agents. J. Appl. Microbiol. 2018, 125, 1238–1252. [Google Scholar] [CrossRef]
- Mukherjee, S.; Zheng, H.; Derebe, M.G.; Callenberg, K.M.; Partch, C.L.; Rollins, D.; Propheter, D.C.; Rizo, J.; Grabe, M.; Jiang, Q.-X.; et al. Antibacterial Membrane Attack by a Pore-Forming Intestinal C-Type Lectin. Nature 2014, 505, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Giese, K.C.; Michalowski, C.B.; Little, J.W. RecA-Dependent Cleavage of LexA Dimers. J. Mol. Biol. 2008, 377, 148–161. [Google Scholar] [CrossRef] [PubMed]
- Mnich, M.E.; van Dalen, R.; van Sorge, N.M. C-Type Lectin Receptors in Host Defense Against Bacterial Pathogens. Front. Cell Infect. Microbiol. 2020, 10, 309. [Google Scholar] [CrossRef] [PubMed]

| Species | Common Name | GenBank Accession number | Identity (%) |
|---|---|---|---|
| Larimichthys crocea | Large yellow croaker | XP_027130834.1 | 100 |
| Nibea albiflora | Yellow drum | MW858251.1 | 95.56 |
| Perca flavescens | Yellow perch | XP_028421130.1 | 91.85 |
| Scophthalmus maximus | Breet | MF797865.1 | 91.85 |
| Paralichthys olivaceus | Bastard Halibut | XP_019962524.1 | 91.85 |
| Amphiprion ocellaris | Clownfish | XP_023142436.1 | 90.30 |
| Siniperca chuatsi | Aucha perch | XM_044167408.1 | 87.41 |
| Cynoglossus semilaevis | Tongue sole | XP_016891389.1 | 84.33 |
| Ctenopharyngodon idella | Grass carp | XP_051772445.1 | 77.04 |
| Oncorhynchus mykiss | Rainbow trout | XP_036818451.1 | 73.76 |
| Mus musculus | Mouse | NM_010705.3 | 50.36 |
| Gallus gallus | Chicken | XM_046917600.1 | 50.00 |
| Equus caballus | Horse | XM_023627775.1 | 49.64 |
| Xenopus laevis | Clawed frog | BC046662.1 | 47.83 |
| Xenopus tropicalis | Xenopus tropicalis | NM_203655.1 | 46.72 |
| Homo sapiens | Human | BAB83625.1 | 43.32 |
| Primer Name | Sequence (5′-3′) | Purpose |
|---|---|---|
| Gal-3-F | CCCCTGGGATCCCCGGAATTCATGTCGGACTTCTCGCTGGC | ORF amplification |
| Gal-3-R | GTCACGATGCGGCCGCTCGAGTCAGATCATGCTCGGGGCG | |
| qGal-3-F | AGGAGTCCATCTTCTGATCATGT | qRT-PCR analysis |
| qGal-3-R | ATCCTGGGTTTGTGGGAGGA | |
| β-actin-F | TTATGAAGGCTATGCCCTGCC | |
| β-actin-R | TGAAGGAGTAGCCACGCTCTGT | |
| sGal-3-F | CTACCGGACTCAGATCTCGAGATGTCGGACTTCTCGCTGGC | subcellular localization |
| sGal-3-R | GTACCGTCGACTGCAGAATTCGGATCATGCTCGGGGCGGC | |
| HFNPR-F | AGAGCCGACGTGGCGTTTTTCAAACGCT | deletion mutations |
| HFNPR-R | AGCGTTTGAAAAACGCCACGTCGGCTCT | |
| PFNPR-F | TTTCCCTTCAACCCGAGGTTCAAACGCTCG | point mutations |
| PFNPR-R | CGAGCGTTTGAACCTCGGGTTGAAGGGAAA | |
| DFNPR-F | GCGTTTGACTTCAACCCGAGGTTCAAACGCT | |
| DFNPR-R | AGCGTTTGAACCTCGGGTTGAAGTCAAACGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Wu, B.; Li, W.; Han, F. Molecular Characterization of Galectin-3 in Large Yellow Croaker Larimichthys crocea Functioning in Antibacterial Activity. Int. J. Mol. Sci. 2023, 24, 11539. https://doi.org/10.3390/ijms241411539
Yang Y, Wu B, Li W, Han F. Molecular Characterization of Galectin-3 in Large Yellow Croaker Larimichthys crocea Functioning in Antibacterial Activity. International Journal of Molecular Sciences. 2023; 24(14):11539. https://doi.org/10.3390/ijms241411539
Chicago/Turabian StyleYang, Yao, Baolan Wu, Wanbo Li, and Fang Han. 2023. "Molecular Characterization of Galectin-3 in Large Yellow Croaker Larimichthys crocea Functioning in Antibacterial Activity" International Journal of Molecular Sciences 24, no. 14: 11539. https://doi.org/10.3390/ijms241411539
APA StyleYang, Y., Wu, B., Li, W., & Han, F. (2023). Molecular Characterization of Galectin-3 in Large Yellow Croaker Larimichthys crocea Functioning in Antibacterial Activity. International Journal of Molecular Sciences, 24(14), 11539. https://doi.org/10.3390/ijms241411539





