Genomic Instability Evolutionary Footprints on Human Health: Driving Forces or Side Effects?
Abstract
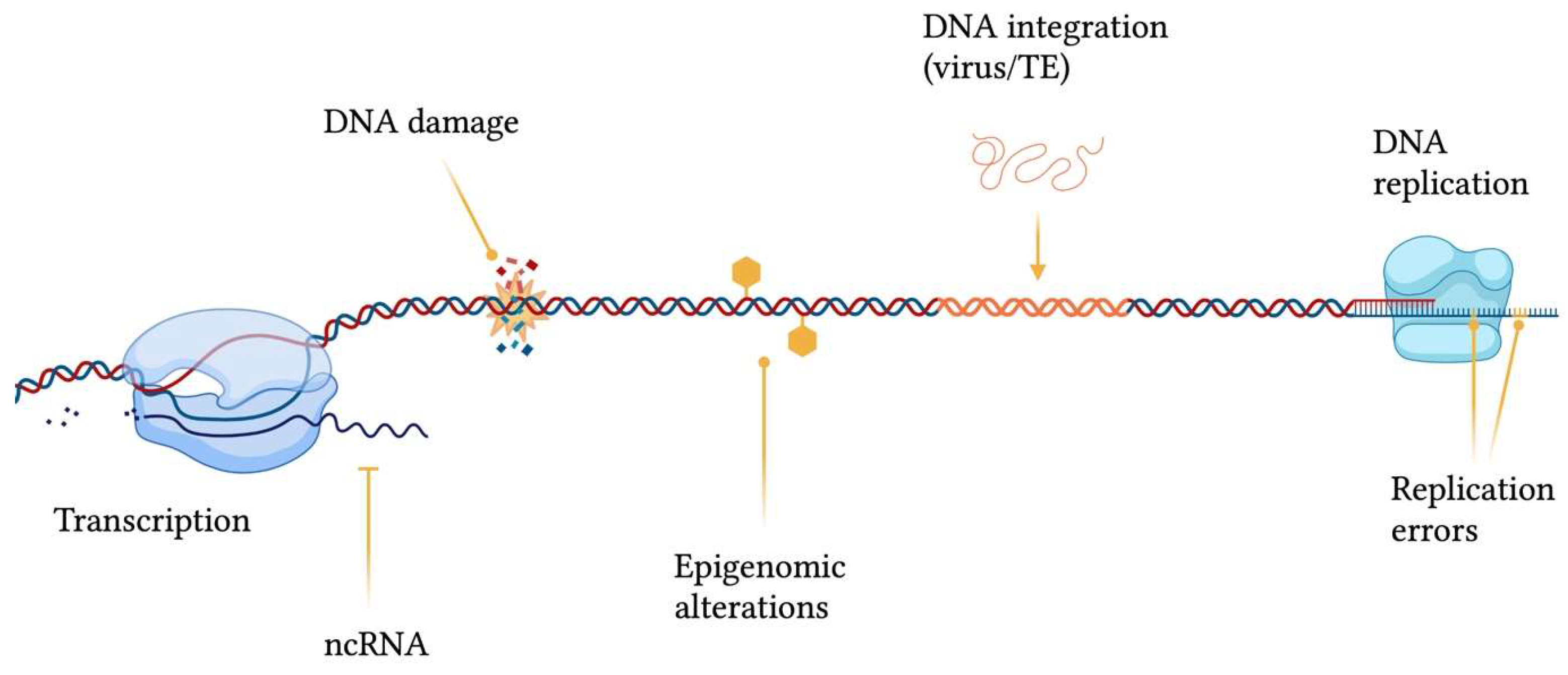
1. Introduction
2. The Balance between Variability and Conservation
3. The Guardians of Genomic Stability across the Tree of Life
3.1. Nucleotide Excision Repair Pathway
3.2. Mismatch Repair System
3.3. Double-Strand Break Repair
3.4. DNA Repair: Going Viral
4. Genomic (in)Stability in Homo sapiens: Just a Matter of Luck?
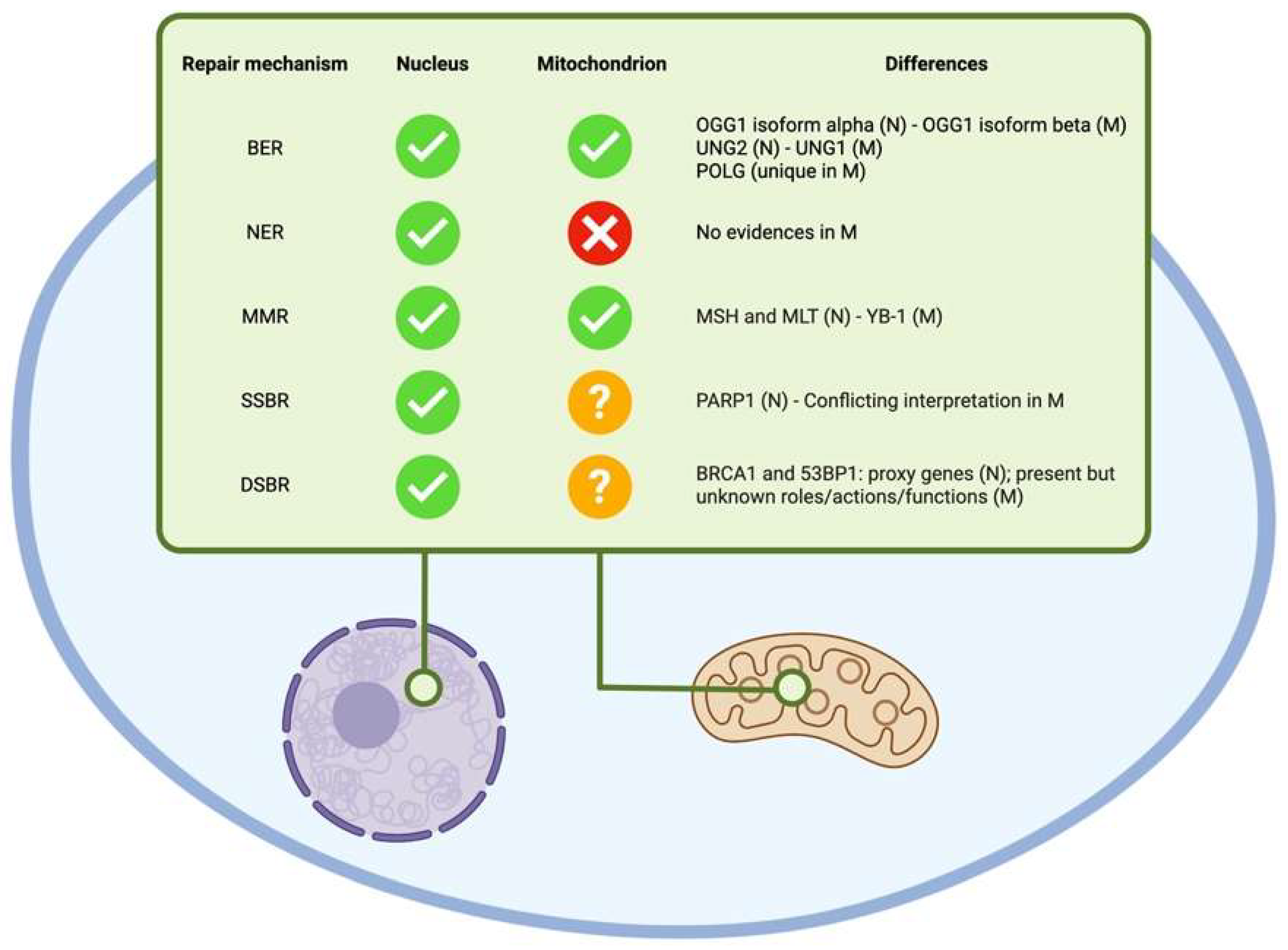
4.1. Nuclear and Mitochondrial DNA
4.2. Evolutionary Insights
5. Genomic Instability, Aging and Late Onset Complex Diseases
5.1. Telomeric Instability
5.2. Microsatellites Structural Maintenance
5.3. Mitochondrial Dysfunction
5.4. Human Networking: The Systemic Complexity of Life
6. The Epigenome: Shedding Light on the Dark Side of the Genome
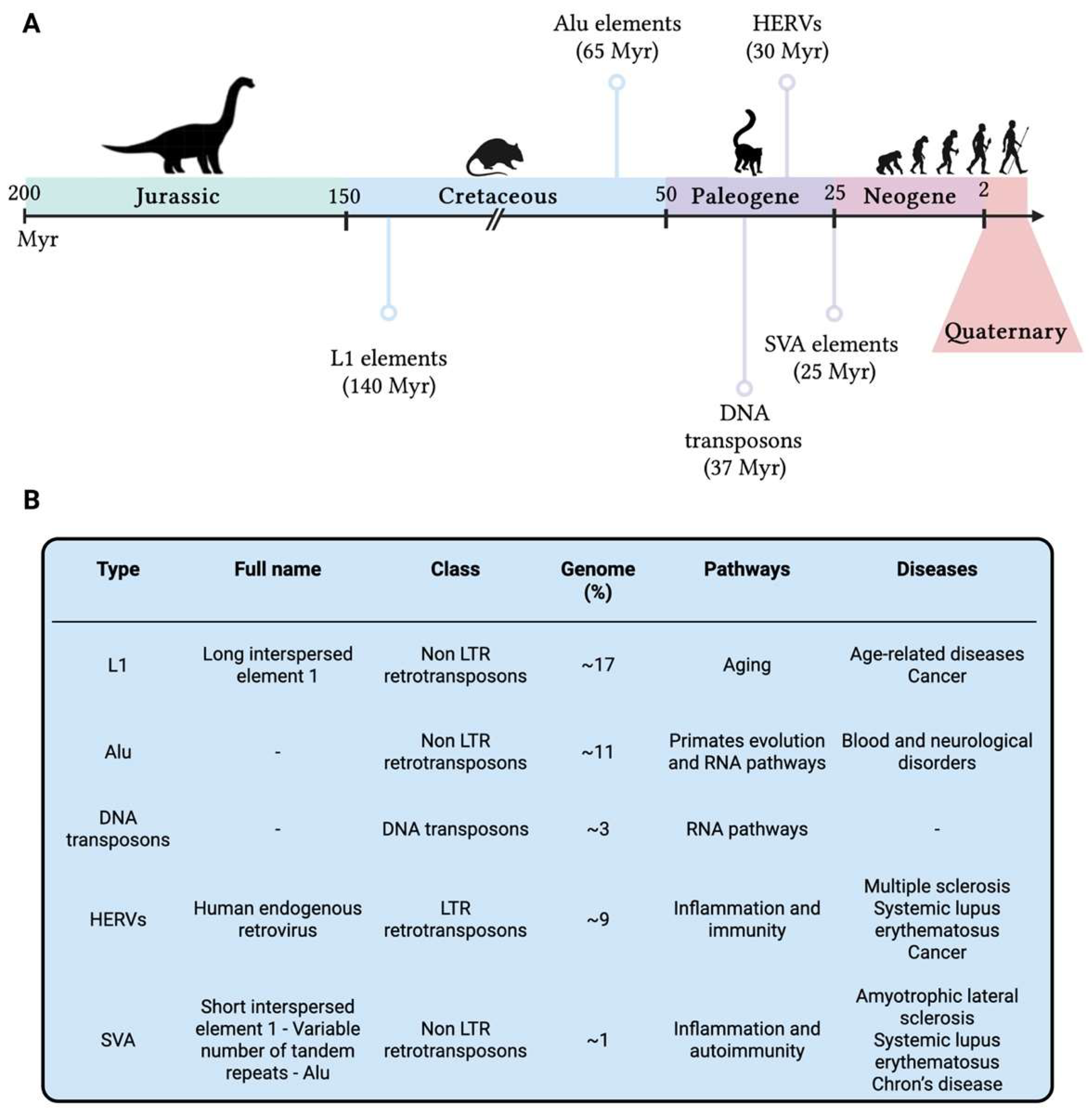
6.1. The Non-Coding Impact on Coding
6.2. The Diamond in the “Junk”
7. Towards a Better Living
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Igamberdiev, A.U. The Drawbridge of Nature: Evolutionary Complexification as a Generation and Novel Interpretation of Coding Systems. Biosystems 2021, 207, 104454. [Google Scholar] [CrossRef] [PubMed]
- Pross, A. On the Emergence of Biological Complexity: Life as a Kinetic State of Matter. Orig. Life Evol. Biosph. 2005, 35, 151–166. [Google Scholar] [CrossRef]
- Voskarides, K.; Dweep, H.; Chrysostomou, C. Evidence That DNA Repair Genes, a Family of Tumor Suppressor Genes, Are Associated with Evolution Rate and Size of Genomes. Hum. Genom. 2019, 13, 26. [Google Scholar] [CrossRef]
- Basu, A.K.; Essigmann, J.M. Establishing Linkages Among DNA Damage, Mutagenesis, and Genetic Diseases. Chem. Res. Toxicol. 2022, 35, 1655–1675. [Google Scholar] [CrossRef]
- Clarke, T.L.; Mostoslavsky, R. DNA Repair as a Shared Hallmark in Cancer and Ageing. Mol. Oncol. 2022, 16, 3352–3379. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.D.; Crick, F.H.C. Genetical Implications of the Structure of Deoxyribonucleic Acid. Nature 1953, 171, 964–967. [Google Scholar] [CrossRef] [PubMed]
- Friedberg, E.C. The Intersection between the Birth of Molecular Biology and the Discovery of DNA Repair. DNA Repair. 2002, 1, 855–867. [Google Scholar] [CrossRef]
- Kelner, A. Effect of Visible Light on the Recovery of Streptomyces Griseus Conidia from Ultra-Violet Irradiation Injury. Proc. Natl. Acad. Sci. USA 1949, 35, 73–79. [Google Scholar] [CrossRef]
- Gibson, D.G.; Glass, J.I.; Lartigue, C.; Noskov, V.N.; Chuang, R.-Y.; Algire, M.A.; Benders, G.A.; Montague, M.G.; Ma, L.; Moodie, M.M.; et al. Creation of a Bacterial Cell Controlled by a Chemically Synthesized Genome. Science 2010, 329, 52–56. [Google Scholar] [CrossRef]
- Callaway, E. ‘Minimal’ Cell Raises Stakes in Race to Harness Synthetic Life. Nature 2016, 531, 557–558. [Google Scholar] [CrossRef]
- Aravind, L.; Walker, D.R.; Koonin, E.V. Conserved Domains in DNA Repair Proteins and Evolution of Repair Systems. Nucleic Acids Res. 1999, 27, 1223–1242. [Google Scholar] [CrossRef]
- Krokan, H.E.; Bjørås, M. Base Excision Repair. Cold Spring Harb. Perspect. Biol. 2013, 5, a012583. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, T.A.; Erie, D.A. Eukaryotic Mismatch Repair in Relation to DNA Replication. Annu. Rev. Genet. 2015, 49, 291–313. [Google Scholar] [CrossRef]
- Caldecott, K.W. Single-Strand Break Repair and Genetic Disease. Nat. Rev. Genet. 2008, 9, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Kowalczykowski, S.C. An Overview of the Molecular Mechanisms of Recombinational DNA Repair. Cold Spring Harb. Perspect. Biol. 2015, 7, a016410. [Google Scholar] [CrossRef] [PubMed]
- Waters, C.A.; Strande, N.T.; Wyatt, D.W.; Pryor, J.M.; Ramsden, D.A. Nonhomologous End Joining: A Good Solution for Bad Ends. DNA Repair. 2014, 17, 39–51. [Google Scholar] [CrossRef]
- Hashimoto, S.; Anai, H.; Hanada, K. Mechanisms of Interstrand DNA Crosslink Repair and Human Disorders. Genes Environ. 2016, 38, 9. [Google Scholar] [CrossRef]
- Christie, S.M.; Fijen, C.; Rothenberg, E. V(D)J Recombination: Recent Insights in Formation of the Recombinase Complex and Recruitment of DNA Repair Machinery. Front. Cell Dev. Biol. 2022, 10, 886718. [Google Scholar] [CrossRef]
- Hanscom, T.; McVey, M. Regulation of Error-Prone DNA Double-Strand Break Repair and Its Impact on Genome Evolution. Cells 2020, 9, 1657. [Google Scholar] [CrossRef]
- Olave, M.C.; Graham, R.P. Mismatch Repair Deficiency: The What, How and Why It Is Important. Genes Chromosomes Cancer 2022, 61, 314–321. [Google Scholar] [CrossRef]
- Gonzalez, D.; Stenzinger, A. Homologous Recombination Repair Deficiency (HRD): From Biology to Clinical Exploitation. Genes Chromosomes Cancer 2021, 60, 299–302. [Google Scholar] [CrossRef]
- Shiomi, T.; Hieda-Shiomi, N.; Sato, K.; Tsuji, H.; Takahashi, E.-I.; Tobari, I. A Mouse-Cell Mutant Sensitive to Ionizing Radiation Is Hypermutable by Low Doses of γ-Radiation. Mutat. Res./Fundam. Mol. Mech. Mutagen. 1981, 83, 107–116. [Google Scholar] [CrossRef]
- Todd, P.A.; Brouwer, J.; Glickman, B.W. Influence of DNA-Repair Deficiencies on MMS- and EMS-Induced Mutagenesis in Escherichia Coli K-12. Mutat. Res./Fundam. Mol. Mech. Mutagen. 1981, 82, 239–250. [Google Scholar] [CrossRef]
- Veschetti, L.; Sandri, A.; Krogh Johansen, H.; Lleò, M.M.; Malerba, G. Hypermutation as an Evolutionary Mechanism for Achromobacter Xylosoxidans in Cystic Fibrosis Lung Infection. Pathogens 2020, 9, 72. [Google Scholar] [CrossRef] [PubMed]
- Campbell, B.B.; Light, N.; Fabrizio, D.; Zatzman, M.; Fuligni, F.; de Borja, R.; Davidson, S.; Edwards, M.; Elvin, J.A.; Hodel, K.P.; et al. Comprehensive Analysis of Hypermutation in Human Cancer. Cell 2017, 171, 1042–1056.e10. [Google Scholar] [CrossRef] [PubMed]
- Grote, A.; Earl, A.M. Within-Host Evolution of Bacterial Pathogens during Persistent Infection of Humans. Curr. Opin. Microbiol. 2022, 70, 102197. [Google Scholar] [CrossRef] [PubMed]
- Tenaillon, O.; Le Nagard, H.; Godelle, B.; Taddei, F. Mutators and Sex in Bacteria: Conflict between Adaptive Strategies. Proc. Natl. Acad. Sci. USA 2000, 97, 10465–10470. [Google Scholar] [CrossRef]
- Janin, N. A Simple Model for Carcinogenesis of Colorectal Cancers with Microsatellite Instability. Adv. Cancer Res. 2000, 77, 189–221. [Google Scholar] [CrossRef]
- Shaver, A.C.; Dombrowski, P.G.; Sweeney, J.Y.; Treis, T.; Zappala, R.M.; Sniegowski, P.D. Fitness Evolution and the Rise of Mutator Alleles in Experimental Escherichia Coli Populations. Genetics 2002, 162, 557–566. [Google Scholar] [CrossRef]
- Breivik, J.; Gaudernack, G. Resolving the Evolutionary Paradox of Genetic Instability: A Cost-Benefit Analysis of DNA Repair in Changing Environments. FEBS Lett. 2004, 563, 7–12. [Google Scholar] [CrossRef]
- Scheuerl, T.; Hopkins, M.; Nowell, R.W.; Rivett, D.W.; Barraclough, T.G.; Bell, T. Bacterial Adaptation Is Constrained in Complex Communities. Nat. Commun. 2020, 11, 754. [Google Scholar] [CrossRef]
- Veschetti, L.; Boaretti, M.; Saitta, G.M.; Passarelli Mantovani, R.; Lleò, M.M.; Sandri, A.; Malerba, G. Achromobacter Spp. Prevalence and Adaptation in Cystic Fibrosis Lung Infection. Microbiol. Res. 2022, 263, 127140. [Google Scholar] [CrossRef]
- Lyons, T.W.; Reinhard, C.T.; Planavsky, N.J. The Rise of Oxygen in Earth’s Early Ocean and Atmosphere. Nature 2014, 506, 307–315. [Google Scholar] [CrossRef]
- Warke, M.R.; Di Rocco, T.; Zerkle, A.L.; Lepland, A.; Prave, A.R.; Martin, A.P.; Ueno, Y.; Condon, D.J.; Claire, M.W. The Great Oxidation Event Preceded a Paleoproterozoic “Snowball Earth”. Proc. Natl. Acad. Sci. USA 2020, 117, 13314–13320. [Google Scholar] [CrossRef]
- Prorok, P.; Grin, I.R.; Matkarimov, B.T.; Ishchenko, A.A.; Laval, J.; Zharkov, D.O.; Saparbaev, M. Evolutionary Origins of DNA Repair Pathways: Role of Oxygen Catastrophe in the Emergence of DNA Glycosylases. Cells 2021, 10, 1591. [Google Scholar] [CrossRef]
- White, M.F.; Allers, T. DNA Repair in the Archaea—An Emerging Picture. FEMS Microbiol. Rev. 2018, 42, 514–526. [Google Scholar] [CrossRef]
- Goosen, N.; Moolenaar, G.F. Repair of UV Damage in Bacteria. DNA Repair. 2008, 7, 353–379. [Google Scholar] [CrossRef] [PubMed]
- Brunette, G.J.; Jamalruddin, M.A.; Baldock, R.A.; Clark, N.L.; Bernstein, K.A. Evolution-Based Screening Enables Genome-Wide Prioritization and Discovery of DNA Repair Genes. Proc. Natl. Acad. Sci. USA 2019, 116, 19593–19599. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, A.; Burroughs, A.M.; Iyer, L.M.; Aravind, L. Unexpected Evolution of Lesion-Recognition Modules in Eukaryotic NER and Kinetoplast DNA Dynamics Proteins from Bacterial Mobile Elements. iScience 2018, 9, 192–208. [Google Scholar] [CrossRef] [PubMed]
- Grogan, D.W. Understanding DNA Repair in Hyperthermophilic Archaea: Persistent Gaps and Other Reasons to Focus on the Fork. Archaea 2015, 2015, e942605. [Google Scholar] [CrossRef]
- Bergoglio, V.; Magnaldo, T. Nucleotide Excision Repair and Related Human Diseases. Genome Dyn. 2006, 1, 35–52. [Google Scholar] [CrossRef]
- Kelman, Z.; White, M.F. Archaeal DNA Replication and Repair. Curr. Opin. Microbiol. 2005, 8, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Ishino, S.; Nishi, Y.; Oda, S.; Uemori, T.; Sagara, T.; Takatsu, N.; Yamagami, T.; Shirai, T.; Ishino, Y. Identification of a Mismatch-Specific Endonuclease in Hyperthermophilic Archaea. Nucleic Acids Res. 2016, 44, 2977–2986. [Google Scholar] [CrossRef] [PubMed]
- Castañeda-García, A.; Prieto, A.I.; Rodríguez-Beltrán, J.; Alonso, N.; Cantillon, D.; Costas, C.; Pérez-Lago, L.; Zegeye, E.D.; Herranz, M.; Plociński, P.; et al. A Non-Canonical Mismatch Repair Pathway in Prokaryotes. Nat. Commun. 2017, 8, 14246. [Google Scholar] [CrossRef]
- Schöniger, S.; Rüschoff, J. Mismatch Repair Deficiency and Microsatellite Instability. Encyclopedia 2022, 2, 1559–1576. [Google Scholar] [CrossRef]
- Fuss, J.O.; Tsai, C.-L.; Ishida, J.P.; Tainer, J.A. Emerging Critical Roles of Fe–S Clusters in DNA Replication and Repair. Biochim. Biophys. Acta (BBA)—Mol. Cell Res. 2015, 1853, 1253–1271. [Google Scholar] [CrossRef]
- Khodour, Y.; Kaguni, L.S.; Stiban, J. Chapter Seven—Iron–Sulfur Clusters in Nucleic Acid Metabolism: Varying Roles of Ancient Cofactors. In The Enzymes; Zhao, L., Kaguni, L.S., Eds.; DNA Repair; Academic Press: Cambridge, MA, USA, 2019; Volume 45, pp. 225–256. [Google Scholar]
- Gates, F.T.; Linn, S. Endonuclease V of Escherichia coli. J. Biol. Chem. 1977, 252, 1647–1653. [Google Scholar] [CrossRef]
- Kiyonari, S.; Egashira, Y.; Ishino, S.; Ishino, Y. Biochemical Characterization of Endonuclease V from the Hyperthermophilic Archaeon, Pyrococcus Furiosus. J. Biochem. 2014, 155, 325–333. [Google Scholar] [CrossRef]
- Bowman, K.K.; Sidik, K.; Smith, C.A.; Taylor, J.-S.; Doetsch, P.W.; Freyer, G.A. A New ATP-Independent DNA Endonuclease from Schizosaccharomyces Pombe That Recognizes Cyclobutane Pyrimidine Dimers and 6–4 Photoproducts. Nucleic Acids Res. 1994, 22, 3026–3032. [Google Scholar] [CrossRef] [PubMed]
- Yajima, H.; Takao, M.; Yasuhira, S.; Zhao, J.H.; Ishii, C.; Inoue, H.; Yasui, A. A Eukaryotic Gene Encoding an Endonuclease That Specifically Repairs DNA Damaged by Ultraviolet Light. EMBO J. 1995, 14, 2393–2399. [Google Scholar] [CrossRef] [PubMed]
- Avery, A.M.; Kaur, B.; Taylor, J.-S.; Mello, J.A.; Essigmann, J.M.; Doetsch, P.W. Substrate Specificity of Ultraviolet DNA Endonuclease (UVDE/Uve1p) from Schizosaccharomyces Pombe. Nucleic Acids Res. 1999, 27, 2256–2264. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi, M.; Ishino, S.; Cann, I.; Ishino, Y. A Functional Endonuclease Q Exists in the Bacterial Domain: Identification and Characterization of Endonuclease Q from Bacillus Pumilus. Biosci. Biotechnol. Biochem. 2017, 81, 931–937. [Google Scholar] [CrossRef] [PubMed]
- White, M.F. Homologous Recombination in the Archaea: The Means Justify the Ends. Biochem. Soc. Trans. 2011, 39, 15–19. [Google Scholar] [CrossRef]
- van Wolferen, M.; Wagner, A.; van der Does, C.; Albers, S.-V. The Archaeal Ced System Imports DNA. Proc. Natl. Acad. Sci. USA 2016, 113, 2496–2501. [Google Scholar] [CrossRef]
- Naor, A.; Altman-Price, N.; Soucy, S.M.; Green, A.G.; Mitiagin, Y.; Turgeman-Grott, I.; Davidovich, N.; Gogarten, J.P.; Gophna, U. Impact of a Homing Intein on Recombination Frequency and Organismal Fitness. Proc. Natl. Acad. Sci. USA 2016, 113, E4654–E4661. [Google Scholar] [CrossRef]
- Weller, G.R.; Kysela, B.; Roy, R.; Tonkin, L.M.; Scanlan, E.; Della, M.; Devine, S.K.; Day, J.P.; Wilkinson, A.; di Fagagna, F. d’Adda; et al. Identification of a DNA Nonhomologous End-Joining Complex in Bacteria. Science 2002, 297, 1686–1689. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, B.; Sui, X.; Zhang, Z.; Liu, T.; Li, Y.; Hu, G.; He, M.; Peng, N. CRISPR-Mediated Host Genomic DNA Damage Is Efficiently Repaired through Microhomology-Mediated End Joining in Zymomonas Mobilis. J. Genet. Genom. 2021, 48, 115–122. [Google Scholar] [CrossRef]
- Bohgaki, T.; Bohgaki, M.; Hakem, R. DNA Double-Strand Break Signaling and Human Disorders. Genome Integr. 2010, 1, 15. [Google Scholar] [CrossRef]
- Luftig, M.A. Viruses and the DNA Damage Response: Activation and Antagonism. Annu. Rev. Virol. 2014, 1, 605–625. [Google Scholar] [CrossRef]
- Weitzman, M.D.; Fradet-Turcotte, A. Virus DNA Replication and the Host DNA Damage Response. Annu. Rev. Virol. 2018, 5, 141–164. [Google Scholar] [CrossRef]
- Weitzman, M.D.; Lilley, C.E.; Chaurushiya, M.S. Genomes in Conflict: Maintaining Genome Integrity During Virus Infection. Annu. Rev. Microbiol. 2010, 64, 61–81. [Google Scholar] [CrossRef]
- Lindahl, T. Instability and Decay of the Primary Structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef]
- Atamna, H.; Cheung, I.; Ames, B.N. A Method for Detecting Abasic Sites in Living Cells: Age-Dependent Changes in Base Excision Repair. Proc. Natl. Acad. Sci. USA 2000, 97, 686–691. [Google Scholar] [CrossRef]
- Martin, L.J. DNA Damage and Repair: Relevance to Mechanisms of Neurodegeneration. J. Neuropathol. Exp. Neurol. 2008, 67, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.J.; Liu, Z. DNA Damage Profiling in Motor Neurons: A Single-Cell Analysis by Comet Assay. Neurochem. Res. 2002, 27, 1093–1104. [Google Scholar] [CrossRef]
- Schumacher, B.; Pothof, J.; Vijg, J.; Hoeijmakers, J.H.J. The Central Role of DNA Damage in the Ageing Process. Nature 2021, 592, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Guachalla, L.M.; Rudolph, K.L. ROS Induced DNA Damage and Checkpoint Responses: Influences on Aging? Cell Cycle 2010, 9, 4058–4060. [Google Scholar] [CrossRef] [PubMed]
- Lees-Miller, S.P. Dysfunction of Lamin A Triggers a DNA Damage Response and Cellular Senescence. DNA Repair. 2006, 5, 286–289. [Google Scholar] [CrossRef] [PubMed]
- Simão, V.A.; Ferder, L.; Manucha, W.; Chuffa, L.G.A. Epigenetic Mechanisms Involved in Inflammaging-Associated Hypertension. Curr. Hypertens. Rep. 2022, 24, 547–562. [Google Scholar] [CrossRef]
- Rusecka, J.; Kaliszewska, M.; Bartnik, E.; Tońska, K. Nuclear Genes Involved in Mitochondrial Diseases Caused by Instability of Mitochondrial DNA. J. Appl. Genet. 2018, 59, 43–57. [Google Scholar] [CrossRef]
- Shimura, T.; Kunugita, N. Mitochondrial Reactive Oxygen Species-Mediated Genomic Instability in Low-Dose Irradiated Human Cells through Nuclear Retention of Cyclin D1. Cell Cycle 2016, 15, 1410–1414. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.-H.; Fang, W.-H.; Lin, S.-W.; Yen, S.-J.; Chou, S.-J.; Yang, Y.-C. Mitochondrial Genomic Instability in Colorectal Cancer: No Correlation to Nuclear Microsatellite Instability and Allelic Deletion of HMSH2, HMLH1, and P53 Genes, but Prediction of Better Survival for Dukes’ Stage C Disease. Ann. Surg. Oncol. 2009, 16, 2918–2925. [Google Scholar] [CrossRef]
- Baulch, J.E. Radiation-Induced Genomic Instability, Epigenetic Mechanisms and the Mitochondria: A Dysfunctional Ménage a Trois? Int. J. Radiat. Biol. 2019, 95, 516–525. [Google Scholar] [CrossRef] [PubMed]
- Groelly, F.J.; Fawkes, M.; Dagg, R.A.; Blackford, A.N.; Tarsounas, M. Targeting DNA Damage Response Pathways in Cancer. Nat. Rev. Cancer 2023, 23, 78–94. [Google Scholar] [CrossRef] [PubMed]
- Fakouri, N.B.; Hou, Y.; Demarest, T.G.; Christiansen, L.S.; Okur, M.N.; Mohanty, J.G.; Croteau, D.L.; Bohr, V.A. Toward Understanding Genomic Instability, Mitochondrial Dysfunction and Aging. FEBS J. 2019, 286, 1058–1073. [Google Scholar] [CrossRef]
- Askeland, G.; Dosoudilova, Z.; Rodinova, M.; Klempir, J.; Liskova, I.; Kuśnierczyk, A.; Bjørås, M.; Nesse, G.; Klungland, A.; Hansikova, H.; et al. Increased Nuclear DNA Damage Precedes Mitochondrial Dysfunction in Peripheral Blood Mononuclear Cells from Huntington’s Disease Patients. Sci. Rep. 2018, 8, 9817. [Google Scholar] [CrossRef]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA Damage, Repair, and Mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef]
- Ronen, A.; Glickman, B.W. Human DNA Repair Genes. Environ. Mol. Mutagen. 2001, 37, 241–283. [Google Scholar] [CrossRef]
- Alexeyev, M.; Shokolenko, I.; Wilson, G.; LeDoux, S. The Maintenance of Mitochondrial DNA Integrity—Critical Analysis and Update. Cold Spring Harb. Perspect. Biol. 2013, 5, a012641. [Google Scholar] [CrossRef]
- Zinovkina, L.A. Mechanisms of Mitochondrial DNA Repair in Mammals. Biochem. Mosc. 2018, 83, 233–249. [Google Scholar] [CrossRef]
- Rong, Z.; Tu, P.; Xu, P.; Sun, Y.; Yu, F.; Tu, N.; Guo, L.; Yang, Y. The Mitochondrial Response to DNA Damage. Front. Cell Dev. Biol. 2021, 9, 669379. [Google Scholar] [CrossRef] [PubMed]
- Palikaras, K.; Lionaki, E.; Tavernarakis, N. Coordination of Mitophagy and Mitochondrial Biogenesis during Ageing in C. Elegans. Nature 2015, 521, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Fang, E.F.; Hou, Y.; Lautrup, S.; Jensen, M.B.; Yang, B.; SenGupta, T.; Caponio, D.; Khezri, R.; Demarest, T.G.; Aman, Y.; et al. NAD+ Augmentation Restores Mitophagy and Limits Accelerated Aging in Werner Syndrome. Nat. Commun. 2019, 10, 5284. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xu, J.; Dai, X.; Shi, J.-B.; Xu, S.; Gao, J.; Yao, Q.; Liu, F. Both DNA Damage and Mitochondrial Dysfunction Are Involved in Novel Oxadiazolo[3,4-d]Pyrimidine Nucleoside Derivatives-Induced Cancer Cell Death. J. Appl. Toxicol. 2009, 29, 489–495. [Google Scholar] [CrossRef]
- Patel, J.; Baptiste, B.A.; Kim, E.; Hussain, M.; Croteau, D.L.; Bohr, V.A. DNA Damage and Mitochondria in Cancer and Aging. Carcinogenesis 2020, 41, 1625–1634. [Google Scholar] [CrossRef]
- LeDoux, S.P.; Driggers, W.J.; Hollensworth, B.S.; Wilson, G.L. Repair of Alkylation and Oxidative Damage in Mitochondrial DNA. Mutat. Res./DNA Repair. 1999, 434, 149–159. [Google Scholar] [CrossRef]
- Kazak, L.; Reyes, A.; Holt, I.J. Minimizing the Damage: Repair Pathways Keep Mitochondrial DNA Intact. Nat. Rev. Mol. Cell Biol. 2012, 13, 659–671. [Google Scholar] [CrossRef]
- Jacobs, A.L.; Schär, P. DNA Glycosylases: In DNA Repair and Beyond. Chromosoma 2012, 121, 1–20. [Google Scholar] [CrossRef]
- Longley, M.J.; Prasad, R.; Srivastava, D.K.; Wilson, S.H.; Copeland, W.C. Identification of 5′-Deoxyribose Phosphate Lyase Activity in Human DNA Polymerase γ and Its Role in Mitochondrial Base Excision Repair in Vitro. Proc. Natl. Acad. Sci. USA 1998, 95, 12244–12248. [Google Scholar] [CrossRef]
- Tahbaz, N.; Subedi, S.; Weinfeld, M. Role of Polynucleotide Kinase/Phosphatase in Mitochondrial DNA Repair. Nucleic Acids Res. 2012, 40, 3484–3495. [Google Scholar] [CrossRef]
- Karikkineth, A.C.; Scheibye-Knudsen, M.; Fivenson, E.; Croteau, D.L.; Bohr, V.A. Cockayne Syndrome: Clinical Features, Model Systems and Pathways. Ageing Res. Rev. 2017, 33, 3–17. [Google Scholar] [CrossRef]
- Mason, P.A.; Matheson, E.C.; Hall, A.G.; Lightowlers, R.N. Mismatch Repair Activity in Mammalian Mitochondria. Nucleic Acids Res. 2003, 31, 1052–1058. [Google Scholar] [CrossRef]
- de Souza-Pinto, N.C.; Mason, P.A.; Hashiguchi, K.; Weissman, L.; Tian, J.; Guay, D.; Lebel, M.; Stevnsner, T.V.; Rasmussen, L.J.; Bohr, V.A. Novel DNA Mismatch-Repair Activity Involving YB-1 in Human Mitochondria. DNA Repair. 2009, 8, 704–719. [Google Scholar] [CrossRef] [PubMed]
- Lyabin, D.N.; Eliseeva, I.A.; Ovchinnikov, L.P. YB-1 Protein: Functions and Regulation. WIREs RNA 2014, 5, 95–110. [Google Scholar] [CrossRef] [PubMed]
- Szczesny, B.; Brunyanszki, A.; Olah, G.; Mitra, S.; Szabo, C. Opposing Roles of Mitochondrial and Nuclear PARP1 in the Regulation of Mitochondrial and Nuclear DNA Integrity: Implications for the Regulation of Mitochondrial Function. Nucleic Acids Res. 2014, 42, 13161–13173. [Google Scholar] [CrossRef] [PubMed]
- Coene, E.D.; Hollinshead, M.S.; Waeytens, A.A.T.; Schelfhout, V.R.J.; Eechaute, W.P.; Shaw, M.K.; Van Oostveldt, P.M.V.; Vaux, D.J. Phosphorylated BRCA1 Is Predominantly Located in the Nucleus and Mitochondria. MBoC 2005, 16, 997–1010. [Google Scholar] [CrossRef] [PubMed]
- Youn, C.K.; Kim, H.B.; Wu, T.T.; Park, S.; Cho, S.I.; Lee, J.-H. 53BP1 Contributes to Regulation of Autophagic Clearance of Mitochondria. Sci. Rep. 2017, 7, 45290. [Google Scholar] [CrossRef]
- McKenzie, G.J.; Rosenberg, S.M. Adaptive Mutations, Mutator DNA Polymerases and Genetic Change Strategies of Pathogens. Curr. Opin. Microbiol. 2001, 4, 586–594. [Google Scholar] [CrossRef]
- Sieber, O.M.; Heinimann, K.; Tomlinson, I.P.M. Genomic Instability—The Engine of Tumorigenesis? Nat. Rev. Cancer 2003, 3, 701–708. [Google Scholar] [CrossRef]
- Little, M.P. Cancer Models, Genomic Instability and Somatic Cellular Darwinian Evolution. Biol. Direct 2010, 5, 19. [Google Scholar] [CrossRef]
- Morgan, W.F. Non-Targeted and Delayed Effects of Exposure to Ionizing Radiation: I. Radiation-Induced Genomic Instability and Bystander Effects In Vitro. Rare 2003, 159, 567–580. [Google Scholar] [CrossRef]
- Nagar, S.; Morgan, W.F. The Death-Inducing Effect and Genomic Instability. Rare 2005, 163, 316–323. [Google Scholar] [CrossRef]
- Baird, D.M. Telomeres and Genomic Evolution. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20160437. [Google Scholar] [CrossRef]
- Cordaux, R.; Batzer, M.A. The Impact of Retrotransposons on Human Genome Evolution. Nat. Rev. Genet. 2009, 10, 691–703. [Google Scholar] [CrossRef]
- McClintock, B. Controlling Elements and the Gene. Cold Spring Harb. Symp. Quant. Biol. 1956, 21, 197–216. [Google Scholar] [CrossRef]
- Williamson, A.R. The Biological Origin of Antibody Diversity. Annu. Rev. Biochem. 1976, 45, 467–500. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial Sequencing and Analysis of the Human Genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.N.; Feschotte, C. A Field Guide to Eukaryotic Transposable Elements. Annu. Rev. Genet. 2020, 54, 539–561. [Google Scholar] [CrossRef]
- Qin, S.; Jin, P.; Zhou, X.; Chen, L.; Ma, F. The Role of Transposable Elements in the Origin and Evolution of MicroRNAs in Human. PLoS ONE 2015, 10, e0131365. [Google Scholar] [CrossRef]
- Ayarpadikannan, S.; Kim, H.-S. The Impact of Transposable Elements in Genome Evolution and Genetic Instability and Their Implications in Various Diseases. Genom. Inf. 2014, 12, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Richardson, S.R.; Doucet, A.J.; Kopera, H.C.; Moldovan, J.B.; Garcia-Perez, J.L.; Moran, J.V. The Influence of LINE-1 and SINE Retrotransposons on Mammalian Genomes. Microbiol. Spectr. 2015, 3, 1165–1208. [Google Scholar] [CrossRef] [PubMed]
- Brouha, B.; Schustak, J.; Badge, R.M.; Lutz-Prigge, S.; Farley, A.H.; Moran, J.V.; Kazazian, H.H. Hot L1s Account for the Bulk of Retrotransposition in the Human Population. Proc. Natl. Acad. Sci. USA 2003, 100, 5280–5285. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Jeong, H.-H.; Hsieh, Y.-C.; Klein, H.-U.; Bennett, D.A.; De Jager, P.L.; Liu, Z.; Shulman, J.M. Tau Activates Transposable Elements in Alzheimer’s Disease. Cell Rep. 2018, 23, 2874–2880. [Google Scholar] [CrossRef]
- Ko, K.; Kananazawa, Y.; Yamada, T.; Kakinuma, D.; Matsuno, K.; Ando, F.; Kuriyama, S.; Matsuda, A.; Yoshida, H. Methylation Status and Long-Fragment Cell-Free DNA Are Prognostic Biomarkers for Gastric Cancer. Cancer Med. 2021, 10, 2003–2012. [Google Scholar] [CrossRef]
- Wallace, N.; Wagstaff, B.J.; Deininger, P.L.; Roy-Engel, A.M. LINE-1 ORF1 Protein Enhances Alu SINE Retrotransposition. Gene 2008, 419, 1–6. [Google Scholar] [CrossRef]
- Weiner, A.M. SINEs and LINEs: The Art of Biting the Hand That Feeds You. Curr. Opin. Cell Biol. 2002, 14, 343–350. [Google Scholar] [CrossRef]
- Kriegs, J.O.; Churakov, G.; Jurka, J.; Brosius, J.; Schmitz, J. Evolutionary History of 7SL RNA-Derived SINEs in Supraprimates. Trends Genet. 2007, 23, 158–161. [Google Scholar] [CrossRef]
- Pace, J.K.; Feschotte, C. The Evolutionary History of Human DNA Transposons: Evidence for Intense Activity in the Primate Lineage. Genome Res. 2007, 17, 422–432. [Google Scholar] [CrossRef]
- Kumar, A. Jump around: Transposons in and out of the Laboratory. F1000Research 2020, 9, F1000 Faculty Rev-135. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, J.; Brosius, J. Exonization of Transposed Elements: A Challenge and Opportunity for Evolution. Biochimie 2011, 93, 1928–1934. [Google Scholar] [CrossRef]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-Interacting RNAs: Small RNAs with Big Functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef] [PubMed]
- Mills, R.E.; Bennett, E.A.; Iskow, R.C.; Devine, S.E. Which Transposable Elements Are Active in the Human Genome? Trends Genet. 2007, 23, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Gilbert, N.; Ooi, S.L.; Lawler, J.F.; Ostertag, E.M.; Kazazian, H.H.; Boeke, J.D.; Moran, J.V. Human L1 Retrotransposition: CisPreference versus Trans Complementation. Mol. Cell. Biol. 2001, 21, 1429–1439. [Google Scholar] [CrossRef]
- Mager, D.L.; Stoye, J.P. Mammalian Endogenous Retroviruses. Microbiol. Spectr. 2015, 3, 1079–1100. [Google Scholar] [CrossRef] [PubMed]
- Balada, E.; Ordi-Ros, J.; Vilardell-Tarrés, M. Molecular mechanisms mediated by human endogenous retroviruses (HERVs) in autoimmunity. Rev. Med. Virol. 2009, 19, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Latifi, T.; Zebardast, A.; Marashi, S.M. The Role of Human Endogenous Retroviruses (HERVs) in Multiple Sclerosis and the Plausible Interplay between HERVs, Epstein–Barr Virus Infection, and Vitamin D. Mult. Scler. Relat. Disord. 2022, 57, 103318. [Google Scholar] [CrossRef]
- Quaglia, M.; Merlotti, G.; De Andrea, M.; Borgogna, C.; Cantaluppi, V. Viral Infections and Systemic Lupus Erythematosus: New Players in an Old Story. Viruses 2021, 13, 277. [Google Scholar] [CrossRef]
- Gao, Y.; Yu, X.-F.; Chen, T. Human Endogenous Retroviruses in Cancer: Expression, Regulation and Function (Review). Oncol. Lett. 2021, 21, 121. [Google Scholar] [CrossRef]
- Hancks, D.C.; Kazazian, H.H. SVA Retrotransposons: Evolution and Genetic Instability. Semin. Cancer Biol. 2010, 20, 234–245. [Google Scholar] [CrossRef]
- Ostertag, E.M.; Goodier, J.L.; Zhang, Y.; Kazazian, H.H. SVA Elements Are Nonautonomous Retrotransposons That Cause Disease in Humans. Am. J. Hum. Genet. 2003, 73, 1444–1451. [Google Scholar] [CrossRef]
- Raiz, J.; Damert, A.; Chira, S.; Held, U.; Klawitter, S.; Hamdorf, M.; Löwer, J.; Strätling, W.H.; Löwer, R.; Schumann, G.G. The Non-Autonomous Retrotransposon SVA Is Trans -Mobilized by the Human LINE-1 Protein Machinery. Nucleic Acids Res. 2012, 40, 1666–1683. [Google Scholar] [CrossRef] [PubMed]
- Savage, A.L.; Wilm, T.P.; Khursheed, K.; Shatunov, A.; Morrison, K.E.; Shaw, P.J.; Shaw, C.E.; Smith, B.; Breen, G.; Al-Chalabi, A.; et al. An Evaluation of a SVA Retrotransposon in the FUS Promoter as a Transcriptional Regulator and Its Association to ALS. PLoS ONE 2014, 9, e90833. [Google Scholar] [CrossRef] [PubMed]
- Lauc, G.; Huffman, J.E.; Pučić, M.; Zgaga, L.; Adamczyk, B.; Mužinić, A.; Novokmet, M.; Polašek, O.; Gornik, O.; Krištić, J.; et al. Loci Associated with N-Glycosylation of Human Immunoglobulin G Show Pleiotropy with Autoimmune Diseases and Haematological Cancers. PLoS Genet. 2013, 9, e1003225. [Google Scholar] [CrossRef] [PubMed]
- Jurka, J.; Kapitonov, V.V.; Kohany, O.; Jurka, M.V. Repetitive Sequences in Complex Genomes: Structure and Evolution. Annu. Rev. Genom. Hum. Genet. 2007, 8, 241–259. [Google Scholar] [CrossRef] [PubMed]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory Activities of Transposable Elements: From Conflicts to Benefits. Nat. Rev. Genet. 2017, 18, 71–86. [Google Scholar] [CrossRef]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory Evolution of Innate Immunity through Co-Option of Endogenous Retroviruses. Science 2016, 351, 1083–1087. [Google Scholar] [CrossRef]
- Gerdes, P.; Richardson, S.R.; Mager, D.L.; Faulkner, G.J. Transposable Elements in the Mammalian Embryo: Pioneers Surviving through Stealth and Service. Genome Biol. 2016, 17, 100. [Google Scholar] [CrossRef]
- Wang, L.; Jordan, I.K. Transposable Element Activity, Genome Regulation and Human Health. Curr. Opin. Genet. Dev. 2018, 49, 25–33. [Google Scholar] [CrossRef]
- Bodega, B.; Orlando, V. Repetitive Elements Dynamics in Cell Identity Programming, Maintenance and Disease. Curr. Opin. Cell Biol. 2014, 31, 67–73. [Google Scholar] [CrossRef]
- Suomalainen, A. Mitochondrial DNA and Disease. Ann. Med. 1997, 29, 235–246. [Google Scholar] [CrossRef]
- Sargurupremraj, M.; Wjst, M. Transposable Elements and Their Potential Role in Complex Lung Disorder. Respir. Res. 2013, 14, 99. [Google Scholar] [CrossRef] [PubMed]
- Gorbunova, V.; Seluanov, A.; Mao, Z.; Hine, C. Changes in DNA Repair during Aging. Nucleic Acids Res. 2007, 35, 7466–7474. [Google Scholar] [CrossRef]
- Niedernhofer, L.J.; Gurkar, A.U.; Wang, Y.; Vijg, J.; Hoeijmakers, J.H.J.; Robbins, P.D. Nuclear Genomic Instability and Aging. Annu. Rev. Biochem. 2018, 87, 295–322. [Google Scholar] [CrossRef] [PubMed]
- Laffon, B.; Bonassi, S.; Costa, S.; Valdiglesias, V. Genomic Instability as a Main Driving Factor of Unsuccessful Ageing: Potential for Translating the Use of Micronuclei into Clinical Practice. Mutat. Res./Rev. Mutat. Res. 2021, 787, 108359. [Google Scholar] [CrossRef] [PubMed]
- Vermeij, W.P.; Hoeijmakers, J.H.J.; Pothof, J. Genome Integrity in Aging: Human Syndromes, Mouse Models, and Therapeutic Options. Annu. Rev. Pharmacol. Toxicol. 2016, 56, 427–445. [Google Scholar] [CrossRef]
- Vijg, J.; Suh, Y. Genome Instability and Aging. Annu. Rev. Physiol. 2013, 75, 645–668. [Google Scholar] [CrossRef]
- Shaposhnikov, M.; Proshkina, E.; Shilova, L.; Zhavoronkov, A.; Moskalev, A. Lifespan and Stress Resistance in Drosophila with Overexpressed DNA Repair Genes. Sci. Rep. 2015, 5, 15299. [Google Scholar] [CrossRef]
- Kanfi, Y.; Naiman, S.; Amir, G.; Peshti, V.; Zinman, G.; Nahum, L.; Bar-Joseph, Z.; Cohen, H.Y. The Sirtuin SIRT6 Regulates Lifespan in Male Mice. Nature 2012, 483, 218–221. [Google Scholar] [CrossRef]
- Salomon, J.A.; Wang, H.; Freeman, M.K.; Vos, T.; Flaxman, A.D.; Lopez, A.D.; Murray, C.J. Healthy Life Expectancy for 187 Countries, 1990–2010: A Systematic Analysis for the Global Burden Disease Study 2010. Lancet 2012, 380, 2144–2162. [Google Scholar] [CrossRef]
- Flatt, T.; Partridge, L. Horizons in the Evolution of Aging. BMC Biol. 2018, 16, 93. [Google Scholar] [CrossRef]
- Finch, C.E. Evolution of the Human Lifespan and Diseases of Aging: Roles of Infection, Inflammation, and Nutrition. Proc. Natl. Acad. Sci. USA 2010, 107, 1718–1724. [Google Scholar] [CrossRef] [PubMed]
- Crimmins, E.M. Lifespan and Healthspan: Past, Present, and Promise. Gerontologist 2015, 55, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, D.K.; Escott-Price, V.; Ziehm, M.; Magwire, M.M.; Mackay, T.F.C.; Partridge, L.; Thornton, J.M. Longevity GWAS Using the Drosophila Genetic Reference Panel. J. Gerontol. Ser. A 2015, 70, 1470–1478. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Maklakov, A.A. Longer Life Span Evolves under High Rates of Condition-Dependent Mortality. Curr. Biol. 2012, 22, 2140–2143. [Google Scholar] [CrossRef]
- Blasco, M.A. Telomeres and Human Disease: Ageing, Cancer and Beyond. Nat. Rev. Genet. 2005, 6, 611–622. [Google Scholar] [CrossRef]
- Palm, W.; de Lange, T. How Shelterin Protects Mammalian Telomeres. Annu. Rev. Genet. 2008, 42, 301–334. [Google Scholar] [CrossRef]
- Ferrucci, L.; Gonzalez-Freire, M.; Fabbri, E.; Simonsick, E.; Tanaka, T.; Moore, Z.; Salimi, S.; Sierra, F.; de Cabo, R. Measuring Biological Aging in Humans: A Quest. Aging Cell 2020, 19, e13080. [Google Scholar] [CrossRef]
- Marion, R.M.; Strati, K.; Li, H.; Tejera, A.; Schoeftner, S.; Ortega, S.; Serrano, M.; Blasco, M.A. Telomeres Acquire Embryonic Stem Cell Characteristics in Induced Pluripotent Stem Cells. Cell Stem Cell 2009, 4, 141–154. [Google Scholar] [CrossRef] [PubMed]
- García-Calzón, S.; Gea, A.; Razquin, C.; Corella, D.; Lamuela-Raventós, R.M.; Martínez, J.A.; Martínez-González, M.A.; Zalba, G.; Marti, A. Longitudinal Association of Telomere Length and Obesity Indices in an Intervention Study with a Mediterranean Diet: The PREDIMED-NAVARRA Trial. Int. J. Obes. 2014, 38, 177–182. [Google Scholar] [CrossRef]
- Mundstock, E.; Sarria, E.E.; Zatti, H.; Mattos Louzada, F.; Kich Grun, L.; Herbert Jones, M.; Guma, F.T.C.R.; Mazzola (in Memoriam), J.; Epifanio, M.; Stein, R.T.; et al. Effect of Obesity on Telomere Length: Systematic Review and Meta-Analysis. Obesity 2015, 23, 2165–2174. [Google Scholar] [CrossRef]
- Osler, M.; Bendix, L.; Rask, L.; Rod, N.H. Stressful Life Events and Leucocyte Telomere Length: Do Lifestyle Factors, Somatic and Mental Health, or Low Grade Inflammation Mediate This Relationship? Results from a Cohort of Danish Men Born in 1953. Brain Behav. Immun. 2016, 58, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Puterman, E.; Gemmill, A.; Karasek, D.; Weir, D.; Adler, N.E.; Prather, A.A.; Epel, E.S. Lifespan Adversity and Later Adulthood Telomere Length in the Nationally Representative US Health and Retirement Study. Proc. Natl. Acad. Sci. USA 2016, 113, E6335–E6342. [Google Scholar] [CrossRef] [PubMed]
- Baragetti, A.; Palmen, J.; Garlaschelli, K.; Grigore, L.; Humphries, S.; Catapano, A.L.; Talmud, P.J.; Giuseppe Danilo, N. Leukocyte Telomere Length, Genetically Determined, Is Causally Associated with the Progression of Carotid Intima-Media Thickness and Incidence of Cardiovascular Events. Atherosclerosis 2016, 252, e252. [Google Scholar] [CrossRef]
- Hammadah, M.; Al Mheid, I.; Wilmot, K.; Ramadan, R.; Abdelhadi, N.; Alkhoder, A.; Obideen, M.; Pimple, P.M.; Levantsevych, O.; Kelli, H.M.; et al. Telomere Shortening, Regenerative Capacity, and Cardiovascular Outcomes. Circ. Res. 2017, 120, 1130–1138. [Google Scholar] [CrossRef] [PubMed]
- Najarro, K.; Nguyen, H.; Chen, G.; Xu, M.; Alcorta, S.; Yao, X.; Zukley, L.; Metter, E.J.; Truong, T.; Lin, Y.; et al. Telomere Length as an Indicator of the Robustness of B- and T-Cell Response to Influenza in Older Adults. J. Infect. Dis. 2015, 212, 1261–1269. [Google Scholar] [CrossRef]
- Martínez, P.; Blasco, M.A. Telomere-Driven Diseases and Telomere-Targeting Therapies. J. Cell Biol. 2017, 216, 875–887. [Google Scholar] [CrossRef]
- Stadler, G.; Rahimov, F.; King, O.D.; Chen, J.C.J.; Robin, J.D.; Wagner, K.R.; Shay, J.W.; Emerson, C.P.; Wright, W.E. Telomere Position Effect Regulates DUX4 in Human Facioscapulohumeral Muscular Dystrophy. Nat. Struct. Mol. Biol. 2013, 20, 671–678. [Google Scholar] [CrossRef]
- Halldorsson, B.V.; Eggertsson, H.P.; Moore, K.H.S.; Hauswedell, H.; Eiriksson, O.; Ulfarsson, M.O.; Palsson, G.; Hardarson, M.T.; Oddsson, A.; Jensson, B.O.; et al. The Sequences of 150,119 Genomes in the UK Biobank. Nature 2022, 607, 732–740. [Google Scholar] [CrossRef]
- Dieringer, D.; Schlötterer, C. Two Distinct Modes of Microsatellite Mutation Processes: Evidence from the Complete Genomic Sequences of Nine Species. Genome Res. 2003, 13, 2242–2251. [Google Scholar] [CrossRef]
- Brinkmann, B.; Klintschar, M.; Neuhuber, F.; Hühne, J.; Rolf, B. Mutation Rate in Human Microsatellites: Influence of the Structure and Length of the Tandem Repeat. Am. J. Hum. Genet. 1998, 62, 1408–1415. [Google Scholar] [CrossRef]
- De’ Angelis, G.L.; Bottarelli, L.; Azzoni, C.; De’ Angelis, N.; Leandro, G.; Di Mario, F.; Gaiani, F.; Negri, F. Microsatellite Instability in Colorectal Cancer. Acta Biomed. 2018, 89, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Puliga, E.; Corso, S.; Pietrantonio, F.; Giordano, S. Microsatellite Instability in Gastric Cancer: Between Lights and Shadows. Cancer Treat. Rev. 2021, 95, 102175. [Google Scholar] [CrossRef] [PubMed]
- Vanoli, A.; Di Sabatino, A.; Martino, M.; Klersy, C.; Grillo, F.; Mescoli, C.; Nesi, G.; Volta, U.; Fornino, D.; Luinetti, O.; et al. Small Bowel Carcinomas in Celiac or Crohn’s Disease: Distinctive Histophenotypic, Molecular and Histogenetic Patterns. Mod. Pathol. 2017, 30, 1453–1466. [Google Scholar] [CrossRef] [PubMed]
- Diniz, G.; Aktas, S.; Cubuk, C.; Ortac, R.; Vergin, C.; Olgun, N. Tissue Expression of MLH1, PMS2, MSH2, and MSH6 Proteins and Prognostic Value of Microsatellite Instability in Wilms Tumor: Experience of 45 Cases. Pediatr. Hematol. Oncol. 2013, 30, 273–284. [Google Scholar] [CrossRef]
- Latham, A.; Srinivasan, P.; Kemel, Y.; Shia, J.; Bandlamudi, C.; Mandelker, D.; Middha, S.; Hechtman, J.; Zehir, A.; Dubard-Gault, M.; et al. Microsatellite Instability Is Associated With the Presence of Lynch Syndrome Pan-Cancer. J. Clin. Oncol. 2019, 37, 286–295. [Google Scholar] [CrossRef]
- Wieland, J.; Buchan, S.; Sen Gupta, S.; Mantzouratou, A. Genomic Instability and the Link to Infertility: A Focus on Microsatellites and Genomic Instability Syndromes. Eur. J. Obstet. Gynecol. Reprod. Biol. 2022, 274, 229–237. [Google Scholar] [CrossRef]
- Harman, D. The Free Radical Theory of Aging. Antioxid. Redox Signal. 2003, 5, 557–561. [Google Scholar] [CrossRef]
- Holloway, G.P.; Holwerda, A.M.; Miotto, P.M.; Dirks, M.L.; Verdijk, L.B.; van Loon, L.J.C. Age-Associated Impairments in Mitochondrial ADP Sensitivity Contribute to Redox Stress in Senescent Human Skeletal Muscle. Cell Rep. 2018, 22, 2837–2848. [Google Scholar] [CrossRef]
- Marazziti, D.; Baroni, S.; Picchetti, M.; Landi, P.; Silvestri, S.; Vatteroni, E.; Catena Dell’Osso, M. Psychiatric Disorders and Mitochondrial Dysfunctions. Eur. Rev. Med. Pharmacol. Sci. 2012, 16, 270–275. [Google Scholar]
- Flippo, K.H.; Strack, S. An Emerging Role for Mitochondrial Dynamics in Schizophrenia. Schizophr. Res. 2017, 187, 26–32. [Google Scholar] [CrossRef]
- Misrani, A.; Tabassum, S.; Yang, L. Mitochondrial Dysfunction and Oxidative Stress in Alzheimer’s Disease. Front. Aging Neurosci. 2021, 13, 57. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Kato, N. Mitochondrial Dysfunction in Bipolar Disorder. Bipolar Disord. 2000, 2, 180–190. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, M.F.; Elwell, C.; Johnson, M.H. Mitochondrial Dysfunction in Autism Spectrum Disorders. Autism Open Access 2016, 6, 1000190. [Google Scholar] [CrossRef]
- Verma, P.; Singh, A.; Nthenge-Ngumbau, D.N.; Rajamma, U.; Sinha, S.; Mukhopadhyay, K.; Mohanakumar, K.P. Attention Deficit-Hyperactivity Disorder Suffers from Mitochondrial Dysfunction. BBA Clin. 2016, 6, 153–158. [Google Scholar] [CrossRef]
- Malpartida, A.B.; Williamson, M.; Narendra, D.P.; Wade-Martins, R.; Ryan, B.J. Mitochondrial Dysfunction and Mitophagy in Parkinson’s Disease: From Mechanism to Therapy. Trends Biochem. Sci. 2021, 46, 329–343. [Google Scholar] [CrossRef]
- Mathers, C.D.; Boerma, T.; Ma Fat, D. Global and Regional Causes of Death. Br. Med. Bull. 2009, 92, 7–32. [Google Scholar] [CrossRef]
- Roslund, M.I.; Parajuli, A.; Hui, N.; Puhakka, R.; Grönroos, M.; Soininen, L.; Nurminen, N.; Oikarinen, S.; Cinek, O.; Kramná, L.; et al. A Placebo-Controlled Double-Blinded Test of the Biodiversity Hypothesis of Immune-Mediated Diseases: Environmental Microbial Diversity Elicits Changes in Cytokines and Increase in T Regulatory Cells in Young Children. Ecotoxicol. Environ. Saf. 2022, 242, 113900. [Google Scholar] [CrossRef]
- Hartstra, A.V.; Bouter, K.E.C.; Bäckhed, F.; Nieuwdorp, M. Insights Into the Role of the Microbiome in Obesity and Type 2 Diabetes. Diabetes Care 2014, 38, 159–165. [Google Scholar] [CrossRef]
- Lee, C.J.; Sears, C.L.; Maruthur, N. Gut Microbiome and Its Role in Obesity and Insulin Resistance. Ann. N. Y. Acad. Sci. 2020, 1461, 37–52. [Google Scholar] [CrossRef]
- Liddicoat, C.; Sydnor, H.; Cando-Dumancela, C.; Dresken, R.; Liu, J.; Gellie, N.J.C.; Mills, J.G.; Young, J.M.; Weyrich, L.S.; Hutchinson, M.R.; et al. Naturally-Diverse Airborne Environmental Microbial Exposures Modulate the Gut Microbiome and May Provide Anxiolytic Benefits in Mice. Sci. Total Environ. 2020, 701, 134684. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Liao, S.; He, Y.; Wang, S.; Xia, G.; Liu, F.; Zhu, J.; You, C.; Chen, Q.; Zhou, L.; et al. Dysbiosis of Gut Microbiota with Reduced Trimethylamine-N-Oxide Level in Patients With Large-Artery Atherosclerotic Stroke or Transient Ischemic Attack. J. Am. Heart Assoc. 2015, 4, e002699. [Google Scholar] [CrossRef] [PubMed]
- Brooks, C.N.; Wight, M.E.; Azeez, O.E.; Bleich, R.M.; Zwetsloot, K.A. Growing Old Together: What We Know about the Influence of Diet and Exercise on the Aging Host’s Gut Microbiome. Front. Sports Act. Living 2023, 5, 1168731. [Google Scholar] [CrossRef] [PubMed]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The Hallmarks of Aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Tyler, J.K. Epigenetics and Aging. Sci. Adv. 2016, 2, e1600584. [Google Scholar] [CrossRef]
- Bernstein, B.E.; Meissner, A.; Lander, E.S. The Mammalian Epigenome. Cell 2007, 128, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Fraga, M.F.; Ballestar, E.; Paz, M.F.; Ropero, S.; Setien, F.; Ballestar, M.L.; Heine-Suñer, D.; Cigudosa, J.C.; Urioste, M.; Benitez, J.; et al. Epigenetic Differences Arise during the Lifetime of Monozygotic Twins. Proc. Natl. Acad. Sci. USA 2005, 102, 10604–10609. [Google Scholar] [CrossRef]
- Kucharski, R.; Maleszka, J.; Foret, S.; Maleszka, R. Nutritional Control of Reproductive Status in Honeybees via DNA Methylation. Science 2008, 319, 1827–1830. [Google Scholar] [CrossRef]
- Kane, A.E.; Sinclair, D.A. Epigenetic Changes during Aging and Their Reprogramming Potential. Crit. Rev. Biochem. Mol. Biol. 2019, 54, 61–83. [Google Scholar] [CrossRef]
- Saul, D.; Kosinsky, R.L. Epigenetics of Aging and Aging-Associated Diseases. Int. J. Mol. Sci. 2021, 22, 401. [Google Scholar] [CrossRef]
- Bahar, R.; Hartmann, C.H.; Rodriguez, K.A.; Denny, A.D.; Busuttil, R.A.; Dollé, M.E.T.; Calder, R.B.; Chisholm, G.B.; Pollock, B.H.; Klein, C.A.; et al. Increased Cell-to-Cell Variation in Gene Expression in Ageing Mouse Heart. Nature 2006, 441, 1011–1014. [Google Scholar] [CrossRef]
- Nicholas, A.; de Magalhaes, J.P.; Kraytsberg, Y.; Richfield, E.K.; Levanon, E.Y.; Khrapko, K. Age-Related Gene-Specific Changes of A-to-I MRNA Editing in the Human Brain. Mech. Ageing Dev. 2010, 131, 445–447. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Duan, J.; Du, Y.; Xie, J.; Zhang, H.; Li, C.; Zhang, W. Long Non-Coding RNA Signatures Associated With Liver Aging in Senescence-Accelerated Mouse Prone 8 Model. Front. Cell Dev. Biol. 2021, 9, 698442. [Google Scholar] [CrossRef] [PubMed]
- Stegeman, R.; Weake, V.M. Transcriptional Signatures of Aging. J. Mol. Biol. 2017, 429, 2427–2437. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.C.; Leitão, A.L.; Enguita, F.J. Noncoding Transcriptional Landscape in Human Aging. Curr. Top Microbiol. Immunol. 2016, 394, 177–202. [Google Scholar] [CrossRef]
- Barth, E.; Srivastava, A.; Stojiljkovic, M.; Frahm, C.; Axer, H.; Witte, O.W.; Marz, M. Conserved Aging-Related Signatures of Senescence and Inflammation in Different Tissues and Species. Aging 2019, 11, 8556–8572. [Google Scholar] [CrossRef]
- Frahm, C.; Srivastava, A.; Schmidt, S.; Mueller, J.; Groth, M.; Guenther, M.; Ji, Y.; Priebe, S.; Platzer, M.; Witte, O.W. Transcriptional Profiling Reveals Protective Mechanisms in Brains of Long-Lived Mice. Neurobiol. Aging 2017, 52, 23–31. [Google Scholar] [CrossRef]
- Boehm, M.; Slack, F. A Developmental Timing MicroRNA and Its Target Regulate Life Span in C. Elegans. Science 2005, 310, 1954–1957. [Google Scholar] [CrossRef]
- Ugalde, A.P.; Kwarciak, A.; Caravia, X.M.; López-Otín, C.; Ramsay, A.J. The Emergence of GeroMIRs: A Group of MicroRNAs Implicated in Aging. In MicroRNAs in Medicine; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2013; pp. 77–89. ISBN 978-1-118-30031-2. [Google Scholar]
- Esteller, M. Non-Coding RNAs in Human Disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Moskalev, A.; Aliper, A.; Smit-McBride, Z.; Buzdin, A.; Zhavoronkov, A. Genetics and Epigenetics of Aging and Longevity. Cell Cycle 2014, 13, 1063–1077. [Google Scholar] [CrossRef]
- Olivieri, F.; Rippo, M.R.; Procopio, A.D.; Fazioli, F. Circulating Inflamma-MiRs in Aging and Age-Related Diseases. Front. Genet. 2013, 4, 121. [Google Scholar] [CrossRef]
- Wang, L.-L.; Huang, Y.; Wang, G.; Chen, S.-D. The Potential Role of MicroRNA-146 in Alzheimer’s Disease: Biomarker or Therapeutic Target? Med. Hypotheses 2012, 78, 398–401. [Google Scholar] [CrossRef]
- DiStefano, J.K. The Emerging Role of Long Noncoding RNAs in Human Disease. In Disease Gene Identification: Methods and Protocols; DiStefano, J.K., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2018; pp. 91–110. ISBN 978-1-4939-7471-9. [Google Scholar]
- Guo, J.; Huang, X.; Dou, L.; Yan, M.; Shen, T.; Tang, W.; Li, J. Aging and Aging-Related Diseases: From Molecular Mechanisms to Interventions and Treatments. Signal Transduct. Target. Ther. 2022, 7, 391. [Google Scholar] [CrossRef] [PubMed]
- Curtin, N.J. DNA Repair Dysregulation from Cancer Driver to Therapeutic Target. Nat. Rev. Cancer 2012, 12, 801–817. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Peng, G. Non-Coding RNAs: An Emerging Player in DNA Damage Response. Mutat. Res./Rev. Mutat. Res. 2015, 763, 202–211. [Google Scholar] [CrossRef]
- Wan, G.; Hu, X.; Liu, Y.; Han, C.; Sood, A.K.; Calin, G.A.; Zhang, X.; Lu, X. A Novel Non-Coding RNA LncRNA-JADE Connects DNA Damage Signalling to Histone H4 Acetylation. EMBO J. 2013, 32, 2833–2847. [Google Scholar] [CrossRef]
- Wei, W.; Ba, Z.; Gao, M.; Wu, Y.; Ma, Y.; Amiard, S.; White, C.I.; Rendtlew Danielsen, J.M.; Yang, Y.-G.; Qi, Y. A Role for Small RNAs in DNA Double-Strand Break Repair. Cell 2012, 149, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Makunin, I.V. Non-Coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef]
- Vicentini, C.; Galuppini, F.; Corbo, V.; Fassan, M. Current Role of Non-Coding RNAs in the Clinical Setting. Non-Coding RNA Res. 2019, 4, 82–85. [Google Scholar] [CrossRef]
- Garbo, S.; Maione, R.; Tripodi, M.; Battistelli, C. Next RNA Therapeutics: The Mine of Non-Coding. Int. J. Mol. Sci. 2022, 23, 7471. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veschetti, L.; Treccani, M.; De Tomi, E.; Malerba, G. Genomic Instability Evolutionary Footprints on Human Health: Driving Forces or Side Effects? Int. J. Mol. Sci. 2023, 24, 11437. https://doi.org/10.3390/ijms241411437
Veschetti L, Treccani M, De Tomi E, Malerba G. Genomic Instability Evolutionary Footprints on Human Health: Driving Forces or Side Effects? International Journal of Molecular Sciences. 2023; 24(14):11437. https://doi.org/10.3390/ijms241411437
Chicago/Turabian StyleVeschetti, Laura, Mirko Treccani, Elisa De Tomi, and Giovanni Malerba. 2023. "Genomic Instability Evolutionary Footprints on Human Health: Driving Forces or Side Effects?" International Journal of Molecular Sciences 24, no. 14: 11437. https://doi.org/10.3390/ijms241411437
APA StyleVeschetti, L., Treccani, M., De Tomi, E., & Malerba, G. (2023). Genomic Instability Evolutionary Footprints on Human Health: Driving Forces or Side Effects? International Journal of Molecular Sciences, 24(14), 11437. https://doi.org/10.3390/ijms241411437








