Polyoxometalate K6[P2Mo18O62] Inactivates Escherichia coli O157:H7 by Inducing recA Expression and Apoptosis-like Bacterial Death
Abstract
1. Introduction
2. Results
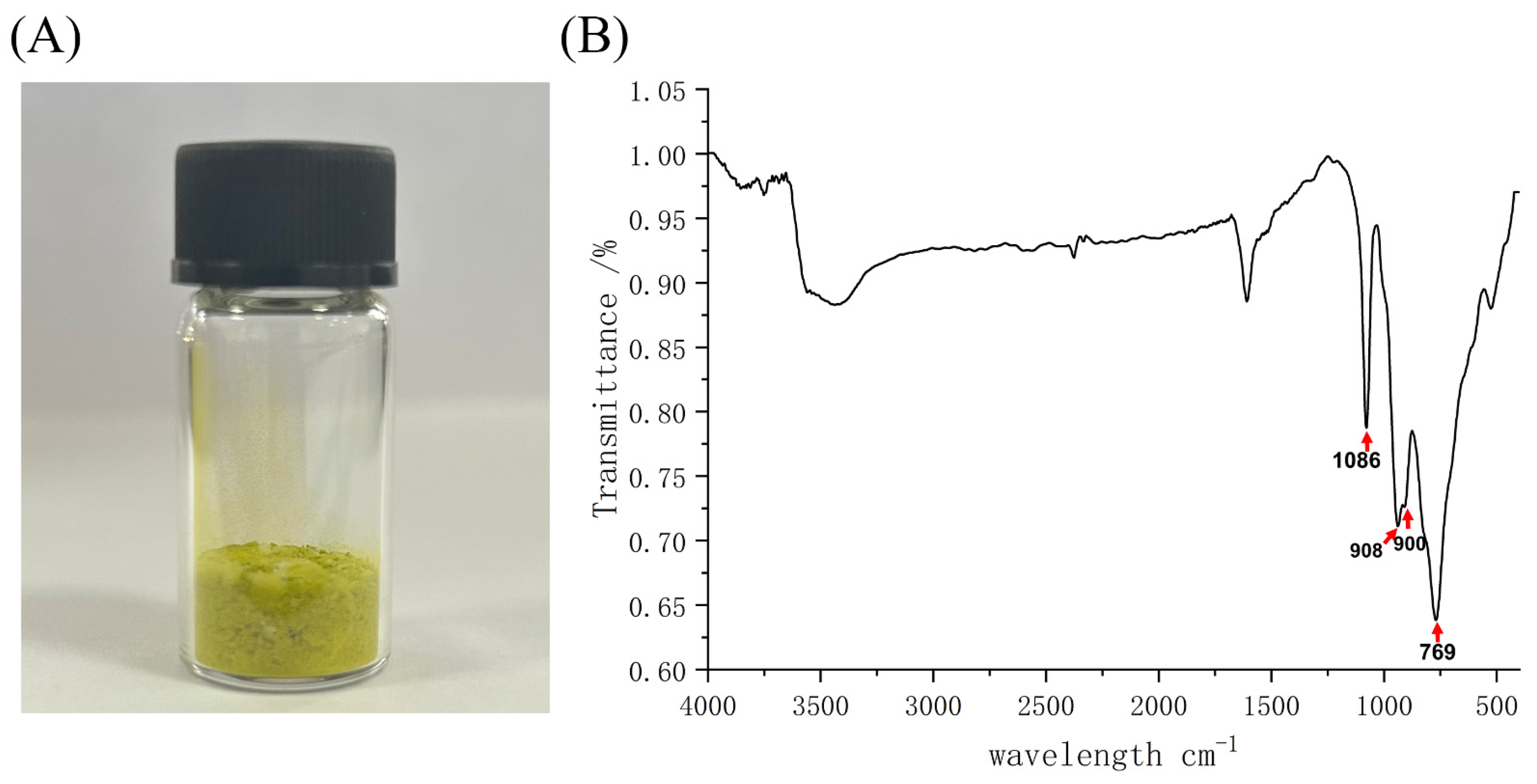
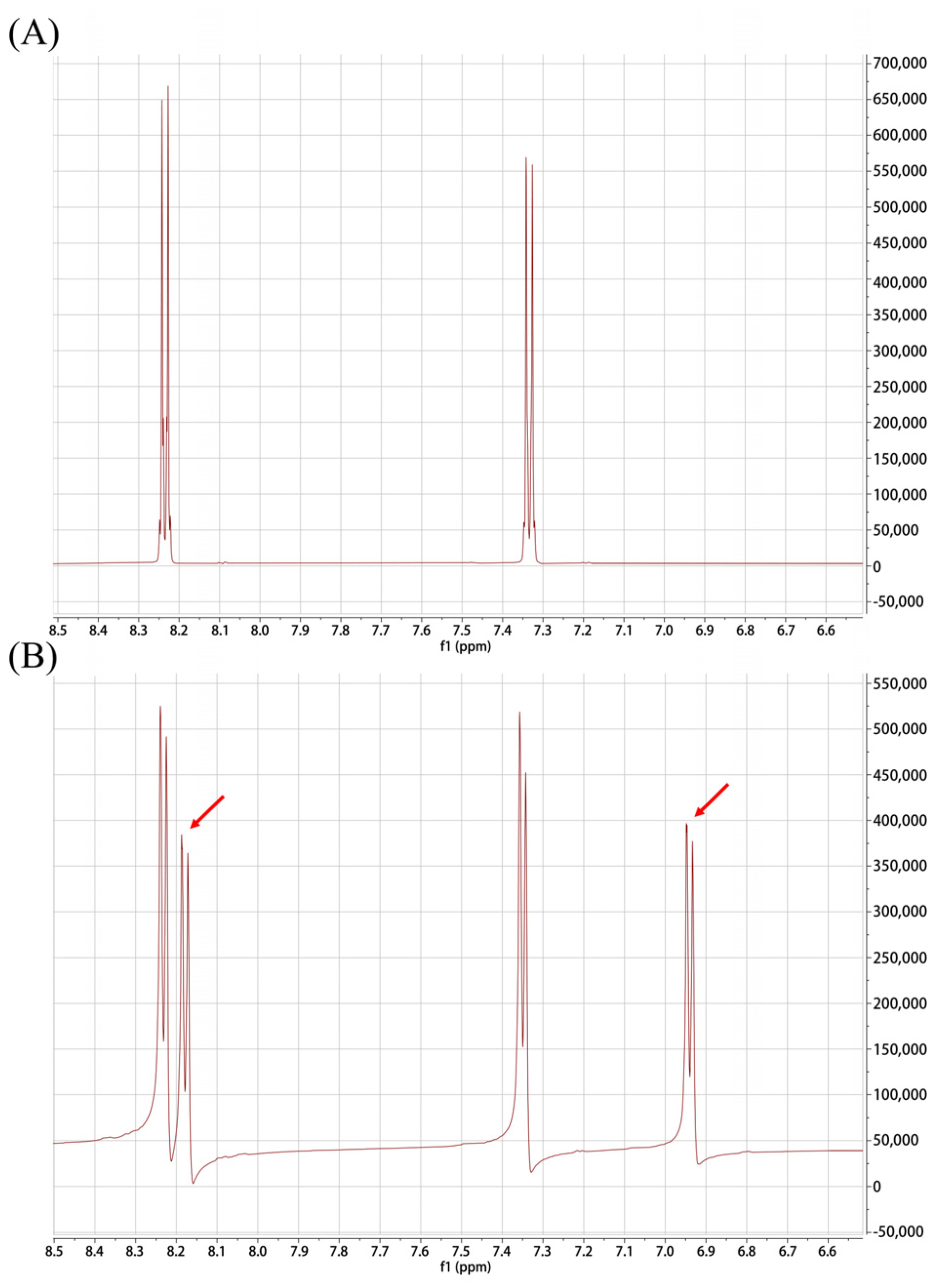
2.1. Synthesis and Characterization of K6[P2Mo18O62]
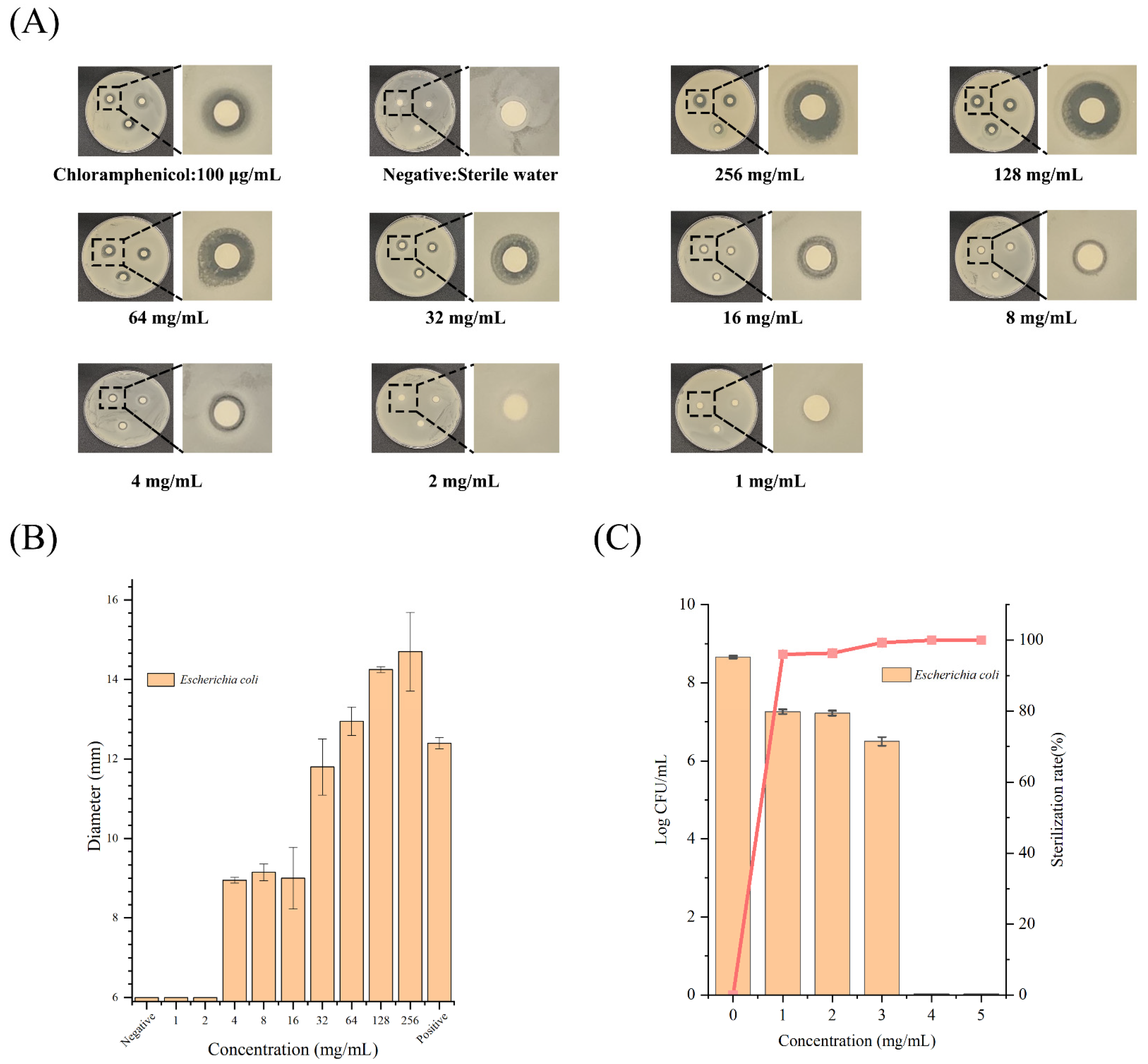
2.2. Antibacterial Effects of K6[P2Mo18O62] against E. coli O157:H7
2.3. K6[P2Mo18O62] Possesses Phosphatase Activity
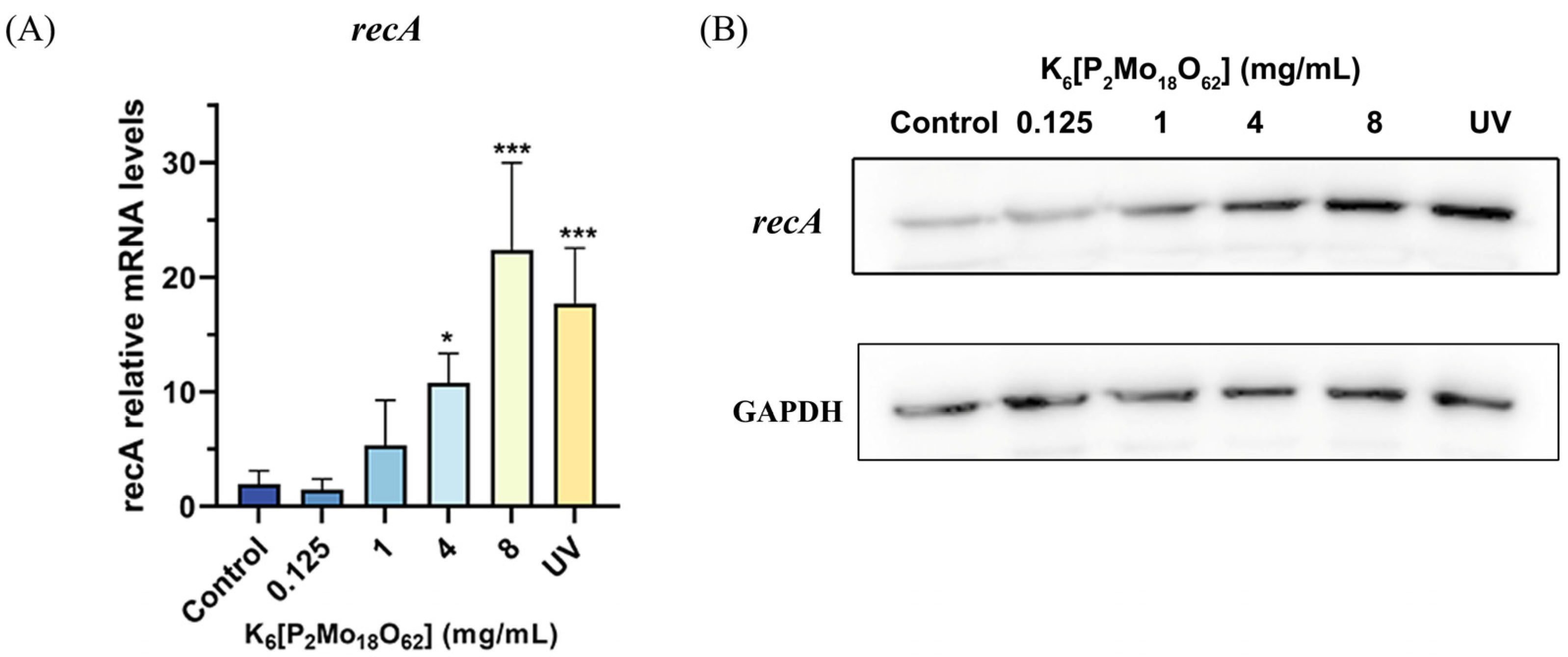
2.4. K6[P2Mo18O62] Treatment Induced a Significant Increase in the recA Expression in E. coli O157:H7
2.5. K6[P2Mo18O62] Treatment Induced Apoptosis-Like Bacterial Death Events in E. coli O157:H7
3. Discussion
4. Materials and Methods
4.1. Chemicals, Reagents and Bacterial Strains
4.2. Synthesis and Characterization of K6[P2Mo18O62]
4.3. Antibacterial Activity Tests
4.3.1. Kirby–Bauer Disk Diffusion Test
4.3.2. Agar Contact Method
4.4. Real-Time PCR
4.5. Western Blot
4.6. Flow Cytometric Analysis
4.6.1. Flow Cytometric Analysis of the TUNEL Assay
4.6.2. Flow Cytometric Analysis of Annexin V-FITC and Propidium Iodide (PI) Staining
4.7. Flow Cytometric Analysis of Annexin V-FITC and Propidium Iodide (PI) Staining
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vaou, N.; Stavropoulou, E.; Voidarou, C.; Tsigalou, C.; Bezirtzoglou, E. Towards advances in medicinal plant antimicrobial activity: A review study on challenges and future perspectives. Microorganisms 2021, 9, 2041. [Google Scholar] [CrossRef]
- Wang, Z.; Koirala, B.; Hernandez, Y.; Zimmerman, M.; Brady, S.F. Bioinformatic prospecting and synthesis of a bifunctional lipopeptide antibiotic that evades resistance. Science 2022, 376, 991–996. [Google Scholar] [CrossRef] [PubMed]
- Yudaev, P.; Mezhuev, Y.; Chistyakov, E. Nanoparticle-containing wound dressing: Antimicrobial and healing effects. Gels 2022, 8, 329. [Google Scholar] [CrossRef] [PubMed]
- Yudaev, P.; Chuev, V.; Klyukin, B.; Kuskov, A.; Mezhuev, Y.; Chistyakov, E. Polymeric dental nanomaterials: Antimicrobial action. Polymers 2022, 14, 864. [Google Scholar] [CrossRef] [PubMed]
- Yudaev, P.; Butorova, I.; Chuev, V.; Posokhova, V.; Klyukin, B.; Chistyakov, E. Wound Gel with Antimicrobial Effects Based on Polyvinyl Alcohol and Functional Aryloxycyclotriphosphazene. Polymers 2023, 15, 2831. [Google Scholar] [CrossRef]
- Swenson, L.S.; Orozco, J.C.; Liu, Y.; Darling, S.B.; Khan, M.I. Novel colloidal materials from functionalized polyoxometalates. Inorg. Chem. Commun. 2017, 84, 20–23. [Google Scholar] [CrossRef]
- Aureliano, M.; Serrano, A.; Martins, J.; Faleiro, L.; Fonseca, C.; Fraqueza, G.; Lagoa, R. Polyoxometalates with anticancer, antibacterial and antiviral activities. In Polyoxometalates; Jenny Stanford Publishing: Singapore, 2022; pp. 309–358. [Google Scholar]
- Tajima, Y. The Sensitizing Effects of Undecatungstocobalto (II) silicate on Methicillin-Resistant Staphylococcusaurens to β-Lactams. Biomed. Res. 2002, 23, 115–125. [Google Scholar] [CrossRef]
- Fukuda, N.; Yamase, T.; Tajima, Y. Inhibitory effect of polyoxotungstates on the production of penicillin-binding proteins and β-lactamase against methicillin-resistant Staphylococcus aureus. Biol. Pharm. Bull. 1999, 22, 463–470. [Google Scholar] [CrossRef]
- Paul, T.J.; Parac-Vogt, T.N.; Quiñonero, D.; Prabhakar, R. Investigating polyoxometalate–protein interactions at chemically distinct binding sites. J. Phys. Chem. B 2018, 122, 7219–7232. [Google Scholar] [CrossRef]
- Bijelic, A.; Aureliano, M.; Rompel, A. The antibacterial activity of polyoxometalates: Structures, antibiotic effects and future perspectives. Chem. Commun. 2018, 54, 1153–1169. [Google Scholar] [CrossRef]
- Vanhaecht, S.; Absillis, G.; Parac-Vogt, T.N. Hydrolysis of DNA model substrates catalyzed by metal-substituted Wells–Dawson polyoxometalates. Dalton Trans. 2012, 41, 10028–10034. [Google Scholar] [CrossRef]
- Luong, T.K.N.; Govaerts, I.; Robben, J.; Shestakova, P.; Parac-Vogt, T. Polyoxometalates as artificial nucleases: Hydrolytic cleavage of DNA promoted by a highly negatively charged Zr IV-substituted Keggin polyanion. Chem. Commun. 2017, 53, 617–620. [Google Scholar] [CrossRef]
- Luong, T.K.N.; Absillis, G.; Shestakova, P.; Parac-Vogt, T.N. Solution Speciation of the Dinuclear ZrIV-Substituted Keggin Polyoxometalate [{α-PW11O39Zr (µ-OH)(H2O)}2]8– and Its Reactivity towards DNA-Model Phosphodiester Hydrolysis. Eur. J. Inorg. Chem. 2014, 2014, 5276–5284. [Google Scholar] [CrossRef]
- Luong, T.K.N.; Shestakova, P.; Parac-Vogt, T.N. Kinetic studies of phosphoester hydrolysis promoted by a dimeric tetrazirconium (IV) Wells–Dawson polyoxometalate. Dalton Trans. 2016, 45, 12174–12180. [Google Scholar] [CrossRef]
- Luong, T.K.N.; Absillis, G.; Shestakova, P.; Parac-Vogt, T.N. Hydrolysis of the RNA model substrate catalyzed by a binuclear Zr IV-substituted Keggin polyoxometalate. Dalton Trans. 2015, 44, 15690–15696. [Google Scholar] [CrossRef]
- Erental, A.; Kalderon, Z.; Saada, A.; Smith, Y.; Engelberg-Kulka, H. Apoptosis-like death, an extreme SOS response in Escherichia coli. MBio 2014, 5, e01426-14. [Google Scholar] [CrossRef]
- Erental, A.; Sharon, I.; Engelberg-Kulka, H. Two programmed cell death systems in Escherichia coli: An apoptotic-like death is inhibited by the mazEF-mediated death pathway. PLoS Biol. 2012, 10, e1001281. [Google Scholar] [CrossRef]
- Casaregola, S.; D’Ari, R.; Huisman, O. Quantitative evaluation of recA gene expression in Escherichia coli. Mol. Gen. Genet. MGG 1982, 185, 430–439. [Google Scholar] [CrossRef]
- Dwyer, D.J.; Camacho, D.M.; Kohanski, M.A.; Callura, J.M.; Collins, J.J. Antibiotic-induced bacterial cell death exhibits physiological and biochemical hallmarks of apoptosis. Mol. Cell 2012, 46, 561–572. [Google Scholar] [CrossRef]
- Choi, H.; Hwang, J.S.; Lee, D.G. Coprisin exerts antibacterial effects by inducing apoptosis-like death in Escherichia coli. IUBMB Life 2016, 68, 72–78. [Google Scholar] [CrossRef]
- Lee, H.; Lee, D.G. Arenicin-1-induced apoptosis-like response requires RecA activation and hydrogen peroxide against Escherichia coli. Curr. Genet. 2019, 65, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, D.G. Nitric oxide–inducing Genistein elicits apoptosis-like death via an intense SOS response in Escherichia coli. Appl. Microbiol. Biotechnol. 2020, 104, 10711–10724. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.Y.; Yoon, J.W.; Hovde, C.J. A brief overview of Escherichia coli O157: H7 and its plasmid O157. J. Microbiol. Biotechnol. 2010, 20, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Tan, S.K.; Lim, M.; Chang, S.H.; Cui, G.; Liu, S.; Narasimhan, K.; New, S.Y.; Wang, X.; Chen, C. Identification of a Wells–Dawson polyoxometalate-based AP-2γ inhibitor with pro-apoptotic activity. Biochem. J. 2018, 475, 1965–1977. [Google Scholar] [CrossRef]
- Gumerova, N.I.; Al-Sayed, E.; Krivosudský, L.; Čipčić-Paljetak, H.; Verbanac, D.; Rompel, A. Antibacterial activity of polyoxometalates against Moraxella catarrhalis. Front. Chem. 2018, 6, 336. [Google Scholar] [CrossRef]
- Inoue, M.; Suzuki, T.; Fujita, Y.; Oda, M.; Matsumoto, N.; Yamase, T. Enhancement of antibacterial activity of β-lactam antibiotics by [P2W18O62]6−, [SiMo12O40]4−, and [PTi2W10O40]7− against methicillin-resistant and vancomycin-resistant Staphylococcus aureus. J. Inorg. Biochem. 2006, 100, 1225–1233. [Google Scholar] [CrossRef]
- Liu, D.; Tan, H.; Chen, W.; Li, Y.; Wang, E. Resolution of chiral polyoxoanion [P2Mo18O62]X with histidine. CrystEngComm 2010, 12, 2044–2046. [Google Scholar] [CrossRef]
- Rhule, J.T.; Hill, C.L.; Judd, D.A.; Schinazi, R.F. Polyoxometalates in medicine. Chem. Rev. 1998, 98, 327–358. [Google Scholar] [CrossRef]
- Thomas, V.C.; Thurlow, L.R.; Boyle, D.; Hancock, L.E. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J. Bacteriol. 2008, 190, 5690–5698. [Google Scholar]
- Shen, G.-Z.; Zou, G.-H.; Li, H.-Y.; Zou, Y.-L. Crystal structure and antibacterial activity of polyoxometalate cobalt-ciprofloxacin complex. J. Mol. Struct. 2019, 1198, 126831. [Google Scholar] [CrossRef]
- Qi, S.; Li, J.; Sun, Y.; Chen, Q.; Le, T. The antifungal effects of cinnamaldehyde against Aspergillus niger and its application in bread preservation. Food Chem. 2020, 317, 126405. [Google Scholar]
- Grinholc, M.; Rodziewicz, A.; Forys, K.; Rapacka-Zdonczyk, A.; Kawiak, A.; Domachowska, A.; Golunski, G.; Wolz, C.; Mesak, L.; Becker, K. Fine-tuning recA expression in Staphylococcus aureus for antimicrobial photoinactivation: Importance of photo-induced DNA damage in the photoinactivation mechanism. Appl. Microbiol. Biotechnol. 2015, 99, 9161–9176. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, S.; Lin, Z.; Zhou, F.; Wang, D.; Zheng, B.; Hu, J. Polyoxometalate K6[P2Mo18O62] Inactivates Escherichia coli O157:H7 by Inducing recA Expression and Apoptosis-like Bacterial Death. Int. J. Mol. Sci. 2023, 24, 11469. https://doi.org/10.3390/ijms241411469
Lin S, Lin Z, Zhou F, Wang D, Zheng B, Hu J. Polyoxometalate K6[P2Mo18O62] Inactivates Escherichia coli O157:H7 by Inducing recA Expression and Apoptosis-like Bacterial Death. International Journal of Molecular Sciences. 2023; 24(14):11469. https://doi.org/10.3390/ijms241411469
Chicago/Turabian StyleLin, Shaoling, Zhongjing Lin, Feng Zhou, Dehua Wang, Baodong Zheng, and Jiamiao Hu. 2023. "Polyoxometalate K6[P2Mo18O62] Inactivates Escherichia coli O157:H7 by Inducing recA Expression and Apoptosis-like Bacterial Death" International Journal of Molecular Sciences 24, no. 14: 11469. https://doi.org/10.3390/ijms241411469
APA StyleLin, S., Lin, Z., Zhou, F., Wang, D., Zheng, B., & Hu, J. (2023). Polyoxometalate K6[P2Mo18O62] Inactivates Escherichia coli O157:H7 by Inducing recA Expression and Apoptosis-like Bacterial Death. International Journal of Molecular Sciences, 24(14), 11469. https://doi.org/10.3390/ijms241411469






