Optical Polymorphism of Liquid–Crystalline Dispersions of DNA at High Concentrations of Crowding Polymer
Abstract
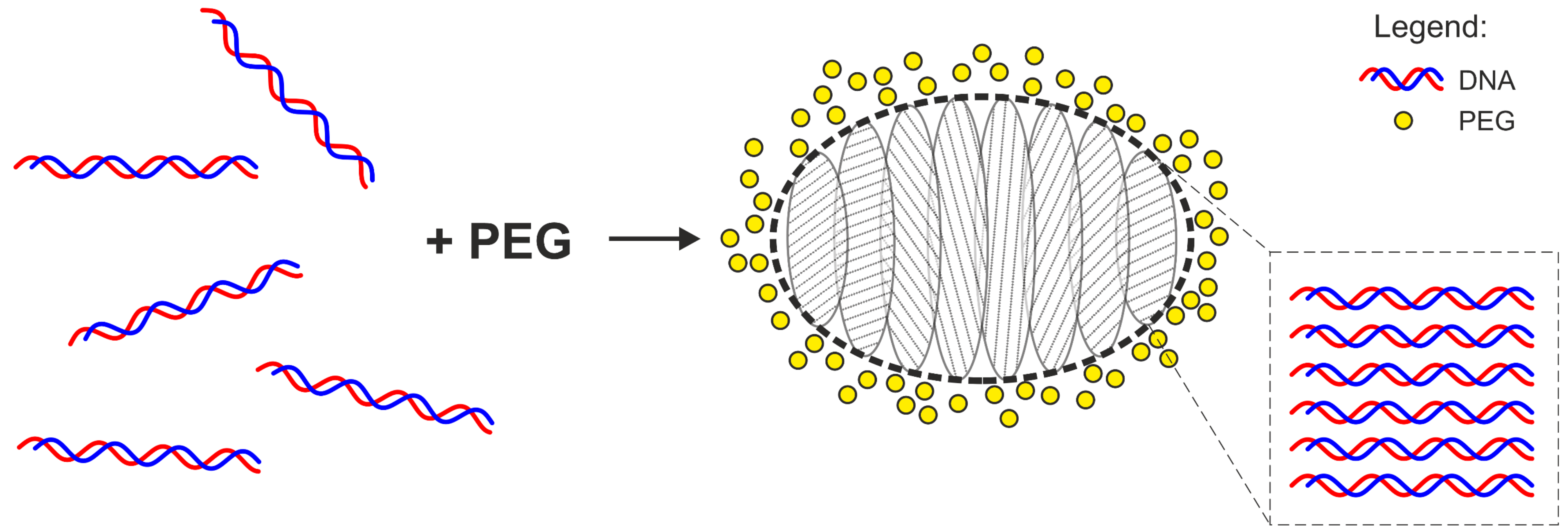
1. Introduction
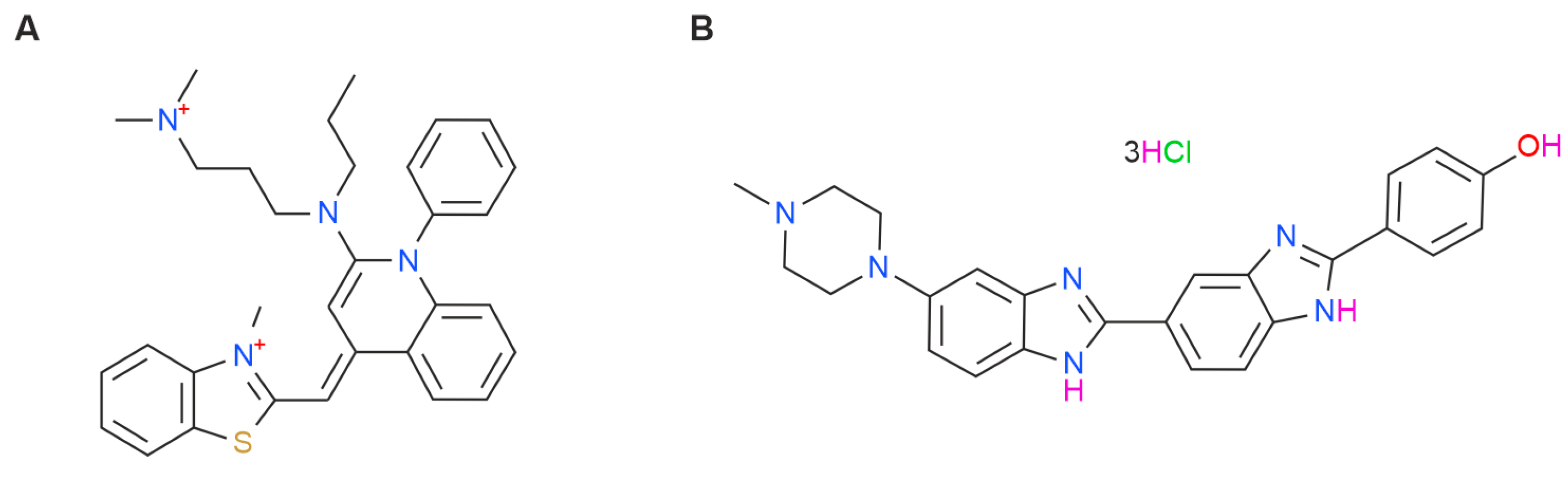
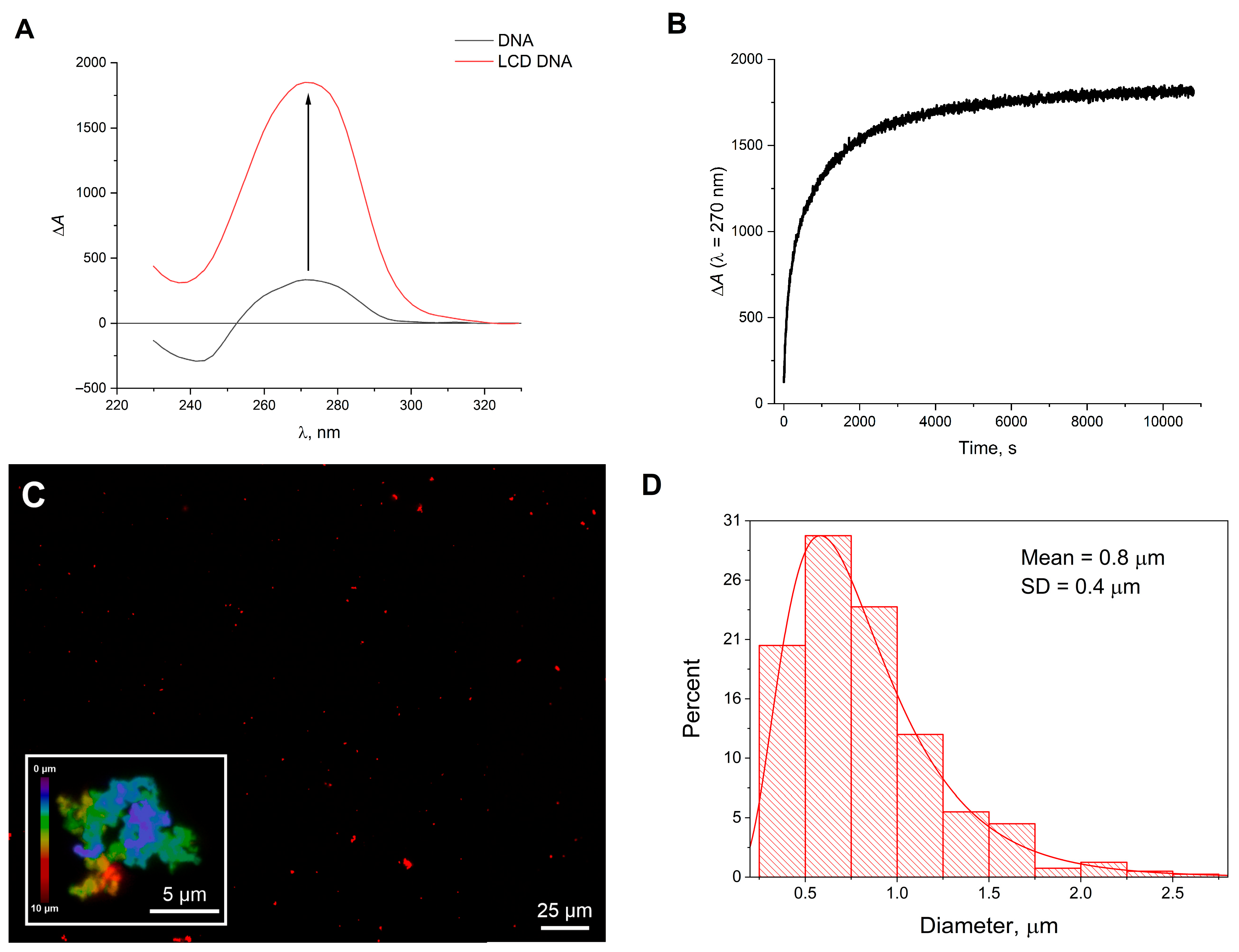
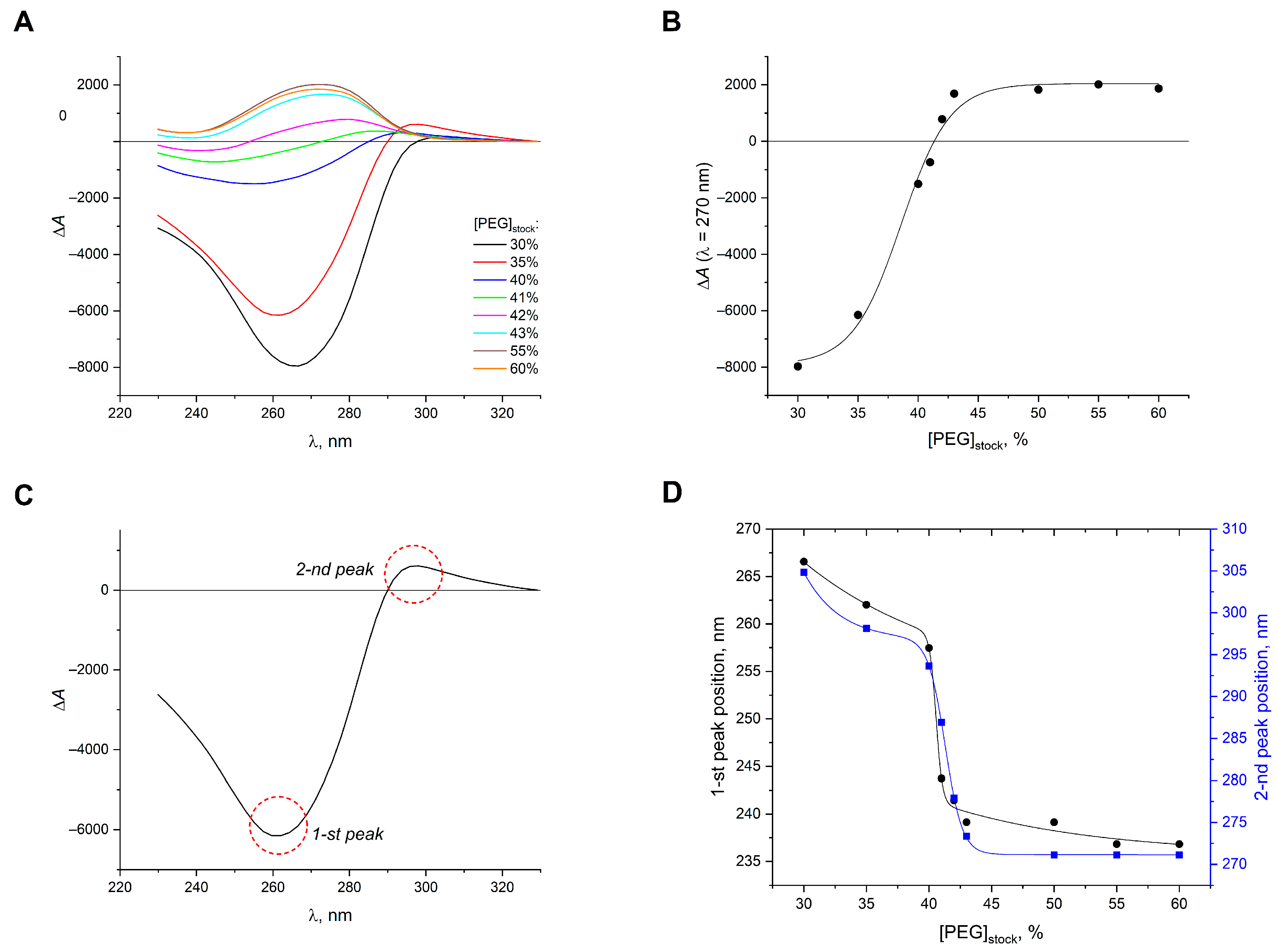
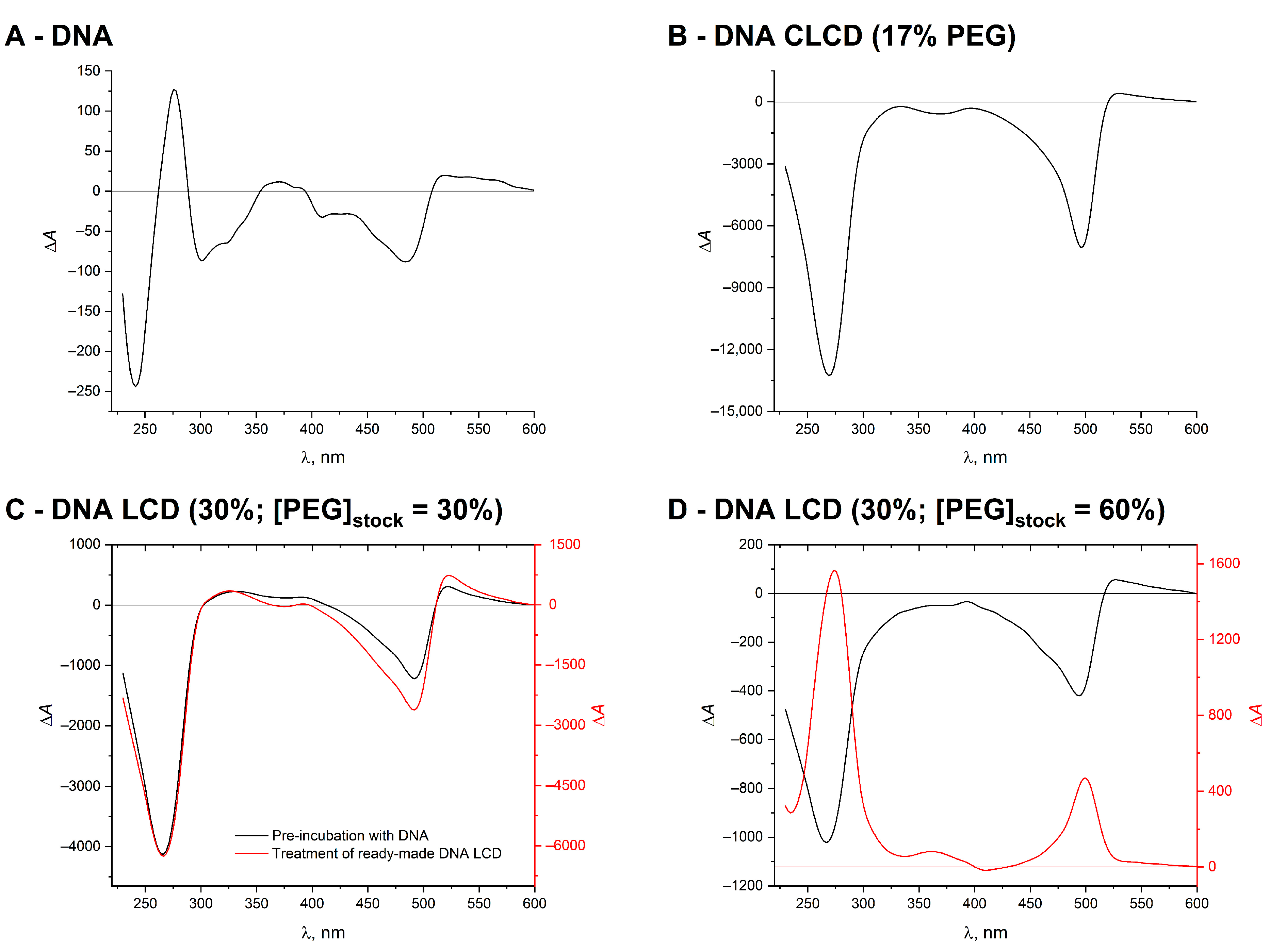
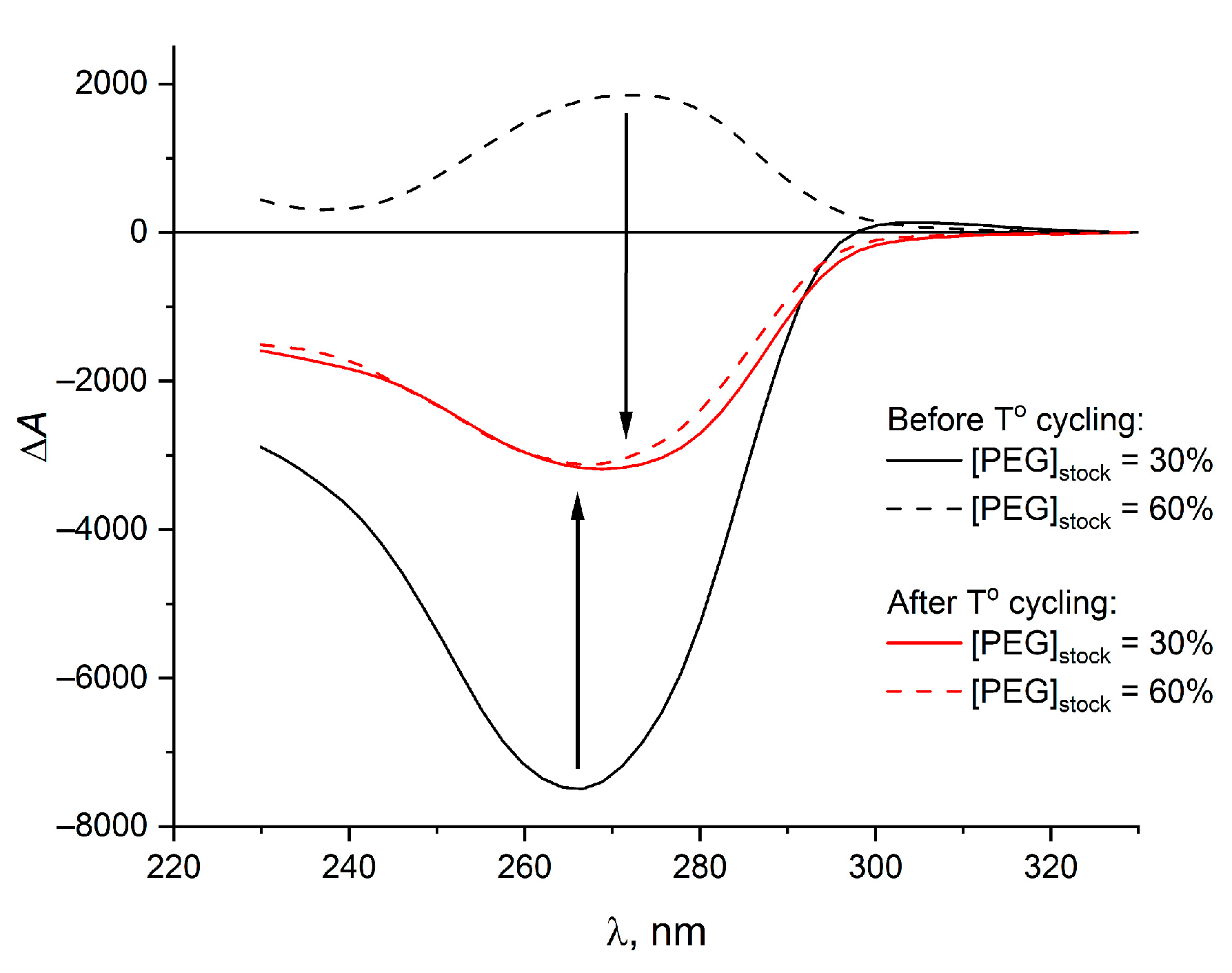
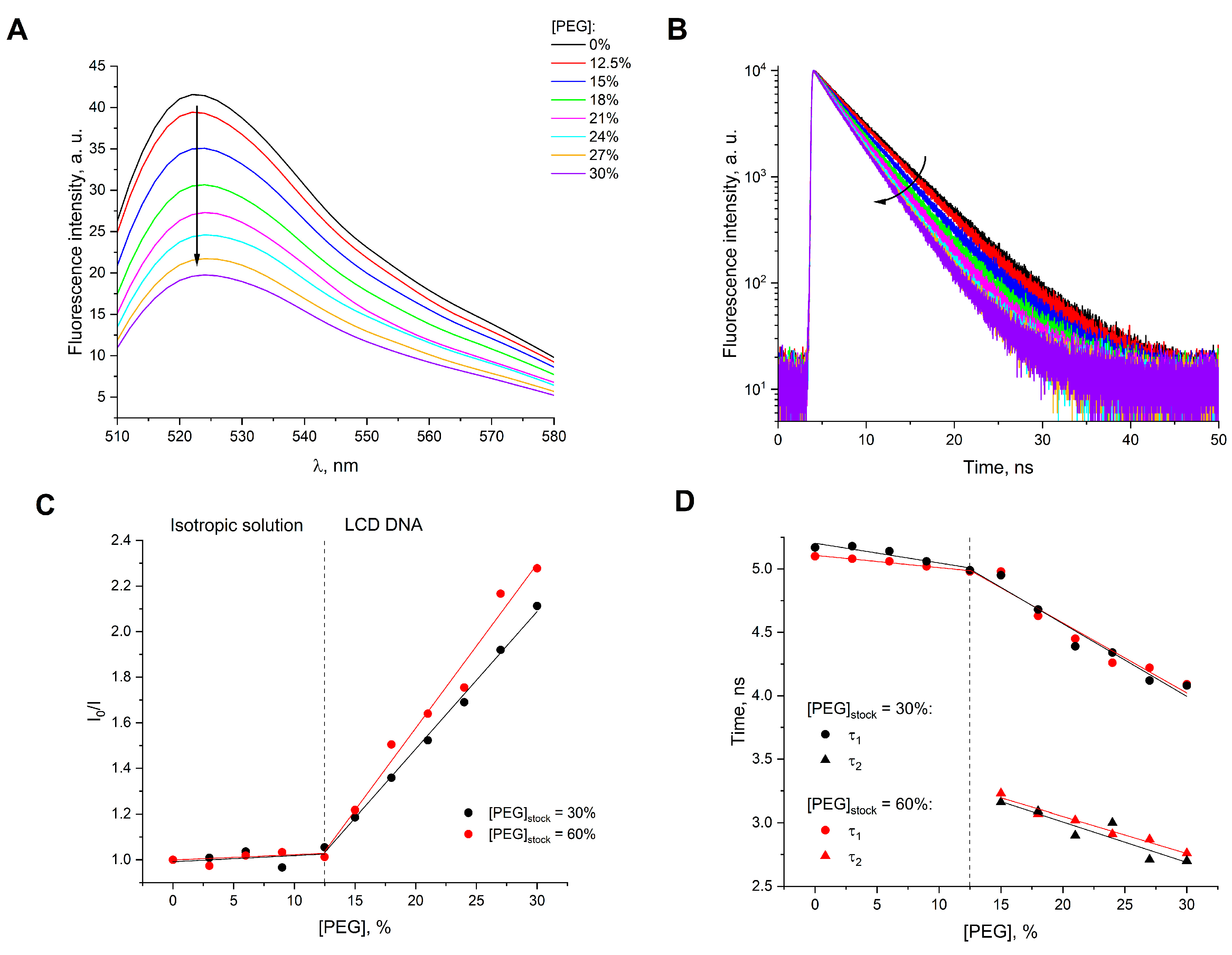
2. Results and Discussion
3. Materials and Methods
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lerman, L.S. A transition to a compact form of DNA in polymer solutions. Proc. Natl. Acad. Sci. USA 1971, 68, 1886–1890. [Google Scholar] [CrossRef] [PubMed]
- Jordan, C.F.; Lerman, L.S.; Venable, J.H. Structure and circular dichroism of DNA in concentrated polymer solutions. Nat. New Biol. 1972, 236, 67–70. [Google Scholar] [CrossRef]
- Katsura, S.; Hirano, K.; Matsuzawa, Y.; Yoshikawa, K.; Mizuno, A. Direct laser trapping of single DNA molecules in the globular state. Nucleic Acids Res. 1998, 26, 4943–4945. [Google Scholar] [CrossRef]
- Zinchenko, A.A.; Yoshikawa, K. Na+ shows a markedly higher potential than K+ in DNA compaction in a crowded environment. Biophys. J. 2005, 88, 4118–4123. [Google Scholar] [CrossRef] [PubMed]
- Yevdokimov, Y.M.; Salyanov, V.I.; Semenov, S.V.; Skuridin, S.G. DNA Liquid-Crystalline Dispersions and Nanoconstructions; CRC Press: Boca Raton, FL, USA; London, UK; New York, NY, USA, 2011. [Google Scholar]
- Nakano, S.I.; Miyoshi, D.; Sugimoto, N. Effects of molecular crowding on the structures, interactions, and functions of nucleic acids. Chem. Rev. 2014, 114, 2733–2758. [Google Scholar] [CrossRef]
- Vasilevskaya, V.V.; Khokhlov, A.R.; Matsuzawa, Y.; Yoshikawa, K. Collapse of single DNA molecule in poly(ethylene glycol) solutions. J. Chem. Phys. 1995, 102, 6595–6602. [Google Scholar] [CrossRef]
- Hirano, K.; Ichikawa, M.; Ishido, T.; Ishikawa, M.; Baba, Y.; Yoshikawa, K. How environmental solution conditions determine the compaction velocity of single DNA molecules. Nucleic Acids Res. 2012, 40, 284–289. [Google Scholar] [CrossRef] [PubMed]
- Ojala, H.; Ziedaite, G.; Wallin, A.E.; Bamford, D.H.; Hæggström, E. Optical tweezers reveal force plateau and internal friction in PEG-induced DNA condensation. Eur. Biophys. J. 2014, 43, 71–79. [Google Scholar] [CrossRef]
- Cheng, C.; Jia, J.L.; Ran, S.Y. Polyethylene glycol and divalent salt-induced DNA reentrant condensation revealed by single molecule measurements. Soft Matter. 2015, 11, 3927–3935. [Google Scholar] [CrossRef]
- Morozov, V.N.; Kolyvanova, M.A.; Dement’eva, O.V.; Rudoy, V.M.; Kuzmin, V.A. Fluorescence superquenching of SYBR Green I in crowded DNA by gold nanoparticles. J. Lumin. 2020, 219, 116898. [Google Scholar] [CrossRef]
- Morozov, V.N.; Kolyvanova, M.A.; Dement’eva, O.V.; Rudoy, V.M.; Kuzmin, V.A. Comparison of quenching efficacy of SYBR Green I and PicoGreen fluorescence by ultrasmall gold nanoparticles in isotropic and liquid-crystalline DNA systems. J. Mol. Liq. 2021, 321, 114751. [Google Scholar] [CrossRef]
- Morozov, V.N.; Klimovich, M.A.; Kostyukov, A.A.; Belousov, A.V.; Kolyvanova, M.A.; Nekipelova, T.D.; Kuzmin, V.A. Förster resonance energy transfer from Hoechst 33258 to SYBR Green I in cholesteric liquid-crystalline DNA. J. Lumin. 2022, 252, 119381. [Google Scholar] [CrossRef]
- Keller, D.; Bustamante, C. Theory of the interaction of light with large inhomogeneous molecular aggregates. II. Psi-type circular dichroism. J. Chem. Phys. 1986, 84, 2972–2980. [Google Scholar] [CrossRef]
- Kolyvanova, M.A.; Klimovich, M.A.; Shibaeva, A.V.; Koshevaya, E.D.; Bushmanov, Y.A.; Belousov, A.V.; Kuzmin, V.A.; Morozov, V.N. Cholesteric liquid-crystalline DNA–a new type of chemical detector of ionizing radiation. Liq. Cryst. 2022, 49, 1359–1366. [Google Scholar] [CrossRef]
- Kolyvanova, M.A.; Klimovich, M.A.; Belousov, A.V.; Kuzmin, V.A.; Morozov, V.N. A principal approach to the detection of radiation-induced DNA damage by circular dichroism spectroscopy and its dosimetric application. Photonics 2022, 9, 787. [Google Scholar] [CrossRef]
- Yevdokimov, Y.; Skuridin, S.; Salyanov, V.; Semenov, S.; Kats, E. Liquid-crystalline dispersions of double-stranded DNA. Crystals 2019, 9, 162. [Google Scholar] [CrossRef]
- Ramos, J.E.B., Jr.; de Vries, R.; Neto, J.R. DNA psi-condensation and reentrant decondensation: Effect of the PEG degree of polymerization. J. Phys. Chem. B 2005, 109, 23661–23665. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.S.; Park, J.H.; Han, S.W.; Kim, S.K.; Lee, Y.A. Retained binding mode of various DNA-binding molecules under molecular crowding condition. J. Biomol. Struct. Dyn. 2018, 36, 3035–3046. [Google Scholar] [CrossRef] [PubMed]
- Goldar, A.; Thomson, H.; Seddon, J.M. Structure of DNA cholesteric spherulitic droplet dispersions. J. Phys. Condens. Matter. 2007, 20, 035102. [Google Scholar] [CrossRef]
- Leonard, M.; Hong, H.; Easwar, N.; Strey, H.H. Soft matter under osmotic stress. Polymer 2001, 42, 5823–5827. [Google Scholar] [CrossRef]
- Stanley, C.B.; Hong, H.; Strey, H.H. DNA cholesteric pitch as a function of density and ionic strength. Biophys. J. 2005, 89, 2552–2557. [Google Scholar] [CrossRef] [PubMed]
- Yevdokimov, Y.M.; Skuridin, S.G.; Salyanov, V.I.; Volkov, V.V.; Dadinova, L.A.; Kompanets, O.N.; Kats, E.I. About the spatial organization of double-stranded DNA molecules in the cholesteric liquid-crystalline phase and dispersion particles of this phase. Biophysics 2015, 60, 708–721. [Google Scholar] [CrossRef]
- Yevdokimov, Y.M.; Skuridin, S.G.; Salyanov, V.I.; Kats, E.I. Temperature-induced changes of the packing of double-stranded linear DNA molecules in particles of liquid-crystalline dispersions. Biophysics 2016, 61, 351–360. [Google Scholar] [CrossRef]
- Sakurai, S.; Jo, K.; Kinoshita, H.; Esumi, M.; Tanaka, M. Guanine damage by singlet oxygen from SYBR Green I in liquid crystalline DNA. Org. Biomol. Chem. 2020, 18, 7183–7187. [Google Scholar] [CrossRef]
- Belyakov, V.A.; Orlov, V.P.; Semenov, S.V.; Skuridin, S.G.; Yevdokimov, Y.M. Comparison of calculated and observed CD spectra of liquid crystalline dispersions formed from double-stranded DNA and from DNA complexes with coloured compounds. Liq. Cryst. 1996, 20, 777–784. [Google Scholar] [CrossRef]
- Skuridin, S.G.; Dubinskaya, V.A.; Rudoy, V.M.; Dement’eva, O.V.; Zakhidov, S.T.; Marshak, T.L.; Kuzmin, V.A.; Popenko, V.I.; Yevdokimov, Y.M. Effect of gold nanoparticles on DNA package in model systems. Dokl. Biochem. Biophys. 2010, 432, 141–143. [Google Scholar] [CrossRef]
- Yevdokimov, Y.M.; Skuridin, S.G.; Salyanov, V.I.; Damaschun, G.; Damaschun, H.; Misselwitz, R.; Kleinwächter, V. Effect of platinum(II) chemotherapeutic agents on properties of DNA liquid crystals. Biophys. Chem. 1990, 35, 143–153. [Google Scholar] [CrossRef]
- Yevdokimov, Y.M.; Skuridin, S.G.; Salyanov, V.I.; Kats, E.I. Anomalous behavior of the DNA liquid-crystalline dispersion particles and their phases. Chem. Phys. Lett. 2018, 707, 154–159. [Google Scholar] [CrossRef]
- Dragan, A.I.; Pavlovic, R.; McGivney, J.B.; Casas-Finet, J.R.; Bishop, E.S.; Strouse, R.J.; Schenerman, M.A.; Geddes, C.D. SYBR Green I: Fluorescence properties and interaction with DNA. J. Fluoresc. 2012, 22, 1189–1199. [Google Scholar] [CrossRef]
- Bazhulina, N.P.; Nikitin, A.M.; Rodin, S.A.; Surovaya, A.N.; Kravatsky, Y.V.; Pismensky, V.F.; Archipova, V.S.; Martin, R.; Gursky, G.V. Binding of Hoechst 33258 and its derivatives to DNA. J. Biomol. Struct. Dyn. 2009, 26, 701–718. [Google Scholar] [CrossRef]
- Morozov, V.N.; Klimovich, M.A.; Kolyvanova, M.A.; Dement’eva, O.V.; Rudoy, V.M.; Kuzmin, V.A. Interaction of gold nanoparticles with cyanine dyes in cholesteric DNA submicroparticles. High Energy Chem. 2021, 55, 341–348. [Google Scholar] [CrossRef]
- Livolant, F. Ordered phases of DNA in vivo and in vitro. Physica A 1991, 176, 117–137. [Google Scholar] [CrossRef]
- Livolant, F.; Leforestier, A. Condensed phases of DNA: Structures and phase transitions. Prog. Polym. Sci. 1996, 21, 1115–1164. [Google Scholar] [CrossRef]
- Durand, D.; Doucet, J.; Livolant, F. A study of the structure of highly concentrated phases of DNA by X-ray diffraction. J. Phys. II Fr. 1992, 2, 1769–1783. [Google Scholar] [CrossRef]
- Budker, V.G.; Slattum, P.M.; Monahan, S.D.; Wolff, J.A. Entrapment and condensation of DNA in neutral reverse micelles. Biophys. J. 2002, 82, 1570–1579. [Google Scholar] [CrossRef]
- Morozov, V.N.; Kuzmin, V.A. Fluorescence self-quenching of Hoechst 33258 and SYBR Green I dyes in a DNA cholesteric liquid-crystalline matrix. High Energy Chem. 2019, 53, 167–169. [Google Scholar] [CrossRef]
- Spink, C.H.; Chaires, J.B. Effects of hydration, ion release, and excluded volume on the melting of triplex and duplex DNA. Biochemistry 1999, 38, 496–508. [Google Scholar] [CrossRef]
- Petraccone, L.; Pagano, B.; Giancola, C. Studying the effect of crowding and dehydration on DNA G-quadruplexes. Methods 2012, 57, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.C.; Buscaglia, R.; Chaires, J.B.; Lane, A.N.; Trent, J.O. Hydration is a major determinant of the G-quadruplex stability and conformation of the human telomere 3′ sequence of d(AG3(TTAG3)3). J. Am. Chem. Soc. 2010, 132, 17105–17107. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.I.; Karimata, H.; Ohmichi, T.; Kawakami, J.; Sugimoto, N. The effect of molecular crowding with nucleotide length and cosolute structure on DNA duplex stability. J. Am. Chem. Soc. 2004, 126, 14330–14331. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, D.; Karimata, H.; Sugimoto, N. Hydration regulates thermodynamics of G-quadruplex formation under molecular crowding conditions. J. Am. Chem. Soc. 2006, 128, 7957–7963. [Google Scholar] [CrossRef]
- Saenger, W. Structure and dynamics of water surrounding biomolecules. Annu. Rev. Biophys. Biophys. Chem. 1987, 16, 93–114. [Google Scholar] [CrossRef]
- Preisler, R.S.; Chen, H.H.; Colombo, M.F.; Choe, Y.; Short, B.J., Jr.; Rau, D.C. The B form to Z form transition of poly(dG-m5dC) is sensitive to neutral solutes through an osmotic stress. Biochemistry 1995, 34, 14400–14407. [Google Scholar] [CrossRef] [PubMed]
- Barciszewski, J.; Jurczak, J.; Porowski, S.; Specht, T.; Erdmann, V.A. The role of water structure in conformational changes of nucleic acids in ambient and high-pressure conditions. Eur. J. Biochem. 1999, 260, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.M.; Sheardy, R.D. Linking temperature, cation concentration and water activity for the B to Z conformational transition in DNA. Molecules 2018, 23, 1806. [Google Scholar] [CrossRef]
- Nakano, S.I.; Wu, L.; Oka, H.; Karimata, H.T.; Kirihata, T.; Sato, Y.; Fujii, S.; Sakai, H.; Kuwahara, M.; Sawai, H.; et al. Conformation and the sodium ion condensation on DNA and RNA structures in the presence of a neutral cosolute as a mimic of the intracellular media. Mol. Biosyst. 2008, 4, 579–588. [Google Scholar] [CrossRef]
- Phromsiri, P.; Gerling, R.R.; Blose, J.M. The effects of a neutral cosolute on the B to Z transition for DNA duplexes incorporating both CG and CA steps. Nucleosides Nucleotides Nucleic Acids 2017, 36, 690–703. [Google Scholar] [CrossRef]
- Yevdokimov, Y.M.; Skuridin, S.G.; Akimenko, N.M. Liquid-crystal microphases of low-molecular weight double stranded nucleic acids and synthetic polynucleotides. Polym. Sci. (USSR) 1984, 26, 2730–2740. [Google Scholar] [CrossRef]
- Bhanjadeo, M.M.; Baral, B.; Subudhi, U. Sequence-specific B-to-Z transition in self-assembled DNA: A biophysical and thermodynamic study. Int. J. Biol. Macromol. 2019, 137, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.M.; Mohr, S.C. Condensed states of nucleic acids. II. Effects of molecular size, base composition, and presence of intercalating agents on the psi transition of DNA. Biopolymers 1975, 14, 663–674. [Google Scholar] [CrossRef]
- Widom, J.; Baldwin, R.L. Inhibition of cation-induced DNA condensation by intercalating dyes. Biopolymers 1983, 22, 1621–1632. [Google Scholar] [CrossRef]
- Rocha, M.S.; Cavalcante, A.G.; Silva, R.; Ramos, E.B. On the effects of intercalators in DNA condensation: A force spectroscopy and gel electrophoresis study. J. Phys. Chem. B 2014, 118, 4832–4839. [Google Scholar] [CrossRef]
- Chaires, J.B. Daunomycin inhibits the B → Z transition in poly d(G-C). Nucleic Acids Res. 1983, 11, 8485–8494. [Google Scholar] [CrossRef]
- Mirau, P.A.; Kearns, D.R. The effect of intercalating drugs on the kinetics of the B to Z transition of poly(dG-dC). Nucleic Acids Res. 1983, 11, 1931–1941. [Google Scholar] [CrossRef]
- Hur, J.H.; Lee, A.R.; Yoo, W.; Lee, J.H.; Kim, K.K. Identification of a new Z-DNA inducer using SYBR green 1 as a DNA conformation sensor. FEBS Lett. 2019, 593, 2628–2636. [Google Scholar] [CrossRef] [PubMed]
- Yoshinaga, N.; Akitaya, T.; Yoshikawa, K. Intercalating fluorescence dye YOYO-1 prevents the folding transition in giant duplex DNA. Biochem. Biophys. Res. Commun. 2001, 286, 264–267. [Google Scholar] [CrossRef] [PubMed]
- Kiser, J.R.; Monk, R.W.; Smalls, R.L.; Petty, J.T. Hydration changes in the association of Hoechst 33258 with DNA. Biochemistry 2005, 44, 16988–16997. [Google Scholar] [CrossRef] [PubMed]
- Anuradha; Alam, M.S.; Chaudhury, N.K. Osmolyte changes the binding affinity and mode of interaction of minor groove binder Hoechst 33258 with calf thymus DNA. Chem. Pharm. Bull. 2010, 58, 1447–1454. [Google Scholar] [CrossRef]
- Tverdislov, V.A.; Malyshko, E.V. On regularities in the spontaneous formation of structural hierarchies in chiral systems of nonliving and living matter. Phys. Usp. 2019, 62, 354–363. [Google Scholar] [CrossRef]
- Kompanets, O.N. Portable optical biosensors for the determination of biologically active and toxic compounds. Phys. Usp. 2004, 47, 630–633. [Google Scholar] [CrossRef]
- Ouameur, A.A.; Tajmir-Riahi, H.A. Structural analysis of DNA interactions with biogenic polyamines and cobalt(III)hexamine studied by Fourier transform infrared and capillary electrophoresis. J. Biol. Chem. 2004, 279, 42041–42054. [Google Scholar] [CrossRef] [PubMed]
- Zipper, H.; Brunner, H.; Bernhagen, J.; Vitzthum, F. Investigations on DNA intercalation and surface binding by SYBR Green I, its structure determination and methodological implications. Nucleic Acids Res. 2004, 32, e103. [Google Scholar] [CrossRef] [PubMed]
- Singhal, G.; Rajeswari, M.R. Molecular aspects of the interaction of Hoechst-33258 with GC-rich promoter region of c-met. DNA Cell Biol. 2010, 29, 91–100. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morozov, V.N.; Klimovich, M.A.; Shibaeva, A.V.; Klimovich, O.N.; Koshevaya, E.D.; Kolyvanova, M.A.; Kuzmin, V.A. Optical Polymorphism of Liquid–Crystalline Dispersions of DNA at High Concentrations of Crowding Polymer. Int. J. Mol. Sci. 2023, 24, 11365. https://doi.org/10.3390/ijms241411365
Morozov VN, Klimovich MA, Shibaeva AV, Klimovich ON, Koshevaya ED, Kolyvanova MA, Kuzmin VA. Optical Polymorphism of Liquid–Crystalline Dispersions of DNA at High Concentrations of Crowding Polymer. International Journal of Molecular Sciences. 2023; 24(14):11365. https://doi.org/10.3390/ijms241411365
Chicago/Turabian StyleMorozov, Vladimir N., Mikhail A. Klimovich, Anna V. Shibaeva, Olga N. Klimovich, Ekaterina D. Koshevaya, Maria A. Kolyvanova, and Vladimir A. Kuzmin. 2023. "Optical Polymorphism of Liquid–Crystalline Dispersions of DNA at High Concentrations of Crowding Polymer" International Journal of Molecular Sciences 24, no. 14: 11365. https://doi.org/10.3390/ijms241411365
APA StyleMorozov, V. N., Klimovich, M. A., Shibaeva, A. V., Klimovich, O. N., Koshevaya, E. D., Kolyvanova, M. A., & Kuzmin, V. A. (2023). Optical Polymorphism of Liquid–Crystalline Dispersions of DNA at High Concentrations of Crowding Polymer. International Journal of Molecular Sciences, 24(14), 11365. https://doi.org/10.3390/ijms241411365





