A 43 Bp-Deletion in the F3′H Gene Reducing Anthocyanins Is Responsible for Keeping Buds Green at Low Temperatures in Broccoli
Abstract
1. Introduction
2. Results
2.1. Anthocyanin Contents
2.2. Mapping of the Green/Purple Bud Trait at Cold Temperatures in Broccoli
2.3. Fine Mapping
2.4. The Application of the KASP Markers
2.5. Expression Profile of Anthocyanin-Related Genes in the Mutant and the Wild Line
2.6. Validation of Anthocyanin-Related Genes by qRT-PCR
2.7. cDNA and Predicted Amino Acid Sequences of the F3′H Gene in HX-11 and HX-16
3. Discussion
4. Materials and Methods
4.1. Construction of F2 and Backcross Populations
4.2. Anthocyanin Extraction and Measurement
4.3. Genomic DNA Extraction and Library Construction
4.4. QTL Mapping
4.5. KASP Primer Designing and Genotyping
4.6. Transcriptome Analysis
4.7. The CDS and Predicted Amino Acid Sequences of F3′H in HX-11 and HX-16
4.8. qRT-PCR Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Springob, K.; Nakajima, J.; Yamazaki, M.; Saito, K. Recent advances in the biosynthesis and accumulation of anthocyanins. Nat. Prod. Rep. 2003, 20, 288–303. [Google Scholar] [PubMed]
- Lepiniec, L.; Debeaujon, I.; Routaboul, J.M.; Baudry, A.; Pourcel, L.; Nesi, N.; Caboche, M. Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 2006, 57, 405–430. [Google Scholar]
- Saito, K.; Yonekura-Sakakibara, K.; Nakabayashi, R.; Higashi, Y.; Yamazaki, M.; Tohge, T.; Fernie, A.R. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol. Biochem. 2013, 72, 21–34. [Google Scholar] [CrossRef]
- Tohge, T.; Fernie, A.R. Leveraging natural variance towards enhanced understanding of phytochemical sunscreens. Trends Plant Sci. 2017, 22, 308–315. [Google Scholar]
- Saigo, T.; Wang, T.; Watanabe, M.; Tohge, T. Diversity of anthocyanin and proanthocyanin biosynthesis in land plants. Curr. Opin. Plant Biol. 2020, 55, 93–99. [Google Scholar] [PubMed]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef]
- Grotewold, E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006, 57, 761–780. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Li, Y.; Zhang, F.; Zhang, G.; Jiang, X.; Yu, H.; Hou, B. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 2017, 89, 85–103. [Google Scholar] [CrossRef]
- Zhang, Y.; Butelli, E.; Martin, C. Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant Biol. 2014, 19, 81–90. [Google Scholar] [CrossRef]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef]
- Spelt, C.; Quattrocchio, F.; Mol, J.N.M.; Koes, R. Anthocyanin1 of petunia encodes a basic helix–loop–helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 2000, 12, 1619–1631. [Google Scholar]
- Lorenc-Kukula, K.; Jafra, S.; Oszmianski, J.; Szopa, J. Ectopic expression of anthocyanin 5-o-glucosyltransferase in potato tuber causes increased resistance to bacteria. J. Agric. Food Chem. 2005, 53, 272–281. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, Y.; Sun, H.; Sun, L.; Zhang, L. Transposon-induced methylation of the RsMYB1 promoter disturbs anthocyanin accumulation in red-fleshed radish. J. Exp. Bot. 2020, 71, 2537–2550. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Sun, Y.; Zhang, X.; Chen, M.; Wu, T.; Zhang, J.; Xing, Y.; Tian, J.; Yao, Y. ROS1 promotes low temperature-induced anthocyanin accumulation in apple by demethylating the promoter of anthocyanin-associated genes. Hortic. Res. 2022, 9, uhac007. [Google Scholar] [CrossRef]
- Ubi, B.E.; Honda, C.; Bessho, H.; Kondo, S.; Wada, M.; Kobayashi, S.; Moriguchi, T. Expression analysis of anthocyanin biosynthetic genes in apple skin: Effect of UV-B and temperature. Plant Sci. 2006, 170, 571–578. [Google Scholar]
- Takos, A.M.; Jaffé, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef]
- Zhou, L.; Li, Y.; Zhang, R.; Zhang, C.; Xie, X.; Zhao, C.; Hao, Y. The small ubiquitin-like modifier E3 ligase MdSIZ1 promotes anthocyanin accumulation by sumoylating MdMYB1 under low-temperature conditions in apple. Plant Cell Environ. 2017, 40, 2068–2080. [Google Scholar]
- Zhang, Y.; Zheng, S.; Liu, Z.; Wang, L.; Bi, Y. Both HY5 and HYH are necessary regulators for low temperature-induced anthocyanin accumulation in Arabidopsis seedlings. J. Plant Physiol. 2010, 47, 934–945. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Hu, Z.; Zhang, Y.; Li, Y.; Zhou, S.; Chen, G. A putative functional MYB transcription factor induced by low temperature regulates anthocyanin biosynthesis in purple kale (Brassica Oleracea var. acephala f. tricolor). Plant Cell Rep. 2012, 31, 281–289. [Google Scholar] [CrossRef]
- Mao, W.; Han, Y.; Chen, Y.; Sun, M.; Feng, Q.; Li, L.; Liu, L.; Zhang, K.; Wei, L.; Han, Z.; et al. Low temperature inhibits anthocyanin accumulation in strawberry fruit by activating FvMAPK3-induced phosphorylation of FvMYB10 and degradation of Chalcone Synthase. Plant Cell 2022, 34, 1226–1249. [Google Scholar]
- Jiang, H.; Zhou, L.; Gao, H.; Wang, X.; Li, Z.; Li, Y. The transcription factor MdMYB2 influences cold tolerance and anthocyanin accumulation by activating SUMO E3 ligase MdSIZ1 in apple. Plant Physiol. 2022, 189, 2044–2060. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhang, B.; Allan, A.C.; Lin-Wang, K.; Zhao, Y.; Wang, K.; Chen, K.; Xu, C. DNA demethylation is involved in the regulation of temperature-dependent anthocyanin accumulation in peach. Plant J. 2020, 102, 965–976. [Google Scholar] [CrossRef] [PubMed]
- Chiu, L.; Zhou, X.; Burke, S.; Wu, X.; Prior, R.L.; Li, L. The Purple Cauliflower Arises from Activation of a MYB Transcription Factor. Plant Physiol. 2010, 154, 1470–1480. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; An, G.; Zhu, T.; Zhang, W.; Zhang, L.; Peng, L.; Chen, J.; Kuang, H. Independent activation of the BoMYB2 gene leading to purple traits in Brassica oleracea. Theor. Appl. Genet. 2019, 132, 895–906. [Google Scholar] [CrossRef]
- Moreno, D.A.; Perez-Balibrea, S.; Ferreres, F.; Gil-Izquierdo, A.; Garcia-Viguera, C. Acylated anthocyanins in Broccoli sprouts. Food Chem. 2010, 123, 358–363. [Google Scholar] [CrossRef]
- Han, Y.; Vimolmangkang, S.; Soria-Guerra, R.E.; Rosales-Mendoza, S.; Zheng, D.; Lygin, A.V.; Korban, S.S. Ectopic Expression of Apple F3′H Genes Contributes to Anthocyanin Accumulation in the Arabidopsis tt7 Mutant Grown Under Nitrogen Stress. Plant Physiol. 2010, 153, 806–820. [Google Scholar] [CrossRef] [PubMed]
- Seitz, C.; Ameres, S.; Forkmann, G. Identification of the molecular basis for the functional difference between flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase. FEBS Lett. 2007, 581, 3429–3434. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, Y.; Li, K.; Yang, D.; Liu, N.; Zhang, L.; Zhao, L.; Zhang, X.; Liu, Y.; Gao, L.; et al. Roles of the 2-Oxoglutarate-Dependent Dioxygenase Superfamily in the Flavonoid Pathway: A Review of the Functional Diversity of F3H, FNS I, FLS, and LDOX/ANS. Molecules 2021, 26, 6745. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Gao, L.; Ma, X.; Guo, F.; Ruan, H.; Bao, Y.; Xia, T.; Wang, Y. Functional analysis of flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylases from tea plant (Camellia sinensis), involved in the B-ring hydroxylation of flavonoids. Gene 2019, 717, 144046. [Google Scholar] [CrossRef]
- Jia, Y.; Li, B.; Zhang, Y.; Zhang, X.; Xu, Y.; Li, C. Evolutionary dynamic analyses on monocot flavonoid 3′-hydroxylase gene family reveal evidence of plant-environment interaction. BMC Plant Biol. 2019, 19, 347. [Google Scholar] [CrossRef]
- Dai, Y.; Zhang, L.; Sun, X.; Li, F.; Zhang, S.; Zhang, H.; Li, G.; Fang, Z.; Sun, R.; Hou, X.; et al. Transcriptome analysis reveals anthocyanin regulation in Chinese cabbage (Brassica rapa L.) at low temperatures. Sci. Rep. 2022, 12, 6308. [Google Scholar] [CrossRef]
- Li, C.; Yu, W.; Xu, J.; Lu, X.; Liu, Y. Anthocyanin Biosynthesis Induced by MYB Transcription Factors in Plants. Int. J. Mol. Sci. 2022, 23, 11701. [Google Scholar] [CrossRef]
- Saito, K.; Kobayashi, M.; Gong, Z.; Tanaka, Y.; Yamazaki, M. Direct evidence for anthocyanidin synthase as a 2-oxoglutaratedependent oxygenase: Molecular cloning and functional expression of cDNA from a red forma of Perilla frutescens. Plant J. 1999, 17, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, J.J.; Sobey, W.J.; Aplin, R.T.; Hassan, A.; Firmin, J.L.; Schofield, C.J.; Prescott, A.G. Are anthocyanidins the immediate products of anthocyanidin synthase? Chem. Commun. 2000, 24, 2473–2474. [Google Scholar] [CrossRef]
- Wellmann, F.; Griesser, M.; Schwab, W.; Martens, S.; Eisenreich, W.; Matern, U.; Lukacin, R. Anthocyanidin synthase from Gerbera hybrida catalyzes the conversion of (+)-catechin to cyanidin and a novel procyanidin. FEBS Lett. 2006, 580, 1642–1648. [Google Scholar] [CrossRef] [PubMed]
- Wilmouth, R.C.; Turnbull, J.J.; Welford, R.W.; Clifton, I.J.; Prescott, A.G.; Schofield, C.J. Structure and mechanism of anthocyanidin synthase from Arabidopsis thaliana. Structure 2002, 10, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Owens, D.K.; Crosby, K.C.; Runac, J.; Howard, B.A.; Winkel, B.S. Biochemical and genetic characterization of Arabidopsis flavanone 3β-hydroxylase. Plant Physiol. Biochem. 2008, 46, 833–843. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, J.J.; Nakajima, J.I.; Welford, R.W.; Yamazaki, M.; Saito, K.; Schofifield, C.J. Mechanistic studies on three 2-oxoglutaratedependent oxygenases of flavonoid biosynthesis: Anthocyanidin synthase, flavonol synthase, and flavanone 3 beta-hydroxylase. J. Biol. Chem. 2004, 279, 1206–1216. [Google Scholar] [CrossRef]
- Liu, C.; Yao, X.; Li, G.; Huang, L.; Wu, X.; Xie, Z. Identifification of Major Loci and Candidate Genes for Anthocyanin Biosynthesis in Broccoli Using QTL-Seq. Horticulturae 2021, 7, 246. [Google Scholar] [CrossRef]
- Tang, Q.; Tian, M.; An, G.; Zhang, W.; Chen, J.; Yan, C. Rapid identification of the purple stem (Ps) gene of Chinese kale (Brassica oleracea var. alboglabra) in a segregation distortion population by bulked segregant analysis and RNA sequencing. Mol. Breed. 2017, 37, 153. [Google Scholar] [CrossRef]
- Zhang, Z.; Kou, X.; Fugal, K.; Mclaughlin, J. Comparison of HPLC Methods for Determination of Anthocyanins and Anthocyanidins in Bilberry Extracts. J. Agric. Food Chem. 2004, 52, 688–691. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Wang, J.; Zhao, Z.; Sheng, X.; Shen, Y.; Branca, F.; Gu, H. Construction of a high-density genetic map and identification of loci related to hollow stem trait in broccoli (Brassic oleracea L. italica). Front. Plant Sci. 2019, 10, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
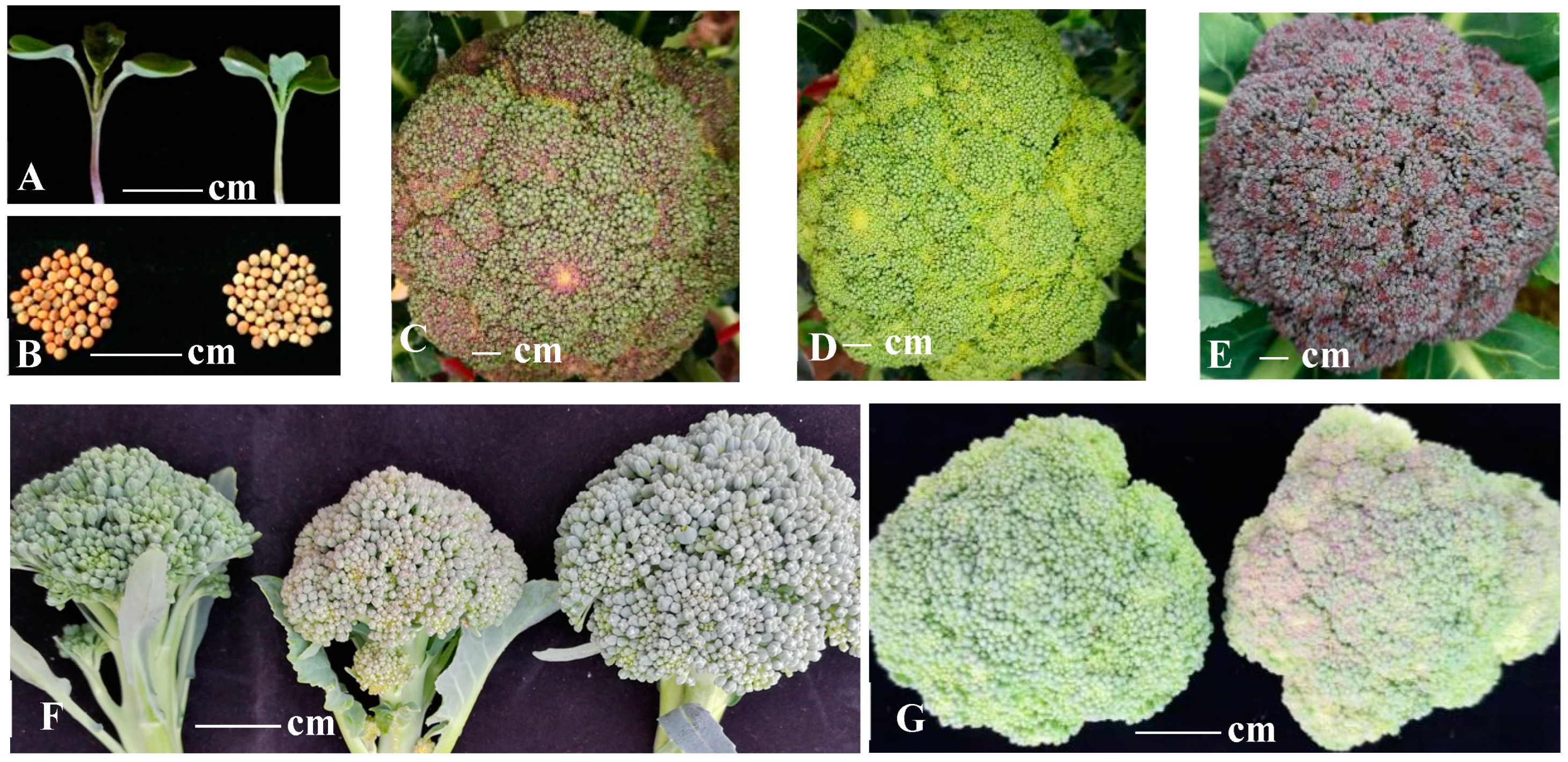

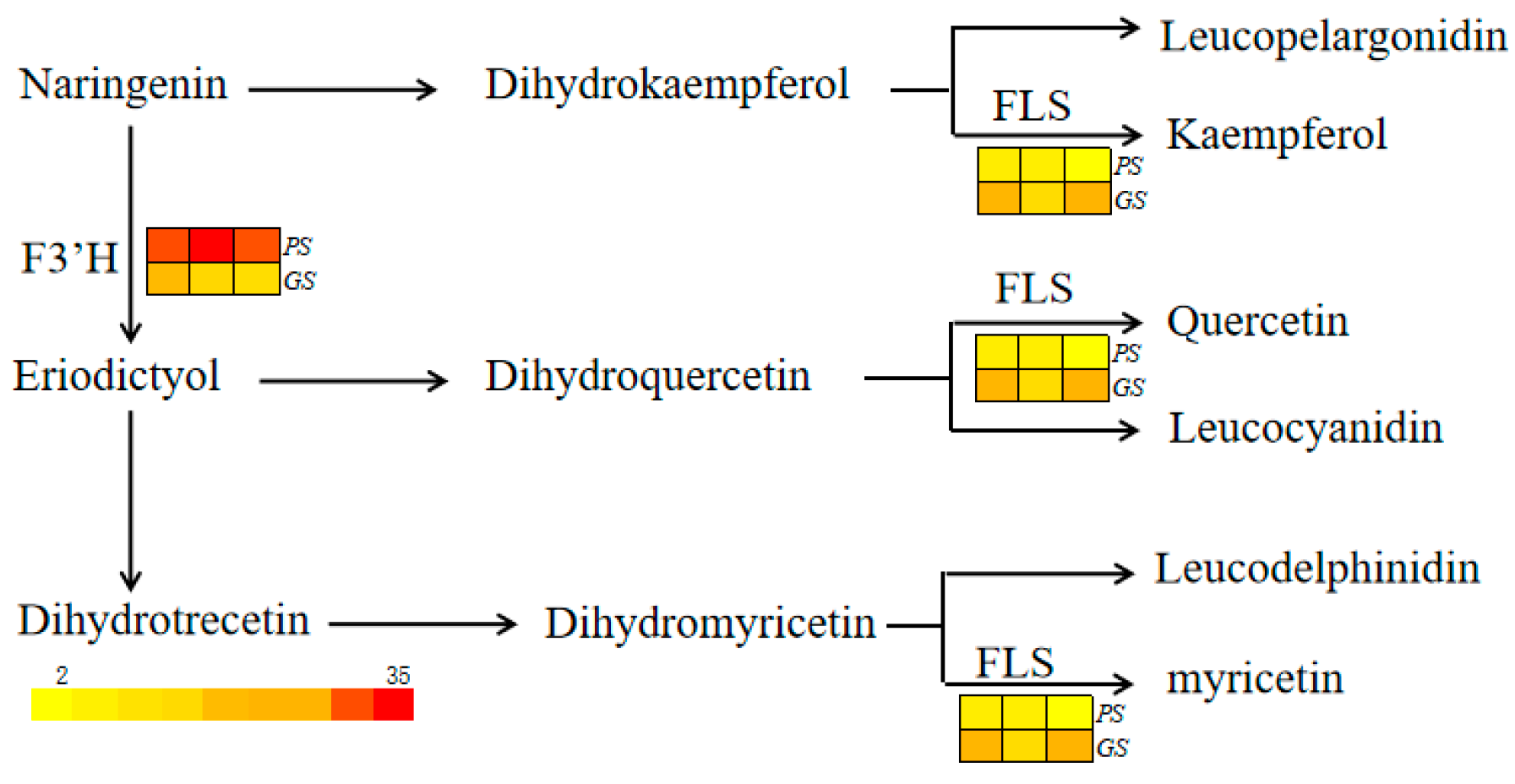


| Total Anthocyanins C µg/g | CC µg/g | DC µg/g | MvC µg/g | PelC µg/g | PeoC µg/g | PtC µg/g | |
|---|---|---|---|---|---|---|---|
| GS | 12.123 ± 0.770 c | 3.830 ± 0.420 b | 4.190 ± 0.210 b | 0.002 ± 0.011 b | 0 | 0 | 0 |
| PS | 20.712 ± 0.378 b | 6.980 ± 0.280 b | 5.690 ± 0.090 b | 0.002 ± 0.012 b | 0 | 0 | 0 |
| PB | 103.138 ± 4.399 a | 87.890 ± 4.480 a | 9.110 ± 1.360 a | 0.881 ± 0.323 a | 0 | 0 | 0 |
| Material Codes | Species | Material Codes | Species | Material Codes | Species |
|---|---|---|---|---|---|
| 1015 | B. oleracea L. var. italica | SG1921 | B. oleracea L. var. italica | SG1911 | B. oleracea L. var. italica |
| 1403 | B. oleracea L. var. italica | SG1614 | B. oleracea L. var. botrytis | SG1912-1 | B. oleracea L. var. italica |
| 1405-1 | B. oleracea L. var. italica | 51932-2 | B. oleracea L. var. italica | SG1920 | B. oleracea L. var. italica |
| 1409 | B. oleracea L. var. italica | 51936-1 | B. oleracea L. var. italica | SG2009-2 | Brassica oleracea L. var. kohlrabi |
| 1502-1 | B. oleracea L. var. italica | 51938-13 | B. oleracea L. var. italica | SG2021 | B. oleracea L. var. acephala |
| 1610-4B | B. oleracea L. var. italica | PR2003 | B. oleracea L. var. botrytis | UC001 | B. oleracea L. var. italica |
| 1910-1 | B. oleracea L. var. italica | 2035 | B. oleracea L. var. italica | UC002 | B. oleracea L. var. botrytis |
| 2035 | B. oleracea L. var. italica | 1947 | B. oleracea L. var. italica | UC003 | B. oleracea L. var. capitata |
| 2057-11 | B. oleracea L. var. italica | VR2003 | B. oleracea L. var. botrytis | UC012 | B. oleracea L. var. gongylodes |
| 5815-6A | B. oleracea L. var. italica | SG1401-1 | B. oleracea L. var. italica | UC018 | B. oleracea L. var. gemmifera |
| K2145 | B. oleracea L. var. acephala | SG1913 | B. oleracea L. var. italica | UC044 | B. oleracea L. var. italica |
| K2150 | B. oleracea L. var. caulorapa | SG1914-1 | B. oleracea L. var. italica | UC019 | B. oleracea L. var. acephala |
| 2018K1M3 | B. oleracea L. var. acephala | SG1917-2 | B. oleracea L. var. italica | UC020 | B. oleracea L. var. gongylodes |
| 2018K1M5 | B. oleracea L. var. acephala | SG1908 | B. oleracea L. var. italica | UC030 | B. oleracea L. var. botrytis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, H.; Wang, J.; Shen, Y.; Sheng, X.; Shaw, R.K.; Branca, F.; Gu, H. A 43 Bp-Deletion in the F3′H Gene Reducing Anthocyanins Is Responsible for Keeping Buds Green at Low Temperatures in Broccoli. Int. J. Mol. Sci. 2023, 24, 11391. https://doi.org/10.3390/ijms241411391
Yu H, Wang J, Shen Y, Sheng X, Shaw RK, Branca F, Gu H. A 43 Bp-Deletion in the F3′H Gene Reducing Anthocyanins Is Responsible for Keeping Buds Green at Low Temperatures in Broccoli. International Journal of Molecular Sciences. 2023; 24(14):11391. https://doi.org/10.3390/ijms241411391
Chicago/Turabian StyleYu, Huifang, Jiansheng Wang, Yusen Shen, Xiaoguang Sheng, Ranjan Kumar Shaw, Ferdinando Branca, and Honghui Gu. 2023. "A 43 Bp-Deletion in the F3′H Gene Reducing Anthocyanins Is Responsible for Keeping Buds Green at Low Temperatures in Broccoli" International Journal of Molecular Sciences 24, no. 14: 11391. https://doi.org/10.3390/ijms241411391
APA StyleYu, H., Wang, J., Shen, Y., Sheng, X., Shaw, R. K., Branca, F., & Gu, H. (2023). A 43 Bp-Deletion in the F3′H Gene Reducing Anthocyanins Is Responsible for Keeping Buds Green at Low Temperatures in Broccoli. International Journal of Molecular Sciences, 24(14), 11391. https://doi.org/10.3390/ijms241411391







