Abstract
Osteosarcoma (OS) is the predominant primary bone tumor in the pediatric and adolescent populations. It has high metastatic potential, with the lungs being the most common site of metastasis. In contrast to many other sarcomas, OS lacks conserved translocations or genetic mutations; instead, it has heterogeneous abnormalities, including somatic DNA copy number alteration, ploidy, chromosomal amplification, and chromosomal loss and gain. Unfortunately, clinical outcomes have not significantly improved in over 30 years. Currently, no effective molecularly targeted therapies are available for this disease. Several genomic studies showed inactivation in the tumor suppressor genes, including p53, RB, and ATRX, and hyperactivation of the tumor promoter genes, including MYC and MDM2, in OS. Alterations in the major signaling pathways, including the PI3K/AKT/mTOR, JAK/STAT, Wnt/β-catenin, NOTCH, Hedgehog/Gli, TGF-β, RTKs, RANK/RANKL, and NF-κB signaling pathways, have been identified in OS development and metastasis. Although OS treatment is currently based on surgical excision and systematic multiagent therapies, several potential targeted therapies are in development. This review focuses on the major signaling pathways of OS, and we propose a biological rationale to consider novel and targeted therapies in the future.
1. Introduction
Osteosarcoma (OS) is a rare cancer arising from the mesenchymal cells forming the bone. It is the most common and highly metastatic bone tumor in children and adolescents [1]. Its incidence is higher in adolescents (0.8–1.1/100,000/year in the age group of 15–19 years), with a second peak in older adults [2,3]. Nearly two-thirds of primary tumors occur near the knee joint, with the most common sites being the distal femur, proximal humerus and proximal tibia [4]. Despite the extensive genomic aberrations, OS has no pathognomonic DNA translocation or targetable mutations [5]. Thus, no effective molecularly targeted therapies for OS are currently available. The diagnosis of OS is based on morphological characteristics since no specific molecular markers or testing are available in clinical practice. The management of OS is challenging and requires a multidisciplinary approach. Surgical excision and systematic multiagent therapy are standard clinical practices for OS treatment. However, there is a pressing need to identify novel therapeutic approaches and biomarkers to manage the disease better, given the high relapse rate and poor prognosis of metastatic disease. One of the critical factors in OS development is chromosomal instability and genetic changes [6]. Oncogenes and tumor suppressor genes are often affected in OS. The immune system also plays a role in regulating tumor growth and propagation, and it is evident that the tumor-infiltrating immune cells contribute to the metastatic cascade [7]. Tumor metastasis is the primary challenge for OS therapy [8]. The five-year survival rate of OS has increased to about 70% since the 1970s, although it is only 20–30% for patients with metastasis [9]. Most OSs infiltrate the surrounding tissue and metastasize to the lung.
A better understanding of the bone microenvironment, the interaction between the tumor and non-tumor cells, and the mechanism of OS metastasis will help find a therapeutic target for OS. Several major signaling pathways have been identified in OS tumor development and metastasis, including the PIK3, JAK/STAT, Wnt/β-catenin, NOTCH, Hedgehog, Ras, TGF-β, MAPK/AKT/mTOR, RANK/RANKL, and NF-κB signaling pathways [10,11]. This review emphasizes the current understanding of the signaling pathways involved in OS tumor development and metastasis. Moreover, we also highlight the role of chromosomal instability and immune regulation in OS tumorigenesis and metastasis.
2. Molecular Abnormalities in Osteosarcoma
2.1. Chromosomal Abnormalities
OS is a genetically complex and heterogeneous tumor characterized by chromosomal instability and genetic alterations that lead to aneuploidy and increased tumor aggressiveness. The high rates of chromosomal rearrangements in OS include structural chromosomal abnormalities, such as translocations, deletions, amplifications, and chromothripsis, an extreme form of chromosomal instability [12]. Chromothripsis is associated with increased genomic instability and tumor progression, and it frequently occurs in highly aggressive tumors, including OS. Although the exact cause of chromothripsis and its role in tumorigenesis remain unclear, recent genomic studies have revealed that it is context-dependent and occurs at an overall incidence of between 2% and 3% in pan-cancer samples but over 77% in OS and 100% in liposarcoma [13]. Some potential mechanisms underlying chromothripsis are emerging, including the generation of DNA breaks and rejoining of the DNA fragments, generation of micronuclei, premature chromosome condensation, breakage–fusion–bridge cycle and telomere dysfunction, and ionizing radiation [14]. While the exact mechanistic cause of chromothripsis is still undefined, Crasta et al. identified the micronuclei, having many features of primary nuclei, formed from the acentric fragments of chromosomes, which produce DNA damage behind the chromothripsis [15]. Zhang et al., by using a combination of live cell imaging and single-cell genome sequencing, demonstrated that micronucleus formation could indeed generate a spectrum of genomic rearrangements, which recapitulate the features of chromothripsis [16]. Gong et al. described how Ran GTPase-activating protein 1 (RanGAP1) is commonly reduced or inactivated in human OS, leading to a high probability of chromothripsis, which drives tumorigenesis through its direct effects on the spindle-assembly checkpoint and decatenation and secondary effects on DNA damage surveillance [17].
2.2. Inactivation of Tumor Suppressor Genes and Amplification of Oncogenes
Inactivation of tumor suppressor genes such as TP53, RB1, ATRX, and DLG2 is frequently observed in OS, which is thought to be involved in OS tumorigenesis [18]. The TP53 gene is OS’s most frequently dysregulated gene [18]. Whole-genome DNA sequencing from OS tumor samples demonstrates multiple somatic chromosomal lesions, including structural variations (SVs) and copy number alterations (CNAs). Kataegis is a single nucleotide variation (SNV) detected in 50% of OS tumors. Chen et al. identified p53 pathway lesions in all OS patients, while it was translocated in around 50% of the patients to the first intron of the TP53 gene, leading to gene inactivation. This mechanism of TP53 gene inactivation is unique to OS among pediatric cancers [18]. The p53 protein is a tumor suppressor protein involved in DNA damage recognition that induces apoptosis, cellular quiescence, or senescence. Another tumor suppressor gene frequently inactivated in OS is RB1, located at chromosome 13q14.2 [19]. RB1 encodes the tumor suppressor protein pRB, which is vital for preventing cell cycle progression. ATRX is an important tumor suppressor in OS, and it is a part of a multiprotein complex that regulates chromatin remodeling, nucleosome assembly, and telomere maintenance. Furthermore, a recent report noted that loss of ATRX promotes OS tumor through increased NF-κB signaling and integrin binding [20]. DLG2 is a tumor suppressor gene, and its copy number loss occurs in 42% of human and 56% of canine OS [21]. Deletion of Dlg2 in a mouse model led to the acceleration of OS development [21].
Hyperactivation of tumor-promoting genes such as MYC and MDM2 is associated with OS tumorigenesis. The gain of the 8q24 chromosomal locus, which harbors the oncogene MYC, has been reported in several OS patients [22,23]. MYC is involved in cell cycle regulation, protein biogenesis, metabolism, signal transduction, transcription, and translation [24,25]. A recently generated Myc knock-in genetically engineered mouse model of an OS tumor not only identified intrinsic Myc-mediated mechanisms of OS tumorigenesis but also identified a novel molecular mechanism through which Myc regulates the profile and function of the OS immune landscape [26,27,28]. The oncoprotein MDM2 is a p53 inhibitor, which promotes p53 degradation and downregulates its transcription. Amplification of MDM2 (chromosome 12q15) is more frequent in OS metastasis and recurrence.
2.3. Epigenetic Modification in OS Progression
Epigenetic modifications, including DNA methylation, histone acetylation, and methylation, are critical in the pathogenesis of several cancers, including OS [29]. The level of histone H3 lysine trimethylation was reported to be lower in human OS tissue and cell lines compared with normal bone tissue and osteoblast cells. Enhancement of H3 methylation after treatment with the histone lysine demethylase inhibitor 5-carboxy-8-hydroxyquinoline (IOX-1) showed inhibition of OS migratory and invasive capabilities. Enhanced histone H3 lysine trimethylation levels sensitized cisplatin against the cisplatin-resistant (MG63-CR) cells [30]. Previous studies showed enhancement of the expression of the lysine-specific demethylases KDM1A, KDM2B, KDM4A, KDM6A, KDM6B in OS progression [31,32,33,34]. Recently, Twenhafel et al. provided a comprehensive review of recent advances in the epigenetics of OS and highlighted the clinical benefits in the field of OS research [35]. Morrow et al. highlighted the genetic and epigenetic defects in OS and emphasized the role of epigenetic dysregulation in tumor suppression and oncogene regulation [6].
3. Major Signaling Pathways in OS Tumor Development and Metastasis
Signaling pathways and molecular networks are highly regulated under normal growth conditions and control critical pro-survival and pro-growth cellular processes. Dysregulation of the signaling pathways contributes to OS development, metastasis, and chemoresistance. This review will focus on the essential signaling pathways of OS that play a vital role in disease development and progression. Figure 1 shows the different signaling pathways involved in OS tumorigenesis and metastasis.
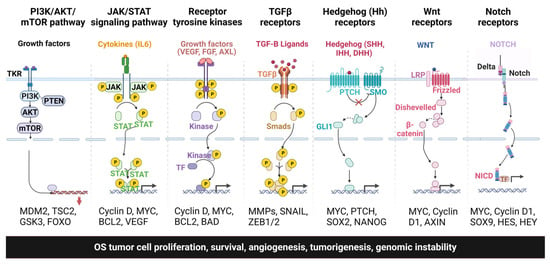
Figure 1.
Different signaling pathways involved in OS tumor progression, metastasis, and chemoresistance.
3.1. PIK3/AKT/mTOR Pathway
The family of lipid kinases known as phosphoinositide 3-kinases (PI3Ks) is vital to almost all cell and tissue biology aspects and central to human cancer, including glioblastoma, sarcoma, skin, respiratory, digestive and breast cancer [36,37]. PI3Ks are multifunctional and regulate a wide range of signaling, membrane trafficking, cell survival, metabolism, and metastasis [38]. An increasing body of evidence has shown that the PI3K pathway is frequently hyperactivated in OS and contributes to tumorigenesis, proliferation, metastasis, invasion, cell cycle progression, inhibition of apoptosis, angiogenesis, and chemoresistance [39]. The PI3K/AKT pathway can be abnormally triggered by interacting with various growth factor receptors, such as vascular endothelial growth factor receptor (VEGFR), epithelial growth factor receptor (EGFR), and fibroblast growth factor receptor (FGFR), and by mutations in the PIK3CA, AKT, PTEN, and mTOR genes [40]. Immunostaining analysis of primary OS shows that activation of the PI3K/AKT signaling pathway facilitates the progression of OS and is also associated with the patient’s poor prognosis. Additionally, the activation of AKT is associated with lung metastasis [41,42,43].
AKT phosphorylates several substrates and downstream effectors, including mTOR, matrix metalloproteinase (MMP), cyclin-dependent kinases (CDKs), and VEGF, associated with tumor progression and metastasis [44,45]. The secretion of VEGF-A and FGF2 from OS cells promotes migration and invasion by activating the PI3K and AKT pathways, eventually leading to MMP9 overexpression [46]. Human epidermal growth factor receptor 4 (HER4), a member of the ERBB family, is upregulated in OS tumor tissue and cell lines, promoting OS progression by inactivating the PTEN-PI3K/AKT pathway [47]. VCAM-1 is increased in metastatic OS cells through the activation of the CXCL1-CXCR2/focal adhesion kinase FAK-PI3K-AKT-NF-κB pathway. CXCL1/CXCR2 pathway activation has been a crucial indicator of lung metastasis in OS, and the higher expression of CXCL1 is positively correlated with the migratory and invasive behavior of OS cells [48]. Other examples highlighting the importance of PI3K-AKT mediated signaling in advanced OS include the increased IHC staining of CDC42 effector protein 3 (CDC42EP3) [49], which was associated with patients’ pathological stage and grade, while the zinc finger CCHC domain containing 12 genes (ZCCHC12) is highly upregulated at the transcriptomic level in OS compared to normal bone tissues and has a vital role in OS cell proliferation and migration [50].
Several non-coding RNAs, including LncRNA H19, LINC00968, LINC00628, LncRNA NDRG1, LncDANCR, and circ_001422, are markedly associated with the OS advanced clinical stage, larger tumor size, higher incidence of metastases, and poorer prognosis, and they have been found to be mediated through the PI3K/AKT signaling pathway [43,51,52,53,54,55].
The PI3K/AKT signaling pathway also contributes to drug resistance in different types of cancers, including lung and esophageal cancer [56]. Several recent reports have shown the involvement of PI3K/AKT pathways in chemoresistance in OS. The zinc transporters Zrt- and Irt-related protein (ZIP/SLC39) ZIP10 is highly expressed in OS, which promotes cell proliferation and chemoresistance that is mediated through the activation of PI3K/AKT signaling [57]. Higher microtubule-affinity regulating kinase 2 (MARK2) expression was associated with the poor prognosis of OS and positively correlated with cisplatin chemoresistance mediated by the activation of the PI3K/AKT/NF-κB signaling pathway [58]. Also, microRNA-22-mediated cisplatin resistance is downstream from the PI3K/AKT/mTOR pathway in OS [59]. EGFR-facilitated tumor progression and gemcitabine resistance in OS were mediated through the PI3K/AKT pathway [60]. Moreover, proanthocyanidin B2 (PB2) inhibited the proliferation and induced the apoptosis of OS cells by suppressing the PI3K/AKT signaling pathway [61], whereas aclidinium bromide inhibited OS cell growth by regulating the PI3K/AKT signaling pathway [62]. As is evident, the PI3K/Akt signaling pathway significantly contributes to the development, progression, and therapeutic responsiveness of OS. The development of drugs targeting PI3K signaling has received much attention; indeed, enormous efforts have been dedicated and several are employed in evaluating clinical trials. The biggest hurdle in developing drugs targeting this pathway is the failure to generate a long-term outcome [63]. Cancer cells can compensate for the alternative pathway and acquire treatment resistance through feedback loops and crosstalk mechanisms. Another challenge is the lack of reliable predictive biomarkers that can identify patients who will most likely benefit from these types of therapies. The future development of promising inhibitors focusing on combined strategies, including the concomitant or sequential blockade of signaling pathways or the generation of less toxic drugs, will help develop novel therapeutic interventions for OS patients.
3.2. JAK/STAT Signaling
There are four members of the JAK family, JAK1, JAK2, JAK3, and TYK2, and seven members of the STAT family, STAT1, STAT2, STAT3, STAT4, STAT5a, STAT5b, and STAT6 [64]. The JAK/STAT signaling pathway has recently gained much attention in relation to malignancies and autoimmune diseases. It regulates most immune regulatory processes, including those involved in tumor cell recognition and tumor-driven immune escape [65]. Antitumor immune responses are largely driven by the STAT1 and STAT2 induction of type I and II interferons (IFNs) and the downstream programs that IFNs potentiate. Conversely, STAT3 has been widely linked to cancer cell survival, immunosuppression, and sustained inflammation in the tumor microenvironment. Interleukin-6 (IL-6) is highly expressed in OS and is considered one of the JAK/STAT pathway activators [66]. Activation of the IL-6/JAK/STAT3 signaling pathway in cancer modulates the expression of several genes that drive the proliferation, metastasis, and survival of tumor cells while suppressing the antitumor immune response. The JAK/STAT pathway is also activated by other inducers in OS. Microarray and bioinformatics analysis shows a higher expression of serglycin, a potential biomarker of OS, which promotes OS proliferation, migration, and invasion by activating the JAK/STAT signaling pathway [67]. Similarly, FAT10 mediates OS development by activating the JAK/STAT signaling pathway [68]. STAT3 overexpression is negatively associated with OS’s five-year overall survival [69].
Several long non-coding RNAs altered in OS mediate tumor progression and metastasis through modulating the JAK/STAT pathway. A microarray analysis showed that LINC01116 promoted OS progression by regulating the IL6R-JAK/STAT signaling pathway [70]. Overexpression of microRNA-101 inhibited OS tumor growth and metastasis by inactivating the PI3K/AKT and JAK/STAT signaling pathways mediated through the downregulation of ROCK1 [71]. Similarly, microRNA-126 inhibits proliferation, migration, invasion, and EMT in OS by targeting ZEB1 through the inactivation of the JNK and JAK1/STAT3 pathways [72].
Studies suggested that the blocking of the JAK/STAT pathway is a potential option for OS treatment [73]. Many JAK inhibitors have achieved efficacy in clinical settings, and more medications are currently being studied, as shown in Table 1. Suppressing STAT5 signaling affects OS growth and stemness. Curculigoside (Cur), a natural component of Curculigo orchioides Gaertn, controls OS growth by downregulating the JAK/STAT and NF-κB pathways, which is an underlying therapeutic option for OS treatment [74]. Similarly, curcumin and the curcumin analog L48H37 suppress human OS cell migration and invasion via inhibition of uPA, as mediated through the JAK/STAT signaling pathway [75,76]. Other targeted agents include the JAK2 inhibitors AG490 and telocinobufagin (TCB), which reduce OS cell proliferation, migration, and invasion by inhibiting the JAK2/STAT3 pathway in in vitro and in vivo set-ups [77].

Table 1.
List of signaling pathway inhibitors and immunotherapeutic agents in clinical trials.
3.3. Wnt/β-Catenin Signaling Pathway
Wnt/β-catenin is an evolutionarily conserved pathway regulating cell fate determination, cell migration, cell polarity, neural patterning, and organogenesis during embryonic development [78]. Mutations or alterations in the Wnt signaling pathway have been reported in several cancers and mediate tumorigenesis by modulating both tumors’ intrinsic and extrinsic properties in the tumor microenvironment mediated through crosstalk between transformed cells and infiltrating immune cells, such as leukocytes [79]. So far, three Wnt signaling pathways have been defined: the canonical Wnt pathway (Wnt/β-catenin), the non-canonical Wnt/PCP pathway (planar cell polarity), and the Wnt/Ca2+ pathway.
The Wnt/β-catenin pathway is activated via the binding of extracellular Wnt ligands to transmembrane receptors by autocrine/paracrine methods. Once activated, the β-catenin is translocated to the nucleus, ultimately facilitating gene expression in cell proliferation, survival, differentiation, and migration [80]. Several reports showed the involvement of the Wnt/beta-catenin signaling pathway in OS tumor development and metastasis [81]. The SP1/Wnt/β-catenin signaling pathway plays a vital role in GABPB1-AS1-mediated OS tumorigenesis [82]. Melittin inhibits lung metastasis of human OS mediated by the Wnt/β-catenin signaling pathway [83]. Chemokine receptor-9 promotes EMT by activating the Wnt/β-catenin pathways to promote OS metastasis [84]. Expression of COL5A2 is elevated in OS patients, and its inhibition suppresses OS cell invasion and metastasis mediated through the TGF-β signaling and Wnt/β-catenin signaling pathways [85].
Several Wnt/β-catenin regulated non-coding RNAs are involved in osteosarcoma development and metastasis. Long non-coding RNA gastric carcinoma proliferation enhancing transcript 1 (lncGHET1) expression was significantly upregulated in OS cell lines, promoting OS development and progression via activating the Wnt/β catenin signaling pathway [86]. Similarly, lncLINC01128 regulates OS development by sponging microRNA-299-3p to mediate MMP2 expression and activate the Wnt/β-catenin signaling pathway [87]. Long non-coding RNA MINCR governs the growth and metastasis of human OS cells via the Wnt/β-catenin signaling pathway [88]. Overexpression of microRNA-135b, an oncogenic microRNA in OS, can promote OS invasion and metastasis in vitro and in vivo by activating the Wnt/β-catenin signaling pathway via directly targeting GSK-3β, APC, β-TrCP, and CK1α [89]. MicroRNA-340-5p suppresses osteosarcoma development by downregulating the Wnt/β-catenin signaling pathway via targeting the STAT3 gene [90]. The circulating RNA Hsa_circ_0087302 affects the progression of osteosarcoma by modulating the Wnt/β-catenin signaling pathway [91].
The Wnt/β-catenin signaling pathway is widely activated in OS and highly related to the invasion and metastasis of OS. Therefore, an increasing number of studies focusing on Wnt/β-catenin signaling pathway inhibition showed a good response. We investigated the antitumor activity of tegavivint, a novel β-catenin/transducin β-like protein 1 (TBL1) inhibitor, against different OS models (in vivo/in vitro/ex vivo). It showed antiproliferative activity against OS cells in vitro and actively reduced micro- and macro-metastatic development ex vivo. We also observed that inherent chemoresistance was suppressed by tegavivint in vivo [92]. Tegavivint is a promising therapeutic agent for the advanced stages of OS, and currently, it is in clinical trials phase I/II. Echinatin inhibits the growth and metastasis of human osteosarcoma cells by regulating the Wnt/β-catenin and p38 signaling pathways [93].
3.4. NOTCH Signaling Pathway
NOTCH signaling is an evolutionarily conserved pathway associated with normal embryonic development, and it is dysregulated in several tumors, including OS [94]. The NOTCH signaling pathways consist of four receptors, including NOTCH-1, -2, -3, and -4, activated by a unique process that includes ligand binding and multistep proteolytic processing. The NOTCH ligands include Delta-like (DLL)-1, -3, -4, Jagged 1, and Jagged 2 in mammals. After binding NOTCH, the intracellular domain of the NOTCH ligand is ubiquitinated via the E3 ligase mind bomb-1, which initiates endocytosis of the NOTCH ligand/NECD complex into the ligand-expressing cell. The forces generated by these endocytosis-related events cause sequential proteolytic cleavage of NOTCH, which allows the NICD to be released into the cytosol and translocated to the nucleus. Higher expression of NOTCH3 and HES1, a downstream target of NOTCH signaling, is correlated with poor OS patient outcomes [95]. The NOTCH target genes, HES1 and HEY2, were significantly upregulated in OS patient samples at the transcript level and associated with a poor prognosis [96]. JAG1 was found to be involved in the activation of various NOTCH receptors, and it is positively associated with the metastasis and recurrence of OS [97]. The NOTCH signaling pathway not only promotes OS tumor progression but is also involved in tumor metastasis. The expressions of NOTCH1, NOTCH2, HES1, and DLL1 were significantly upregulated in a highly invasive and metastatic LM7 cell compared to normal human osteoblasts and the SaOS-2 cell line, which have lower metastatic potential [98,99]. JAG1 expression was significantly higher in the highly metastatic F5M2 cell lines compared to the less metastatic F4 OS cells [97]. NOTCH3 could also mediate the invasion and metastasis of OS cells by upregulating the downstream target genes HES1 and MMP7 [95].
Cell migration-inducing protein (CEMIP) was overexpressed in OS tissues compared to non-tumor tissues, and its expression was positively associated with a poor prognosis. CEMIP promoted OS growth and metastasis by activating the NOTCH/JAG1/HES1 signaling pathway both in vivo and in vitro [100]. NOTCH-targeted therapy has shown potential for the treatment of OS in clinical research. CB-103 is a small molecule protein–protein interaction (PPI) inhibitor able to target the assembly of the NOTCH transcription complex and downregulate NOTCH target genes (c-MYC, CCND1, HES1) and the inhibition of NOTCH signaling. CB-103 has advanced into the first-in-human phase 1–2A study targeting pan-NOTCH signaling in advanced solid tumors and blood malignancies [101]. RO4929097, a NOTCH signaling inhibitor, was well tolerated by solid tumor patients, including sarcoma, and showed clinical antitumor activity, meaning further studies are warranted [102].
3.5. Receptor Tyrosine Kinase Pathway
Protein kinases are enzymes that catalyze the transfer of a phosphate group from ATP to target proteins and play a crucial role in signal transduction and other cellular processes. Receptor tyrosine kinases (RTKs) are cell-surface growth-factor receptors with tyrosine-kinase activity. They regulate critical cellular processes, including cell proliferation, differentiation, cell survival, metabolism, and cell migration [103,104]. There are 58 known RTKs in humans, grouped into 20 subfamilies. All the RTKs have a similar molecular architecture, with a glycosylated N-terminal extracellular domain with a high number of disulfide bonds, which are involved in the ligand recognition, a cytoplasmic region that contains the protein tyrosine kinase (TK) domain, and a single transmembrane helix that plays a key role in the formation and stabilization of the dimer of the receptor chains. Several tyrosine kinase receptors have been associated with OS development and metastasis, including vascular endothelial growth factor receptor (VEGFR), platelet-derived growth factor receptor (PDGFR), insulin-like growth factor receptor (IGF), fibroblast growth factor receptor (FGFR), and AXL receptor tyrosine kinase (AXL). AXL RTK is highly expressed in most OS tissues and cell lines, and its higher mRNA expression correlates with poor clinical outcomes [52,105]. The knockdown of AXL inhibits the proliferation and induces the apoptosis of OS cells [106]. An increased expression of VEGF is associated with a lower overall OS patient survival [107]. However, using bevacizumab, a VEGF inhibitor, showed unfavorable outcomes [108], suggesting VEGF/VEGFR is limited and should be combined with the blockade of other pathways. FGFR amplification has been associated with OS resistance to chemotherapy [109] and the development of lung metastases [110]. Several RTKs inhibitors, including cabozantinib, bevacizumab, apatinib, sorafenib, and many more listed in Table 1, have shown promising effects and are at different stages of clinical trials.
3.6. RANK/RANKL/OPG Pathway
The receptor activator of the nuclear factor κB ligand (RANKL) is a homotrimeric transmembrane protein member of the tumor necrosis factor (TNF) cytokine family expressed by osteoblast cells and tumor cells. In contrast, RANK and osteoprotegerin (OPG) are transmembrane receptor members of the TNF receptor (TNFR) family expressed on the surface of osteoclasts. The RANK/RANKL signaling pathway is crucial for bone homeostasis, osteoclast survival, differentiation, and function. OPG counterbalances RANKL and prevents RANK/RANKL interaction [111]. Several studies have shown the dysregulation of this pathway in OS and its association with metastasis and chemoresistance [112]. Numerous human OS cell lines express higher levels of RANKL, and the ratio of RANKL/OPG was shifted in favor of RANKL in blood samples derived from OS patients [113,114]. RANK was negatively associated with patient survival and chemotherapy response in 91 human samples [115]. OPG administration in OS mouse models indirectly affected tumor progression, diminished tumor growth, and increased survival [116]. Punzo et al. tested denosumab, a human monoclonal antibody with high binding affinity and specificity to RANKL, alone and in combination with doxorubicin but found a discouraging response as it worsened the effect of standard chemotherapy [117]. As reported previously, osteoclast cell abundance in the OS tumor microenvironment prevents metastasis as well as improves the chemotherapeutic response. Additional preclinical studies are needed before blocking the RANK/RANKL pathways in OS.
3.7. Hedgehog/Gli Signaling Pathway
Hedgehog (Hh)/Gli signaling is a conserved signal transduction pathway with a key regulatory function in physiological processes, including embryonic development, tissue differentiation, and cell growth. Dysregulation of the Hedgehog/Gli pathway is considered a critical factor in the development and progression of multiple cancers. Several Hedgehog/GLI pathway-associated genes are candidate targets for tumor therapy [118]. The pathway is unique in that it is comprised of both tumor suppressor genes and oncogenes. The signaling pathway is associated with three ligands, Sonic hedgehog (SHH), Indian hedgehog (IHH), and Desert hedgehog (DHH), and additional components of the pathway include 12-transmembrane patched proteins (PTCH1 and PTCH2), 5-zinc finger transcription factors GLI1, GLI2, GLI3 (glioma-associated oncogene homologs), and the 7-transmembrane protein smoothened (SMO). The Hh/Gli pathway can promote OS metastasis by interacting with other signaling pathways, such as the PI3K/AKT and Wnt pathway.
Lo et al. evaluated the Hh pathway in 42 human OS samples and found higher expression levels of genes encoding the IHH, PTCH1, and GLI in the tumors [119]. Hirotsu et al. demonstrated that SHH, DHH, PTCH1, GLI1, GLI2, and SMO were overexpressed human OS cell lines [120]. They also reported that SMO and GLI activation are vital for OS progression and that their inhibition suppresses OS cell proliferation both in vivo and in vitro [121]. Hedgehog inhibition also prevented migration and metastasis in mouse models [122]. Several Hh-pathways inhibitors, including the SMO inhibitors (cyclopamine, IPI-926 (saridegib), GDC-0449 (vismodegib), and LDE225 (erismodegib), and Gli inhibitors (arsenic trioxide (ATO), Gli antagonists (GANTs)) have been assessed in preclinical models for OS [123]. Furthermore, taladegib is presently in a phase II clinical trial for advanced solid tumors with PTCH1 mutations (NCT05199584). However, more research is needed to discover the broad biological effects of Hedgehog pathway inhibition on OS tumor development and metastasis.
3.8. Transforming Growth Factor-β Signaling Pathway
Transforming growth factor-β (TGF-β) represents an evolutionarily conserved family of secreted polypeptide factors that mediate a diverse range of embryonic and adult signaling functions. In contrast with the dual effects of TGF-βs on tumor progression, TGF-βs seem to have a pro-tumoral effect on sarcomas, specifically in OS. TGF-βs expression is increased in OS patient sera compared to the healthy donors [124] and associated with lung metastases [125,126]. The higher expression of TGF-βs in OS shows a poor response to chemotherapy [127]. COL5A2, which is highly expressed in metastatic OS patients and associated with poor outcomes, is mediated through the TGF-β and Wnt/β-catenin signaling pathways [85]. The IHC staining and qPCR analysis of OS patient samples showing higher expression of the six-transmembrane epithelial antigen of prostate 1 (STEAP1) correlated with poor outcomes is also mediated through the Wnt/β-catenin and TGF-β/Smad2/3 pathways [128]. OS patients with high serum levels of growth and differentiation factors 15 (GDF15) exhibited significantly decreased overall survival and pulmonary metastasis-free survival (PMFS). The knockdown of GDF15 attenuated the migration and invasion of OS cells mediated through the TGF β signaling pathway [129]. TGF-β has been found to protect OS cells from chemotherapeutic cytotoxicity in a succinate dehydrogenase (SDH)/HIF1α-dependent manner [130]. Gamabufotalin, a natural derivative of the Chinese medicine Chansu, suppressed OS stem cells through the TGF-β/periostin/PI3K/AKT pathway [131]. MicroRNA-181c suppresses OS cell progression by targeting SMAD7 and regulating the TGFβ signaling pathway [132]. microRNA-522 was highly expressed in OS cells and presented carcinogenic function by contributing to cell proliferation, migration, and EMT progression mediated through the TGF-β/Smad pathway [133]. Several TGF-β inhibitors are being used in the preclinical set-up, i.e., RepSox, a TGF-β inhibitor, suppresses OS proliferation and EMT and promotes apoptosis by inhibiting the JNK/Smad3 signaling pathway. Oridonin, a bioactive diterpenoid, inhibits EMT and TGF-β1-induced EMT by inhibiting the OS’s TGF-β1/Smad2/3 signaling pathway [134]. The TGF-βR1 inhibitor vactosertib significantly inhibited OS proliferation in vitro and in vivo. It repressed c-Myc expression, inhibited immune suppressor cells (M2-like TAM, MDSC), and enhanced immune effectors (IFNγ + CD8 + cells and NK cells) in the OS tumor microenvironment [135].
4. Conclusions and Future Directions
OS is the most common and highly metastatic primary bone tumor in children. The long tubular bones are the most commonly affected bones, along with the spine, pelvis, and sacrum areas. This tumor is highly metastatic and invasive, and the primary site of metastasis is the lung, with a high mortality rate. Contrasting with many other sarcomas, OS lacks an established translocation or genetic mutation, and the causative factor in most cases remains unclear. Clinical outcomes for this disease have not progressed in over 30 years due to its complex and heterogeneous nature. The progressive accumulation of TP53 and the RB mutation (tumor suppressor genes) are currently attributed to the disease development. Several tumor-promoting genes, including MYC and MDM2, are thought to be involved in the disease progression. We and others are trying to understand the role of MYC in OS disease progression and chemoresistance. We recently developed a Myc knock-in mouse model and are trying to understand its role in disease progression.
The current standard care for OS management is surgical excision and systemic multiagent therapy. However, it is insufficient and there is a pressing need to identify novel therapeutic approaches for this highly aggressive and metastatic disease. Signaling pathways are highly dysregulated and contribute to OS development, metastasis, and chemoresistance. Enormous efforts have been dedicated to identifying the development of new drugs targeting signaling pathways. Several drugs targeting these pathways are in clinical trials and show promising effects, as shown in Table 1. The challenge associated with these targeted therapies is that cancer cells can compensate for the alternative routes and acquire treatment resistance through feedback loops and crosstalk mechanisms. Developing drugs focusing on combined strategies, including the concomitant or sequential blockade of signaling pathways or the generation of less toxic drugs, will help develop novel therapeutic interventions for OS patients. Immunotherapy has gained much attention in relation to cancer treatment but is still ineffective in OS disease management. The poor infiltration of the immune cells to the tumor microenvironment (TME) of OS, low activity from the available T cells, a lack of immune-stimulating neoantigens, and activation of multiple immune-suppressing pathways all combine to dampen responses to immunotherapy. Several approaches are currently being tested, including enhancing NK cells’ mediated natural immunity and using IL-2 to activate the effector T cells, as examples to enhance the efficacy of immunotherapy. Several other approaches, including non-specific adoptive T cell immunotherapy and CART therapy, are also being tried in preclinical and clinical practices. Non-specific immunotherapies stimulate or boost the immune system but do not target cancer cells directly through the use of certain cytokines, including IL2, IL-7, IL-12, IL-21, and IFN-α/β/γ, and the checkpoint inhibitors PD-1, PD-L1, and CTLA-4, and immuno-stimulatory agents such as CpG oligonucleotides, and agonistic CD40 are an example of nonspecific immune therapy and have sometimes led to a better immune response against cancer cells. In adoptive T-cell (ATC) therapy, autologous or allogenic T-cells are infused into patients with cancer, which has shown considerable promise in recent years. Recently, CART therapy has generated substantial excitement among researchers and oncologists. In CART therapy, T-cells are collected from the patient and re-engineered in the laboratory to produce proteins on their surface called chimeric antigen receptors, or CARs. The CARs can identify and bind to the antigens on the surface of cancer cells. Chemotherapy in combination with immune modulators and checkpoint inhibitors represents an attractive area of research and shows promising effects in OS management. Table 1 shows past and running clinical trials with immunotherapy or combination therapies. Much work is still needed to develop a novel effective immunotherapy against the OS tumor. We and others strongly believe there is much potential in immunotherapy that can revolutionize the treatment of OS in the future.
Author Contributions
All the authors contributed to performing the literature search and writing the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
J.T.Y. was supported by National Institute of Health grants 1R01EB026453, 1R01 CA21554, and 1R21CA267914 and the Osteosarcoma Institute.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lindsey, B.A.; Markel, J.E.; Kleinerman, E.S. Osteosarcoma Overview. Rheumatol. Ther. 2017, 4, 25–43. [Google Scholar] [CrossRef] [PubMed]
- Strauss, S.J.; Frezza, A.M.; Abecassis, N.; Bajpai, J.; Bauer, S.; Biagini, R.; Bielack, S.; Blay, J.Y.; Bolle, S.; Bonvalot, S.; et al. Bone sarcomas: ESMO-EURACAN-GENTURIS-ERN PaedCan Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2021, 32, 1520–1536. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.; Gianferante, D.M.; Zhu, B.; Mirabello, L. Osteosarcoma: A Surveillance, Epidemiology, and End Results program-based analysis from 1975 to 2017. Cancer 2022, 128, 2107–2118. [Google Scholar] [CrossRef] [PubMed]
- Heare, T.; Hensley, M.A.; Dell’Orfano, S. Bone tumors: Osteosarcoma and Ewing’s sarcoma. Curr. Opin. Pediatr. 2009, 21, 365–372. [Google Scholar] [CrossRef]
- Martin, J.W.; Squire, J.A.; Zielenska, M. The Genetics of Osteosarcoma. Sarcoma 2012, 2012, 627254. [Google Scholar] [CrossRef]
- Morrow, J.J.; Khanna, C. Osteosarcoma Genetics and Epigenetics: Emerging Biology and Candidate Therapies. Crit. Rev. Oncog. 2015, 20, 173–197. [Google Scholar] [CrossRef]
- Louise, M.E.J.; Emma, E.R.; Craig, D.L.; Willem, W.O. The immune system in cancer metastasis: Friend or foe? J. ImmunoTherapy Cancer 2017, 5, 79. [Google Scholar]
- Bielack, S.S.; Kempf-Bielack, B.; Delling, G.; Exner, G.U.; Flege, S.; Helmke, K.; Kotz, R.; Salzer-Kuntschik, M.; Werner, M.; Winkelmann, W.; et al. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: An analysis of 1702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J. Clin. Oncol. 2002, 20, 776–790. [Google Scholar] [CrossRef]
- Mirabello, L.; Troisi, R.J.; Savage, S.A. Osteosarcoma incidence and survival rates from 1973 to 2004. Cancer 2009, 115, 1531–1543. [Google Scholar] [CrossRef]
- Adamopoulos, C.; Gargalionis, A.N.; Basdra, E.K.; Papavassiliou, A.G. Deciphering signaling networks in osteosarcoma pathobiology. Exp. Biol. Med. 2016, 241, 1296–1305. [Google Scholar] [CrossRef]
- Li, S.; Zhang, H.; Liu, J.; Shang, G. Targeted therapy for osteosarcoma: A review. J. Cancer Res. Clin. Oncol. 2023. [Google Scholar] [CrossRef]
- Leibowitz, M.L.; Zhang, C.Z.; Pellman, D. Chromothripsis: A New Mechanism for Rapid Karyotype Evolution. Annu. Rev. Genet. 2015, 49, 183–211. [Google Scholar] [CrossRef]
- Isidro, C.-C.; June-Koo, L.; Ruibin, X.; Dhawal, J.; Youngsook, L.J.; Lixing, Y.; Dmitry, G.; Leszek, J.K.; Cheng-Zhong, Z.; David, S.P.; et al. Comprehensive analysis of chromothripsis in 2658 human cancers using whole-genome sequencing. Nat. Genet. 2020, 52, 331–341. [Google Scholar]
- Rode, A.; Maass, K.K.; Willmund, K.V.; Lichter, P.; Ernst, A. Chromothripsis in cancer cells: An update. Int. J. Cancer 2016, 138, 2322–2333. [Google Scholar] [CrossRef]
- Crasta, K.; Ganem, N.J.; Dagher, R.; Lantermann, A.B.; Ivanova, E.V.; Pan, Y.; Nezi, L.; Protopopov, A.; Chowdhury, D.; Pellman, D. DNA breaks and chromosome pulverization from errors in mitosis. Nature 2012, 482, 53–58. [Google Scholar] [CrossRef]
- Zhang, C.Z.; Spektor, A.; Cornils, H.; Francis, J.M.; Jackson, E.K.; Liu, S.; Meyerson, M.; Pellman, D. Chromothripsis from DNA damage in micronuclei. Nature 2015, 522, 179–184. [Google Scholar] [CrossRef]
- Gong, Y.; Zou, S.; Deng, D.; Wang, L.; Hu, H.; Qiu, Z.; Wei, T.; Yang, P.; Zhou, J.; Zhang, Y.; et al. Loss of RanGAP1 drives chromosome instability and rapid tumorigenesis of osteosarcoma. Dev. Cell 2023, 58, 192–210.e11. [Google Scholar] [CrossRef]
- Chen, X.; Bahrami, A.; Pappo, A.; Easton, J.; Dalton, J.; Hedlund, E.; Ellison, D.; Shurtleff, S.; Wu, G.; Wei, L.; et al. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. 2014, 7, 104–112. [Google Scholar] [CrossRef]
- Wong, F.L.; Boice, J.D., Jr.; Abramson, D.H.; Tarone, R.E.; Kleinerman, R.A.; Stovall, M.; Goldman, M.B.; Seddon, J.M.; Tarbell, N.; Fraumeni, J.F., Jr.; et al. Cancer incidence after retinoblastoma. Radiation dose and sarcoma risk. JAMA 1997, 278, 1262–1267. [Google Scholar] [CrossRef]
- Bartholf DeWitt, S.; Hoskinson Plumlee, S.; Brighton, H.E.; Sivaraj, D.; Martz, E.J.; Zand, M.; Kumar, V.; Sheth, M.U.; Floyd, W.; Spruance, J.V.; et al. Loss of ATRX promotes aggressive features of osteosarcoma with increased NF-κB signaling and integrin binding. JCI Insight 2022, 7, e151583. [Google Scholar] [CrossRef]
- Shao, Y.W.; Wood, G.A.; Lu, J.; Tang, Q.-L.; Liu, J.; Molyneux, S.; Chen, Y.; Fang, H.; Adissu, H.; McKee, T.; et al. Cross-species genomics identifies DLG2 as a tumor suppressor in osteosarcoma. Oncogene 2019, 38, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Smida, J.; Baumhoer, D.; Rosemann, M.; Walch, A.; Bielack, S.; Poremba, C.; Remberger, K.; Korsching, E.; Scheurlen, W.; Dierkes, C.; et al. Genomic Alterations and Allelic Imbalances Are Strong Prognostic Predictors in Osteosarcoma. Clin. Cancer Res. 2010, 16, 4256. [Google Scholar] [CrossRef] [PubMed]
- Rao, P.H.; Zhao, S.; Zhao, Y.J.; Yu, A.; Rainusso, N.; Trucco, M.; Allen-Rhoades, W.; Satterfield, L.; Fuja, D.; Borra, V.J.; et al. Coamplification of Myc/Pvt1 and homozygous deletion of Nlrp1 locus are frequent genetics changes in mouse osteosarcoma. Genes Chromosomes Cancer 2015, 54, 796–808. [Google Scholar] [CrossRef] [PubMed]
- Sayles, L.C.; Breese, M.R.; Koehne, A.L.; Leung, S.G.; Lee, A.G.; Liu, H.-Y.; Spillinger, A.; Shah, A.T.; Tanasa, B.; Straessler, K.; et al. Genome-Informed Targeted Therapy for Osteosarcoma. Cancer Discov. 2019, 9, 46. [Google Scholar] [CrossRef]
- Grisanzio, C.; Freedman, M.L. Chromosome 8q24-Associated Cancers and MYC. Genes Cancer 2010, 1, 555–559. [Google Scholar] [CrossRef]
- Nirala, B.K.; Patel, T.D.; Kurenbekova, L.; Shuck, R.; Dasgupta, A.; Rainusso, N.; Coarfa, C.; Yustein, J.T. MYC regulates CSF-1 expression via microRNA 17/20a to modulate tumor-associated macrophages in osteosarcoma. JCI Insight 2023, 8, e164947. [Google Scholar] [CrossRef]
- Nirala, B.K.; Kurenbekova, L.; Shuck, R.L.; Patel, T.; Rajapakshe, K.; Yustein, J.T. Development and characterization of a c-Myc-driven preclinical mouse model of osteosarcoma to investigate the tumor immune microenvironment. Cancer Res. 2022, 82 (Suppl. 12), 1668. [Google Scholar] [CrossRef]
- Nirala, B.K.; Kurenbekova, L.; Patel, T.; Shuck, R.L.; Dasgupta, A.; Rainusso, N.C.; Yustein, J.T. Abstract 6713: Myc-regulated miR17, 20a modulate RANK expression in osteosarcoma. Cancer Res. 2023, 83 (Suppl. 7), 6713. [Google Scholar] [CrossRef]
- Lu, Y.; Chan, Y.-T.; Tan, H.-Y.; Li, S.; Wang, N.; Feng, Y. Epigenetic regulation in human cancer: The potential role of epi-drug in cancer therapy. Mol. Cancer 2020, 19, 79. [Google Scholar] [CrossRef]
- Chang, S.L.; Lee, C.W.; Yang, C.Y.; Lin, Z.C.; Peng, K.T.; Liu, S.C.; Wang, S.W.; Tsai, H.C.; Fong, Y.C.; Lai, C.Y.; et al. IOX-1 suppresses metastasis of osteosarcoma by upregulating histone H3 lysine trimethylation. Biochem. Pharmacol. 2023, 210, 115472. [Google Scholar] [CrossRef]
- Kurmasheva, R.T.; Erickson, S.W.; Han, R.; Teicher, B.A.; Smith, M.A.; Roth, M.; Gorlick, R.; Houghton, P.J. In vivo evaluation of the lysine-specific demethylase (KDM1A/LSD1) inhibitor SP-2577 (Seclidemstat) against pediatric sarcoma preclinical models: A report from the Pediatric Preclinical Testing Consortium (PPTC). Pediatr. Blood Cancer 2021, 68, e29304. [Google Scholar] [CrossRef]
- Shi, X.; Fan, M. Tip60-dependent acetylation of KDM2B promotes osteosarcoma carcinogenesis. J. Cell. Mol. Med. 2019, 23, 6154–6163. [Google Scholar] [CrossRef]
- He, C.; Sun, J.; Liu, C.; Jiang, Y.; Hao, Y. Elevated H3K27me3 levels sensitize osteosarcoma to cisplatin. Clin. Epigenetics 2019, 11, 8. [Google Scholar] [CrossRef]
- Chen, M.; Jiang, Y.; Sun, Y. KDM4A-mediated histone demethylation of SLC7A11 inhibits cell ferroptosis in osteosarcoma. Biochem. Biophys. Res. Commun. 2021, 550, 77–83. [Google Scholar] [CrossRef]
- Twenhafel, L.; Moreno, D.; Punt, T.; Kinney, M.; Ryznar, R. Epigenetic Changes Associated with Osteosarcoma: A Comprehensive Review. Cells 2023, 12, 1595. [Google Scholar] [CrossRef]
- Jiang, N.; Dai, Q.; Su, X.; Fu, J.; Feng, X.; Peng, J. Role of PI3K/AKT pathway in cancer: The framework of malignant behavior. Mol. Biol. Rep. 2020, 47, 4587–4629. [Google Scholar] [CrossRef]
- Li, X.; Wu, C.; Chen, N.; Gu, H.; Yen, A.; Cao, L.; Wang, E.; Wang, L. PI3K/Akt/mTOR signaling pathway and targeted therapy for glioblastoma. Oncotarget 2016, 7, 33440–33450. [Google Scholar] [CrossRef]
- Jean, S.; Kiger, A.A. Classes of phosphoinositide 3-kinases at a glance. J. Cell Sci. 2014, 127 Pt 5, 923–928. [Google Scholar] [CrossRef]
- Zhang, J.; Yu, X.-H.; Yan, Y.-G.; Wang, C.; Wang, W.-J. PI3K/Akt signaling in osteosarcoma. Clin. Chim. Acta 2015, 444, 182–192. [Google Scholar] [CrossRef]
- Thorpe, L.M.; Yuzugullu, H.; Zhao, J.J. PI3K in cancer: Divergent roles of isoforms, modes of activation and therapeutic targeting. Nat. Rev. Cancer 2015, 15, 7–24. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, S.; Chen, J.; Liu, H.; Lu, J.; Jiang, H.; Huang, A.; Chen, Y. Fibulin-4 promotes osteosarcoma invasion and metastasis by inducing epithelial to mesenchymal transition via the PI3K/Akt/mTOR pathway. Int. J. Oncol. 2017, 50, 1513–1530. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; Jin, Y.Y.; Tang, Y.L.; Yang, H.J.; Zhou, X.Q.; Lei, Z. GPNMB silencing suppresses the proliferation and metastasis of osteosarcoma cells by blocking the PI3K/Akt/mTOR signaling pathway. Oncol. Rep. 2018, 39, 3034–3040. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Wang, Q.; Zhou, X.; Fu, C.; Cheng, M.; Guo, R.; Liu, H.; Zhang, B.; Dai, M. Celastrol negatively regulates cell invasion and migration ability of human osteosarcoma via downregulation of the PI3K/Akt/NF-κB signaling pathway in vitro. Oncol. Lett. 2016, 12, 3423–3428. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Cheng, H.; Roberts, T.M.; Zhao, J.J. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discov. 2009, 8, 627–644. [Google Scholar] [CrossRef]
- Cidado, J.; Park, B.H. Targeting the PI3K/Akt/mTOR pathway for breast cancer therapy. J. Mammary Gland Biol. Neoplasia 2012, 17, 205–216. [Google Scholar] [CrossRef]
- Doppelt-Flikshtain, O.; Younis, A.; Tamari, T.; Ginesin, O.; Shentzer-Kutiel, T.; Nikomarov, D.; Bar-Sela, G.; Coyac, B.R.; Assaraf, Y.G.; Zigdon-Giladi, H. Endothelial Progenitor Cells Promote Osteosarcoma Progression and Invasiveness via AKT/PI3K Signaling. Cancers 2023, 15, 1818. [Google Scholar] [CrossRef]
- Ma, K.; Zhang, C. HER4 Promotes Osteosarcoma Progression and Predicts Poor Prognosis through the PTEN-PI3K/AKT Pathway. J. Cancer 2022, 13, 290–303. [Google Scholar] [CrossRef]
- Lee, C.W.; Chiang, Y.C.; Yu, P.A.; Peng, K.T.; Chi, M.C.; Lee, M.H.; Fang, M.L.; Lee, K.H.; Hsu, L.F.; Liu, J.F. A Role of CXCL1 Drives Osteosarcoma Lung Metastasis via VCAM-1 Production. Front. Oncol. 2021, 11, 735277. [Google Scholar] [CrossRef]
- Xu, P.; Li, X.; Tang, C.; Wang, T.; Xu, J. Deduction of CDC42EP3 suppress development and progression of osteosarcoma. Exp. Cell Res. 2022, 412, 113018. [Google Scholar] [CrossRef]
- Cui, Y.; Dong, Y.Y. ZCCHC12 promotes the progression of osteosarcoma via PI3K/AKT pathway. Aging 2022, 14, 7505–7516. [Google Scholar] [CrossRef]
- He, R.; Wu, J.X.; Zhang, Y.; Che, H.; Yang, L. LncRNA LINC00628 overexpression inhibits the growth and invasion through regulating PI3K/Akt signaling pathway in osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 5857–5866. [Google Scholar]
- Jiang, N.; Wang, X.; Xie, X.; Liao, Y.; Liu, N.; Liu, J.; Miao, N.; Shen, J.; Peng, T. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 2017, 405, 46–55. [Google Scholar] [CrossRef]
- Yang, C.; Tian, Y.; Zhao, F.; Chen, Z.; Su, P.; Li, Y.; Qian, A. Bone Microenvironment and Osteosarcoma Metastasis. Int. J. Mol. Sci. 2020, 21, 6985. [Google Scholar] [CrossRef]
- Yang, B.; Li, L.; Tong, G.; Zeng, Z.; Tan, J.; Su, Z.; Liu, Z.; Lin, J.; Gao, W.; Chen, J.; et al. Circular RNA circ_001422 promotes the progression and metastasis of osteosarcoma via the miR-195-5p/FGF2/PI3K/Akt axis. J. Exp. Clin. Cancer Res. 2021, 40, 235. [Google Scholar] [CrossRef]
- Wang, Z.; Wei, Y.; Zhu, H.; Yu, L.; Zhu, J.; Han, Q.; Liu, Z.; Huang, J.; Zhu, Y.; Fan, G.; et al. LncRNA NDRG1 aggravates osteosarcoma progression and regulates the PI3K/AKT pathway by sponging miR-96-5p. BMC Cancer 2022, 22, 728. [Google Scholar] [CrossRef]
- Rascio, F.; Spadaccino, F.; Rocchetti, M.T.; Castellano, G.; Stallone, G.; Netti, G.S.; Ranieri, E. The Pathogenic Role of PI3K/AKT Pathway in Cancer Onset and Drug Resistance: An Updated Review. Cancers 2021, 13, 3939. [Google Scholar] [CrossRef]
- Li, H.; Shen, X.; Ma, M.; Liu, W.; Yang, W.; Wang, P.; Cai, Z.; Mi, R.; Lu, Y.; Zhuang, J.; et al. ZIP10 drives osteosarcoma proliferation and chemoresistance through ITGA10-mediated activation of the PI3K/AKT pathway. J. Exp. Clin. Cancer Res. 2021, 40, 340. [Google Scholar] [CrossRef]
- Wei, X.; Xu, L.; Jeddo, S.F.; Li, K.; Li, X.; Li, J. MARK2 enhances cisplatin resistance via PI3K/AKT/NF-κB signaling pathway in osteosarcoma cells. Am. J. Transl. Res. 2020, 12, 1807–1823. [Google Scholar]
- Meng, C.Y.; Zhao, Z.Q.; Bai, R.; Zhao, W.; Wang, Y.X.; Xue, H.Q.; Sun, L.; Sun, C.; Feng, W.; Guo, S.B. MicroRNA-22 mediates the cisplatin resistance of osteosarcoma cells by inhibiting autophagy via the PI3K/Akt/mTOR pathway. Oncol. Rep. 2020, 43, 1169–1186. [Google Scholar] [CrossRef]
- Wang, S.; Wei, H.; Huang, Z.; Wang, X.; Shen, R.; Wu, Z.; Lin, J. Epidermal growth factor receptor promotes tumor progression and contributes to gemcitabine resistance in osteosarcoma. Acta Biochim. Biophys. Sin. 2021, 53, 317–324. [Google Scholar] [CrossRef]
- Wu, X.; Yu, H.; Zhou, H.; Li, Z.; Huang, H.; Xiao, F.; Xu, S.; Yang, Y. Proanthocyanidin B2 inhibits proliferation and induces apoptosis of osteosarcoma cells by suppressing the PI3K/AKT pathway. J. Cell. Mol. Med. 2020, 24, 11960–11971. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Z.; Wang, Y.L.; Yu, Y.H.; Xing, Y.L.; Ji, X.F. Aclidinium bromide inhibits proliferation of osteosarcoma cells through regulation of PI3K/Akt pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 105–112. [Google Scholar] [PubMed]
- Hanker, A.B.; Kaklamani, V.; Arteaga, C.L. Challenges for the Clinical Development of PI3K Inhibitors: Strategies to Improve Their Impact in Solid Tumors. Cancer Discov. 2019, 9, 482–491. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT signaling pathway: From bench to clinic. Signal Transduct. Target. Ther. 2021, 6, 402. [Google Scholar] [CrossRef] [PubMed]
- Owen, K.L.; Brockwell, N.K.; Parker, B.S. JAK-STAT Signaling: A Double-Edged Sword of Immune Regulation and Cancer Progression. Cancers 2019, 11, 2002. [Google Scholar] [CrossRef]
- Liu, Y.; Liao, S.; Bennett, S.; Tang, H.; Song, D.; Wood, D.; Zhan, X.; Xu, J. STAT3 and its targeting inhibitors in osteosarcoma. Cell Prolif. 2021, 54, e12974. [Google Scholar] [CrossRef]
- Lv, B.; Gao, G.; Guo, Y.; Zhang, Z.; Liu, R.; Dai, Z.; Ju, C.; Liang, Y.; Tang, X.; Tang, M.; et al. Serglycin promotes proliferation, migration, and invasion via the JAK/STAT signaling pathway in osteosarcoma. Aging 2021, 13, 21142–21154. [Google Scholar] [CrossRef]
- Shi, F.; Li, L.; Cheng, Y. FAT10 stimulates the development of osteosarcoma by regulating the JAK/STAT signaling pathway. J. BUON 2021, 26, 2090–2096. [Google Scholar]
- Wu, P.; Wu, D.; Zhao, L.; Huang, L.; Shen, G.; Huang, J.; Chai, Y. Prognostic role of STAT3 in solid tumors: A systematic review and meta-analysis. Oncotarget 2016, 7, 19863–19883. [Google Scholar] [CrossRef]
- Zhang, B.; Yu, L.; Han, N.; Hu, Z.; Wang, S.; Ding, L.; Jiang, J. LINC01116 targets miR-520a-3p and affects IL6R to promote the proliferation and migration of osteosarcoma cells through the Jak-stat signaling pathway. Biomed. Pharmacother. 2018, 107, 270–282. [Google Scholar] [CrossRef]
- Jiang, R.; Zhang, C.; Liu, G.; Gu, R.; Wu, H. MicroRNA-101 inhibits proliferation, migration and invasion in osteosarcoma cells by targeting ROCK1. Am. J. Cancer Res. 2017, 7, 88–97. [Google Scholar]
- Jiang, R.; Zhang, C.; Liu, G.; Gu, R.; Wu, H. MicroRNA-126 Inhibits Proliferation, Migration, Invasion, and EMT in Osteosarcoma by Targeting ZEB1. J. Cell Biochem. 2017, 118, 3765–3774. [Google Scholar] [CrossRef]
- Subramaniam, D.; Angulo, P.; Ponnurangam, S.; Dandawate, P.; Ramamoorthy, P.; Srinivasan, P.; Iwakuma, T.; Weir, S.J.; Chastain, K.; Anant, S. Suppressing STAT5 signaling affects osteosarcoma growth and stemness. Cell Death Dis. 2020, 11, 149. [Google Scholar] [CrossRef]
- Guo, H.; Zheng, L.; Guo, Y.; Han, L.; Yu, J.; Lai, F. Curculigoside Represses the Proliferation and Metastasis of Osteosarcoma via the JAK/STAT and NF-κB Signaling Pathways. Biol. Pharm. Bull. 2022, 45, 1466–1475. [Google Scholar] [CrossRef]
- Lu, K.H.; Wu, H.H.; Lin, R.C.; Lin, Y.C.; Lu, P.W.; Yang, S.F.; Yang, J.S. Curcumin Analogue L48H37 Suppresses Human Osteosarcoma U2OS and MG-63 Cells’ Migration and Invasion in Culture by Inhibition of uPA via the JAK/STAT Signaling Pathway. Molecules 2020, 26, 30. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, L.; Wang, Y.; He, A.; Hu, H.; Zhang, J.; Han, M.; Huang, Y. Curcumin inhibits the proliferation and invasion of MG-63 cells through inactivation of the p-JAK2/p-STAT3 pathway. OncoTargets Ther. 2019, 12, 2011–2021. [Google Scholar] [CrossRef]
- Ma, X.; Xu, W.; Jin, X.; Mu, H.; Wang, Z.; Hua, Y.; Cai, Z.; Zhang, T. Telocinobufagin inhibits osteosarcoma growth and metastasis by inhibiting the JAK2/STAT3 signaling pathway. Eur. J. Pharmacol. 2023, 942, 175529. [Google Scholar] [CrossRef]
- Komiya, Y.; Habas, R. Wnt signal transduction pathways. Organogenesis 2008, 4, 68–75. [Google Scholar] [CrossRef]
- Patel, S.; Alam, A.; Pant, R.; Chattopadhyay, S. Wnt Signaling and Its Significance Within the Tumor Microenvironment: Novel Therapeutic Insights. Front. Immunol. 2019, 10, 2872. [Google Scholar] [CrossRef]
- Liu, J.; Xiao, Q.; Xiao, J.; Niu, C.; Li, Y.; Zhang, X.; Zhou, Z.; Shu, G.; Yin, G. Wnt/β-catenin signalling: Function, biological mechanisms, and therapeutic opportunities. Signal Transduct. Target. Ther. 2022, 7, 3. [Google Scholar] [CrossRef]
- Danieau, G.; Morice, S.; Rédini, F.; Verrecchia, F.; Royer, B.B. New Insights about the Wnt/β-Catenin Signaling Pathway in Primary Bone Tumors and Their Microenvironment: A Promising Target to Develop Therapeutic Strategies? Int. J. Mol. Sci. 2019, 20, 3751. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Bian, M.; Pan, L.; Liu, C.; Yang, H. GABPB1-AS1 Promotes the Development of Osteosarcoma by Targeting SP1 and Activating the Wnt/β-Catenin Pathway. J. Oncol. 2022, 2022, 8468896. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Li, Y.; Zhu, H.; Chen, L.; Chen, D.; Lin, S.; Fan, T. Melittin Inhibits Growth of Human Osteosarcoma 143B Cells through Induction of Apoptosis via Suppressing the Wnt/β-catenin Signaling Pathway. Anticancer Agents Med. Chem. 2022, 22, 3172–3181. [Google Scholar] [PubMed]
- Kong, H.; Yu, W.; Chen, Z.; Li, H.; Ye, G.; Hong, J.; Xie, Z.; Chen, K.; Wu, Y.; Shen, H. CCR9 initiates epithelial–mesenchymal transition by activating Wnt/β-catenin pathways to promote osteosarcoma metastasis. Cancer Cell Int. 2021, 21, 648. [Google Scholar] [CrossRef]
- Han, Y.L.; Luo, D.; Habaxi, K.; Tayierjiang, J.; Zhao, W.; Wang, W.; Aikebaier, W.; Wang, L. COL5A2 Inhibits the TGF-β and Wnt/β-Catenin Signaling Pathways to Inhibit the Invasion and Metastasis of Osteosarcoma. Front. Oncol. 2022, 12, 813809. [Google Scholar] [CrossRef]
- Chen, X.; Zhao, W.; Fan, W. Long non-coding RNA GHET1 promotes osteosarcoma development and progression via Wnt/β-catenin signaling pathway. Oncol. Rep. 2020, 44, 349–359. [Google Scholar] [CrossRef]
- Yao, Q.; Chen, T. LINC01128 regulates the development of osteosarcoma by sponging miR-299-3p to mediate MMP2 expression and activating Wnt/β-catenin signalling pathway. J. Cell. Mol. Med. 2020, 24, 14293–14305. [Google Scholar] [CrossRef]
- Bai, S.; Li, Y.; Wang, Y.; Zhou, G.; Liu, C.; Xiong, W.; Chen, J. Long non-coding RNA MINCR regulates the growth and metastasis of human osteosarcoma cells via Wnt/β-catenin signaling pathway. Acta Biochim. Pol. 2022, 69, 551–557. [Google Scholar] [CrossRef]
- Jin, H.; Luo, S.; Wang, Y.; Liu, C.; Piao, Z.; Xu, M.; Guan, W.; Li, Q.; Zou, H.; Tan, Q.Y.; et al. miR-135b Stimulates Osteosarcoma Recurrence and Lung Metastasis via Notch and Wnt/β-Catenin Signaling. Mol. Ther. Nucleic Acids 2017, 8, 111–122. [Google Scholar] [CrossRef]
- Rongxin, S.; Pengfei, L.; Li, S.; Xiaochen, J.; Yihe, H. MicroRNA-340-5p suppresses osteosarcoma development by down-regulating the Wnt/β-catenin signaling pathway via targeting the STAT3 gene. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 982–991. [Google Scholar]
- Peng, L.; Liu, Q.; Wu, T.; Li, P.; Cai, Y.; Wei, X.; Zeng, Y.; Ye, J.; Chen, P.; Huang, J.; et al. Hsa_circ_0087302, a circular RNA, affects the progression of osteosarcoma via the Wnt/β-catenin signaling pathway. Int. J. Med. Sci. 2022, 19, 1377–1387. [Google Scholar] [CrossRef]
- Nomura, M.; Rainusso, N.; Lee, Y.C.; Dawson, B.; Coarfa, C.; Han, R.; Larson, J.L.; Shuck, R.; Kurenbekova, L.; Yustein, J.T. Tegavivint and the β-Catenin/ALDH Axis in Chemotherapy-Resistant and Metastatic Osteosarcoma. J. Natl. Cancer Inst. 2019, 111, 1216–1227. [Google Scholar] [CrossRef]
- Lu, Q.; Huang, H.; Wang, X.; Luo, L.; Xia, H.; Zhang, L.; Xu, J.; Huang, Y.; Luo, X.; Luo, J. Echinatin inhibits the growth and metastasis of human osteosarcoma cells through Wnt/β-catenin and p38 signaling pathways. Pharmacol. Res. 2023, 191, 106760. [Google Scholar] [CrossRef]
- Capaccione, K.M.; Pine, S.R. The Notch signaling pathway as a mediator of tumor survival. Carcinogenesis 2013, 34, 1420–1430. [Google Scholar] [CrossRef]
- Tang, X.F.; Cao, Y.; Peng, D.B.; Zhao, G.S.; Zeng, Y.; Gao, Z.R.; Lv, Y.F.; Guo, Q.N. Overexpression of Notch3 is associated with metastasis and poor prognosis in osteosarcoma patients. Cancer Manag. Res. 2019, 11, 547–559. [Google Scholar] [CrossRef]
- Engin, F.; Bertin, T.; Ma, O.; Jiang, M.M.; Wang, L.; Sutton, R.E.; Donehower, L.A.; Lee, B. Notch signaling contributes to the pathogenesis of human osteosarcomas. Hum. Mol. Genet. 2009, 18, 1464–1470. [Google Scholar] [CrossRef]
- Zhang, J.; Li, N.; Lu, S.; Chen, Y.; Shan, L.; Zhao, X.; Xu, Y. The role of Notch ligand Jagged1 in osteosarcoma proliferation, metastasis, and recurrence. J. Orthop. Surg. Res. 2021, 16, 226. [Google Scholar] [CrossRef]
- Zhang, Z.; Wu, W.; Shao, Z. NOTCH Signaling in Osteosarcoma. Curr. Issues Mol. Biol. 2023, 45, 2266–2283. [Google Scholar] [CrossRef]
- Hughes, D.P. How the NOTCH pathway contributes to the ability of osteosarcoma cells to metastasize. Pediatr. Adolesc. Osteosarcoma 2009, 152, 479–496. [Google Scholar]
- Cheng, J.; Zhang, Y.; Wan, R.; Zhou, J.; Wu, X.; Fan, Q.; He, J.; Tan, W.; Deng, Y. CEMIP Promotes Osteosarcoma Progression and Metastasis through Activating Notch Signaling Pathway. Front. Oncol. 2022, 12, 919108. [Google Scholar] [CrossRef]
- Garcia, J.M.P.; Cortés, J.; Stathis, A.; Mous, R.; López-Miranda, E.; Azaro, A.; Genta, S.; Nuciforo, P.; Vivancos, A.; Ferrarotto, R.; et al. First-in-human phase 1-2A study of CB-103, an oral Protein-Protein Interaction Inhibitor targeting pan-NOTCH signalling in advanced solid tumors and blood malignancies. J. Clin. Oncol. 2018, 36 (Suppl. 15), TPS2619. [Google Scholar] [CrossRef]
- Tolcher, A.W.; Messersmith, W.A.; Mikulski, S.M.; Papadopoulos, K.P.; Kwak, E.L.; Gibbon, D.G.; Patnaik, A.; Falchook, G.S.; Dasari, A.; Shapiro, G.I.; et al. Phase I study of RO4929097, a gamma secretase inhibitor of Notch signaling, in patients with refractory metastatic or locally advanced solid tumors. J. Clin. Oncol. 2012, 30, 2348–2353. [Google Scholar] [CrossRef] [PubMed]
- Blume-Jensen, P.; Hunter, T. Oncogenic kinase signalling. Nature 2001, 411, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, A.; Schlessinger, J. Signal transduction by receptors with tyrosine kinase activity. Cell 1990, 61, 203–212. [Google Scholar] [CrossRef]
- Gay, C.M.; Balaji, K.; Byers, L.A. Giving AXL the axe: Targeting AXL in human malignancy. Br. J. Cancer 2017, 116, 415–423. [Google Scholar] [CrossRef]
- Zhang, Y.; Tang, Y.-J.; Man, Y.; Pan, F.; Li, Z.-H.; Jia, L.-S. Knockdown of AXL Receptor Tyrosine Kinase in Osteosarcoma Cells Leads to Decreased Proliferation and Increased Apoptosis. Int. J. Immunopathol. Pharmacol. 2013, 26, 179–188. [Google Scholar] [CrossRef]
- Chen, D.; Zhang, Y.J.; Zhu, K.W.; Wang, W.C. A systematic review of vascular endothelial growth factor expression as a biomarker of prognosis in patients with osteosarcoma. Tumour. Biol. 2013, 34, 1895–1899. [Google Scholar] [CrossRef]
- Navid, F.; Santana, V.M.; Neel, M.; McCarville, M.B.; Shulkin, B.L.; Wu, J.; Billups, C.A.; Mao, S.; Daryani, V.M.; Stewart, C.F.; et al. A phase II trial evaluating the feasibility of adding bevacizumab to standard osteosarcoma therapy. Int. J. Cancer 2017, 141, 1469–1477. [Google Scholar] [CrossRef]
- Fernanda Amary, M.; Ye, H.; Berisha, F.; Khatri, B.; Forbes, G.; Lehovsky, K.; Frezza, A.M.; Behjati, S.; Tarpey, P.; Pillay, N.; et al. Fibroblastic growth factor receptor 1 amplification in osteosarcoma is associated with poor response to neo-adjuvant chemotherapy. Cancer Med. 2014, 3, 980–987. [Google Scholar] [CrossRef]
- Weekes, D.; Kashima, T.G.; Zandueta, C.; Perurena, N.; Thomas, D.P.; Sunters, A.; Vuillier, C.; Bozec, A.; El-Emir, E.; Miletich, I.; et al. Regulation of osteosarcoma cell lung metastasis by the c-Fos/AP-1 target FGFR1. Oncogene 2016, 35, 2852–2861. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, X. Receptor activator of nuclear factor-κB ligand (RANKL)/RANK/osteoprotegerin system in bone and other tissues (review). Mol. Med. Rep. 2015, 11, 3212–3218. [Google Scholar] [CrossRef]
- Mori, K.; Le Goff, B.; Berreur, M.; Riet, A.; Moreau, A.; Blanchard, F.; Chevalier, C.; Guisle-Marsollier, I.; Léger, J.; Guicheux, J.; et al. Human osteosarcoma cells express functional receptor activator of nuclear factor-kappa B. J. Pathol. 2007, 211, 555–562. [Google Scholar] [CrossRef]
- Branstetter, D.; Rohrbach, K.; Huang, L.Y.; Soriano, R.; Tometsko, M.; Blake, M.; Jacob, A.P.; Dougall, W.C. RANK and RANK ligand expression in primary human osteosarcoma. J. Bone Oncol. 2015, 4, 59–68. [Google Scholar] [CrossRef]
- Grimaud, E.; Soubigou, L.; Couillaud, S.; Coipeau, P.; Moreau, A.; Passuti, N.; Gouin, F.; Redini, F.; Heymann, D. Receptor activator of nuclear factor kappaB ligand (RANKL)/osteoprotegerin (OPG) ratio is increased in severe osteolysis. Am. J. Pathol. 2003, 163, 2021–2031. [Google Scholar] [CrossRef]
- Bago-Horvath, Z.; Schmid, K.; Rössler, F.; Nagy-Bojarszky, K.; Funovics, P.; Sulzbacher, I. Impact of RANK signalling on survival and chemotherapy response in osteosarcoma. Pathology 2014, 46, 411–415. [Google Scholar] [CrossRef]
- Lamoureux, F.; Richard, P.; Wittrant, Y.; Battaglia, S.; Pilet, P.; Trichet, V.; Blanchard, F.; Gouin, F.; Pitard, B.; Heymann, D.; et al. Therapeutic relevance of osteoprotegerin gene therapy in osteosarcoma: Blockade of the vicious cycle between tumor cell proliferation and bone resorption. Cancer Res. 2007, 67, 7308–7318. [Google Scholar] [CrossRef]
- Punzo, F.; Tortora, C.; Argenziano, M.; Pinto, D.D.; Pota, E.; Martino, M.D.; Paola, A.D.; Rossi, F. Can Denosumab be used in combination with Doxorubicin in Osteosarcoma? Oncotarget 2020, 11, 2763–2773. [Google Scholar] [CrossRef]
- Nguyen, N.M.; Cho, J. Hedgehog Pathway Inhibitors as Targeted Cancer Therapy and Strategies to Overcome Drug Resistance. Int. J. Mol. Sci. 2022, 23, 1733. [Google Scholar] [CrossRef]
- Lo, W.W.; Pinnaduwage, D.; Gokgoz, N.; Wunder, J.S.; Andrulis, I.L. Aberrant hedgehog signaling and clinical outcome in osteosarcoma. Sarcoma 2014, 2014, 261804. [Google Scholar] [CrossRef]
- Hirotsu, M.; Setoguchi, T.; Sasaki, H.; Matsunoshita, Y.; Gao, H.; Nagao, H.; Kunigou, O.; Komiya, S. Smoothened as a new therapeutic target for human osteosarcoma. Mol. Cancer 2010, 9, 5. [Google Scholar] [CrossRef]
- Nagao, H.; Ijiri, K.; Hirotsu, M.; Ishidou, Y.; Yamamoto, T.; Nagano, S.; Takizawa, T.; Nakashima, K.; Komiya, S.; Setoguchi, T. Role of GLI2 in the growth of human osteosarcoma. J. Pathol. 2011, 224, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Warzecha, J.; Göttig, S.; Chow, K.U.; Brüning, C.; Percic, D.; Boehrer, S.; Brude, E.; Kurth, A. Inhibition of osteosarcoma cell proliferation by the Hedgehog-inhibitor cyclopamine. J. Chemother. 2007, 19, 554–561. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.M.R.; Fuchs, B. Hedgehog Signaling Inhibitors as Anti-Cancer Agents in Osteosarcoma. Cancers 2015, 7, 784–794. [Google Scholar] [CrossRef] [PubMed]
- Lamora, A.; Talbot, J.; Bougras, G.; Amiaud, J.; Leduc, M.; Chesneau, J.; Taurelle, J.; Stresing, V.; Le Deley, M.C.; Heymann, M.F.; et al. Overexpression of smad7 blocks primary tumor growth and lung metastasis development in osteosarcoma. Clin. Cancer Res. 2014, 20, 5097–5112. [Google Scholar] [CrossRef]
- Yang, R.S.; Wu, C.T.; Lin, K.H.; Hong, R.L.; Liu, T.K.; Lin, K.S. Relation between histological intensity of transforming growth factor-beta isoforms in human osteosarcoma and the rate of lung metastasis. Tohoku J. Exp. Med. 1998, 184, 133–142. [Google Scholar] [CrossRef]
- Xu, S.; Yang, S.; Sun, G.; Huang, W.; Zhang, Y. Transforming growth factor-beta polymorphisms and serum level in the development of osteosarcoma. DNA Cell Biol. 2014, 33, 802–806. [Google Scholar] [CrossRef]
- Mintz, M.B.; Sowers, R.; Brown, K.M.; Hilmer, S.C.; Mazza, B.; Huvos, A.G.; Meyers, P.A.; Lafleur, B.; McDonough, W.S.; Henry, M.M.; et al. An expression signature classifies chemotherapy-resistant pediatric osteosarcoma. Cancer Res. 2005, 65, 1748–1754. [Google Scholar] [CrossRef]
- Zhang, D.; Han, S.; Pan, X.; Li, H.; Zhao, H.; Gao, X.; Wang, S. EFEMP1 binds to STEAP1 to promote osteosarcoma proliferation and invasion via the Wnt/β-catenin and TGF-β/Smad2/3 signal pathways. J. Bone Oncol. 2022, 37, 100458. [Google Scholar] [CrossRef]
- Chen, G.; Wang, M.; Liu, X. GDF15 promotes osteosarcoma cell migration and invasion by regulating the TGF-β signaling pathway. Mol. Med. Rep. 2019, 20, 4262–4270. [Google Scholar] [CrossRef]
- Xu, Y.; Li, Y.; Chen, X.; Xiang, F.; Deng, Y.; Li, Z.; Wei, D. TGF-β protects osteosarcoma cells from chemotherapeutic cytotoxicity in a SDH/HIF1α dependent manner. BMC Cancer 2021, 21, 1200. [Google Scholar] [CrossRef]
- Ma, K.; Zhang, C.; Li, W. Gamabufotalin suppressed osteosarcoma stem cells through the TGF-β/periostin/PI3K/AKT pathway. Chem. Biol. Interact. 2020, 331, 109275. [Google Scholar] [CrossRef]
- Fu, Y.; Tang, Y.; Wang, J.; Guo, Z. MicroRNA-181c Suppresses the Biological Progression of Osteosarcoma via Targeting SMAD7 and Regulating Transforming Growth Factor-β (TGF-β) Signaling Pathway. Med. Sci. Monit. 2019, 25, 4801–4810. [Google Scholar] [CrossRef]
- Xu, X.; Liu, M. miR-522 stimulates TGF-β/Smad signaling pathway and promotes osteosarcoma tumorigenesis by targeting PPM1A. J. Cell. Biochem. 2019, 120, 18425–18434. [Google Scholar] [CrossRef]
- Sun, Y.; Jiang, X.; Lu, Y.; Zhu, J.; Yu, L.; Ma, B.; Zhang, Q. Oridonin prevents epithelial-mesenchymal transition and TGF-β1-induced epithelial-mesenchymal transition by inhibiting TGF-β1/Smad2/3 in osteosarcoma. Chem. Biol. Interact. 2018, 296, 57–64. [Google Scholar] [CrossRef]
- Choi, S.H.; Myers, J.; Tomchuck, S.; Bonner, M.; Eid, S.; Kingsley, D.; VanHeyst, K.; Kim, S.J.; Kim, B.G.; Huang, A. Oral TGF-βR1 inhibitor Vactosertib promotes osteosarcoma regression by targeting tumor proliferation and enhancing anti-tumor immunity. Res. Sq. 2023. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).