The Metabolism of Reactive Oxygen Species and Their Effects on Lipid Biosynthesis of Microalgae
Abstract
1. Introduction
2. ROS Production in Main ETCs of Microalgae
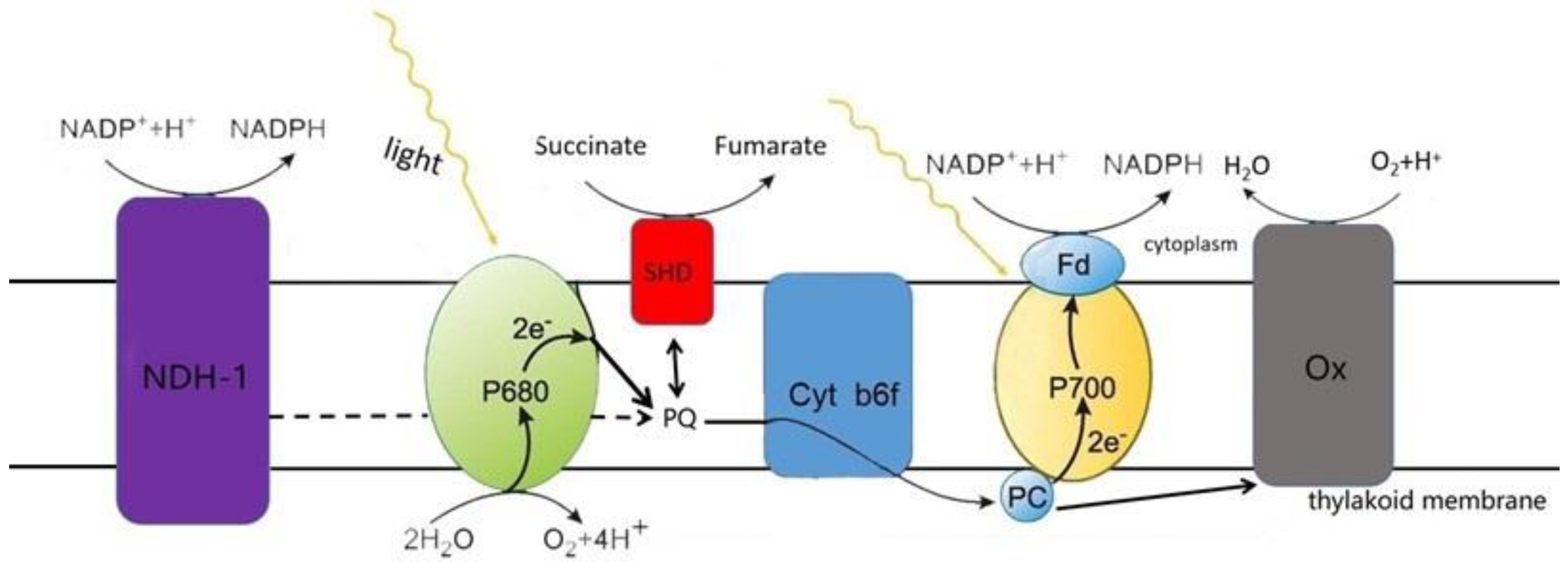
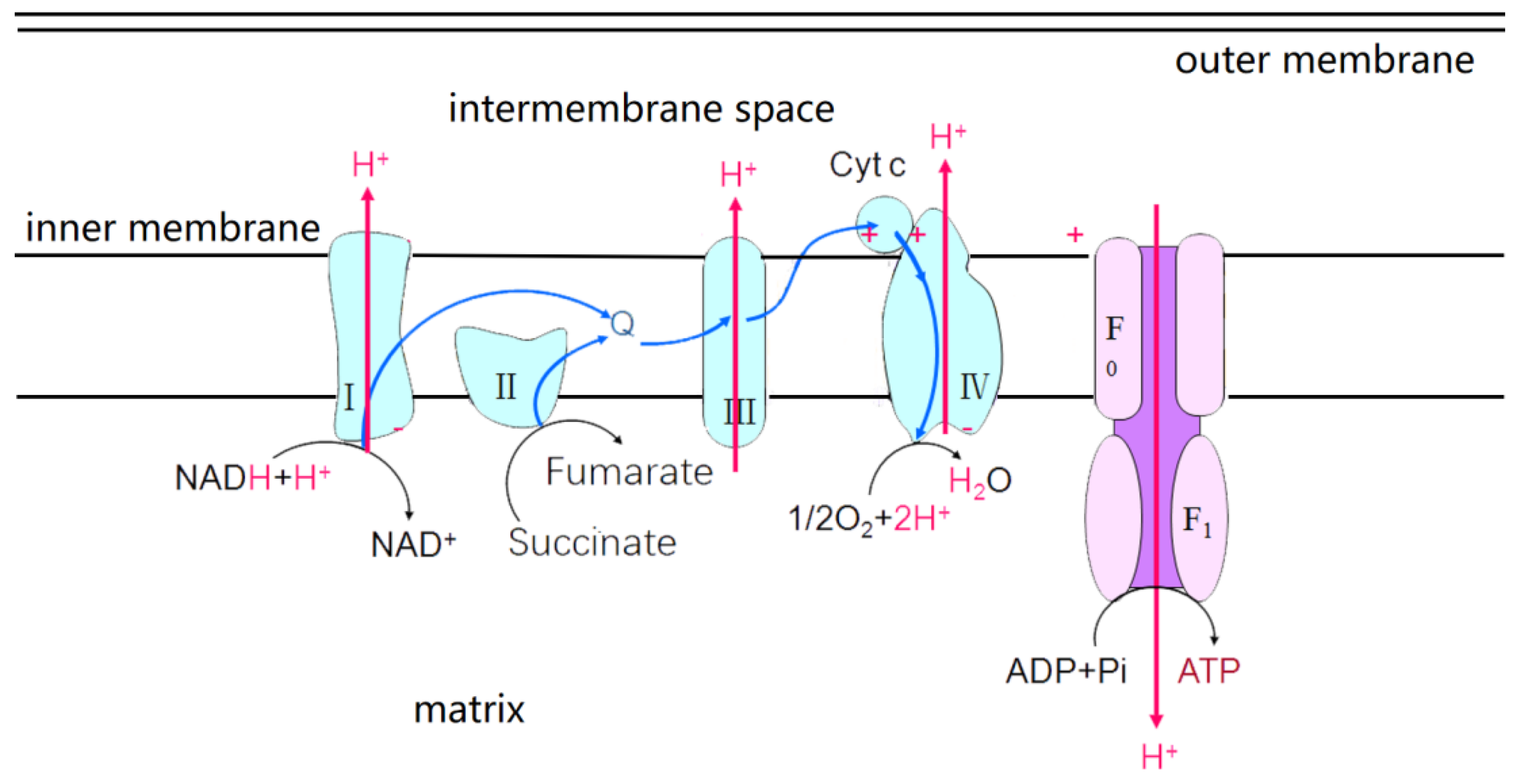
2.1. Introduction to the Main ETCs
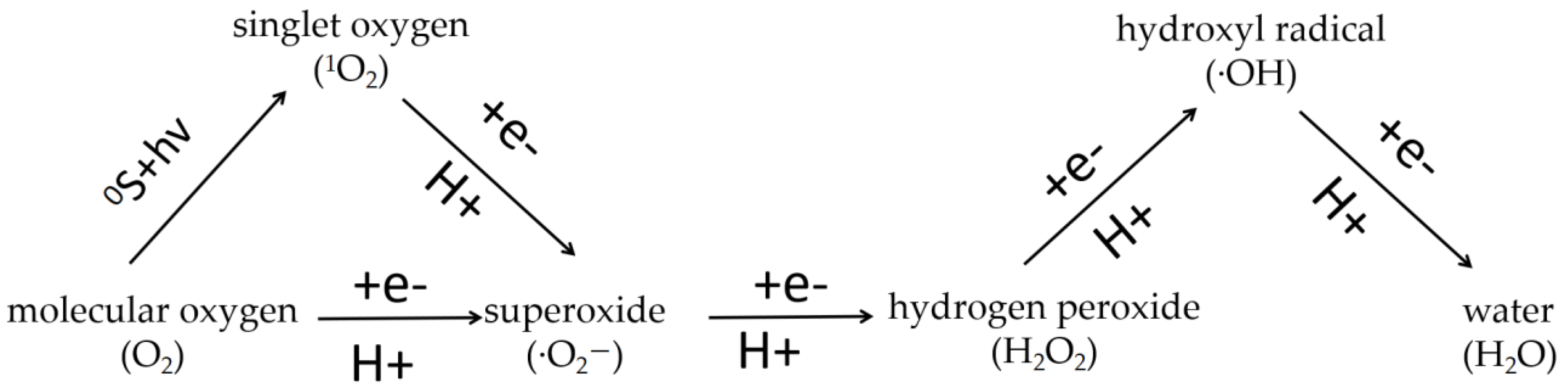
2.2. ROS Production in the Main ETCs
3. ROS Elimination Systems
3.1. Enzyme Scavenging System
3.2. Nonenzymatic Antioxidants
4. Key Pathways of Lipid Biosynthesis in Microalgae
4.1. Fatty Acid Biosynthesis in Chloroplasts of Microalgae
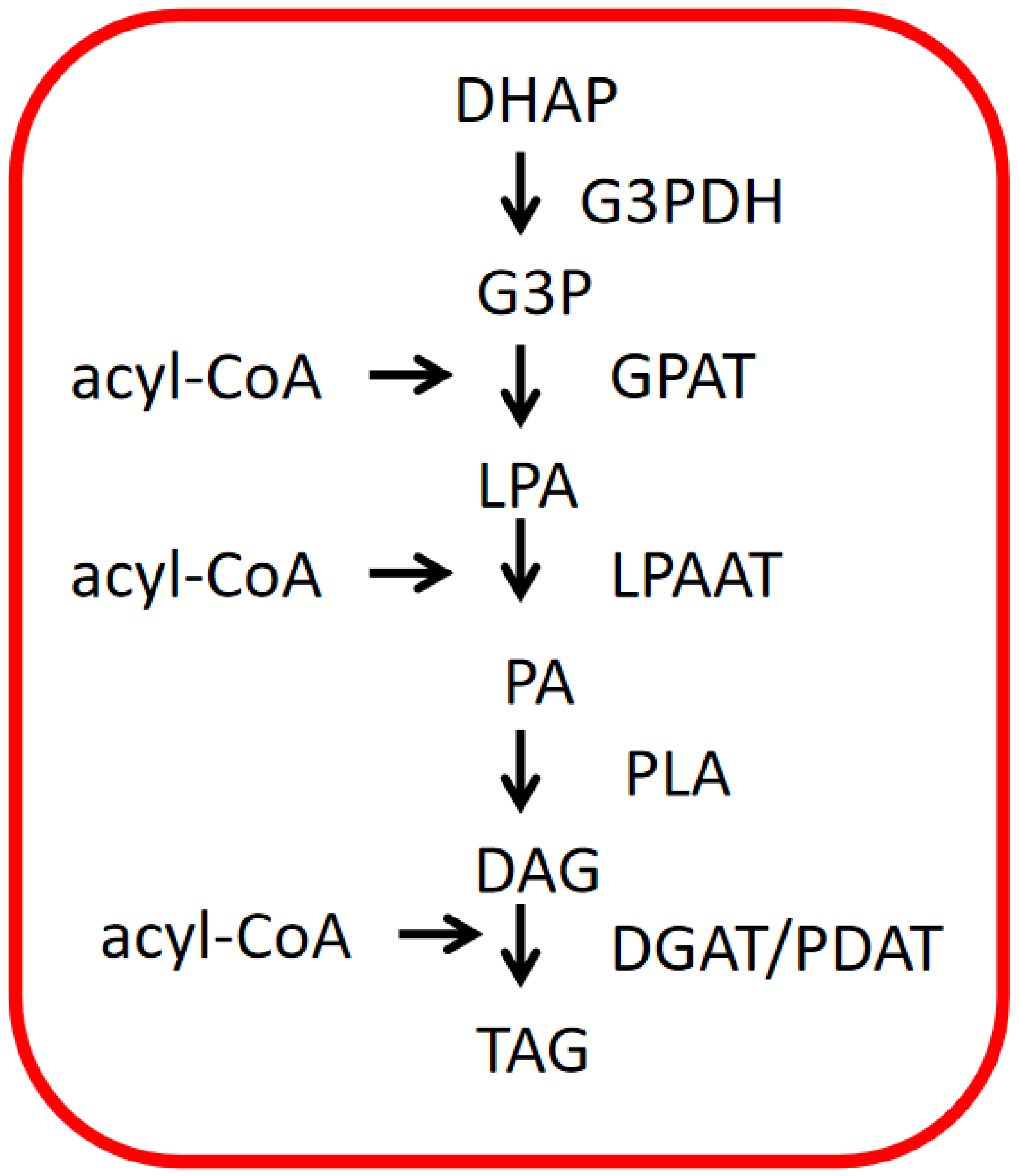
4.2. TAG Biosynthesis in ER of Microalgae
5. The Roles of ROS in Microalgal Lipid Biosynthesis Signaling Pathway
5.1. ROS Sensing by Receptor Proteins
5.2. ROS Inhibition of PPs
5.3. ROS Activation of TFs
6. Strategies and Applications of ROS-Induced Lipid Biosynthesis in Microalgae
7. Future Perspectives
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singh, U.B.; Ahluwalia, A.S. Microalgae: A promising tool for carbon sequestration. Mitig. Adapt. Strat. Glob. Chang. 2013, 18, 73–95. [Google Scholar] [CrossRef]
- Duchoud, F. Towards Improving Carbon Fixation in Plants: Cyanobacteria as a Model Organism. Ph.D. Dissertation, University of California, Los Angeles, CA, USA, 2016. [Google Scholar]
- Eduardo, J.; Zepka, L.Q.; Queiroz, M.I. Cyanobacteria and Carbon Sequestration; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2013. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, Z. Advances in the biological fixation of carbon dioxide by microalgae. J. Chem. Technol. Biotechnol. 2021, 96, 1475–1495. [Google Scholar] [CrossRef]
- Demirbas, A. Biodiesel from oilgae, biofixation of carbon dioxide by microalgae: A solution to pollution problems. Appl. Energy 2011, 88, 3541–3547. [Google Scholar] [CrossRef]
- Chisti, Y. Biodiesel from microalgae. Biotechnol. Adv. 2007, 25, 294–306. [Google Scholar] [CrossRef]
- Brennan, L.; Owende, P. Biofuels from microalgae—A review of technologies for production, processing, and extractions of biofuels and co-products. Renew. Sustain. Energy Rev. 2010, 14, 557–577. [Google Scholar] [CrossRef]
- Tocher, D.R. Omega-3 long-chain polyunsaturated fatty acids and aquaculture in perspective. Aquaculture 2015, 449, 94–107. [Google Scholar] [CrossRef]
- Møller, I.M. Plant mitochondria and oxidative stress: Electron transport, nadph turnover, and metabolism of reactive oxygen species. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 561–591. [Google Scholar] [CrossRef]
- Kärkönen, A.; Kuchitsu, K. Reactive oxygen species in cell wall metabolism and development in plants. Phytochemistry 2015, 112, 22–32. [Google Scholar] [CrossRef]
- Inupakutika, M.A.; Sengupta, S.; Devireddy, A.R.; Azad, R.K.; Mittler, R. The evolution of reactive oxygen species metabolism. J. Exp. Bot. 2016, 67, 5933–5943. [Google Scholar] [CrossRef]
- Qiao, T.; Zhao, Y.; Zhong, D.-B.; Yu, X. Hydrogen peroxide and salinity stress act synergistically to enhance lipids production in microalga by regulating reactive oxygen species and calcium. Algal Res. 2020, 53, 102017. [Google Scholar] [CrossRef]
- Chokshi, K.; Pancha, I.; Ghosh, A.; Mishra, S. Salinity induced oxidative stress alters the physiological responses and improves the biofuel potential of green microalgae Acutodesmus dimorphus. Bioresour. Technol. 2017, 244, 1376–1383. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Zhang, Z. Reactive oxygen species derived from NADPH oxidase regulate astaxanthin and total fatty acid accumulation in Chromochloris zofingiensis. J. Appl. Phycol. 2021, 33, 819–827. [Google Scholar] [CrossRef]
- Zhao, Y.; Yue, C.; Ding, W.; Li, T.; Xu, J.-W.; Zhao, P.; Ma, H.; Yu, X. Butylated hydroxytoluene induces astaxanthin and lipid production in Haematococcus pluvialis under high-light and nitrogen-deficiency conditions. Bioresour. Technol. 2018, 266, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Song, X.; Yu, L.; Han, B.; Li, T.; Yu, X. Influence of cadmium stress on the lipid production and cadmium bioresorption by Monoraphidium sp. QLY-1. Energy Convers. Manag. 2019, 188, 76–85. [Google Scholar] [CrossRef]
- Sivaramakrishnan, R.; Incharoensakdi, A. Enhancement of lipid production in Scenedesmus sp. by UV mutagenesis and hydrogen peroxide treatment. Bioresour. Technol. 2017, 235, 366–370. [Google Scholar] [CrossRef]
- Anand, V.; Kashyap, M.; Sharma, M.P.; Bala, K. Impact of hydrogen peroxide on microalgae cultivated in varying salt-nitrate-phosphate conditions. J. Environ. Chem. Eng. 2021, 9, 105814. [Google Scholar] [CrossRef]
- Zhang, L.; Liao, C.; Yang, Y.; Wang, Y.-Z.; Ding, K.; Huo, D.; Hou, C. Response of lipid biosynthesis in Chlorella pyrenoidosa to intracellular reactive oxygen species level under stress conditions. Bioresour. Technol. 2019, 287, 121414. [Google Scholar] [CrossRef]
- Latifi, A.; Ruiz, M.; Zhang, C.-C. Oxidative stress in Cyanobacteria. FEMS Microbiol. Rev. 2010, 33, 258–278. [Google Scholar] [CrossRef]
- Ruffing, A.; Trahan, C.A.; Jones, H.D.T. Genetic Engineering of Cyanobacteria as Biodiesel Feedstock, Office of Scientific & Technical Information Technical Reports; Sandia National Lab. (SNL-NM): Albuquerque, NM, USA, 2010. [Google Scholar] [CrossRef]
- Scandalios, J.G. Oxidative stress: Molecular perception and transduction of signals triggering antioxidant gene defenses. Braz. J. Med. Biol. Res. 2005, 38, 995–1014. [Google Scholar] [CrossRef]
- Peschek, G.A. Photosynthesis and Respiration of Cyanobacteria; Springer: Berlin/Heidelberg, Germany, 1999; pp. 201–209. [Google Scholar] [CrossRef]
- Schmetterer, G. Cyanobacterial respiration. In The Molecular Biology of Cyanobacteria; Bryant, D.A., Ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1994; pp. 409–435. [Google Scholar]
- Nikkanen, L.; Solymosi, D.; Jokel, M.; Allahverdiyeva, Y. Regulatory electron transport pathways of photosynthesis in cyanobacteria and microalgae: Recent advances and biotechnological prospects. Physiol. Plant. 2021, 173, 514–525. [Google Scholar] [CrossRef]
- Borowitzka, M.A.; Beardall, J.; Raven, J.A. The Physiology of Microalgae. In Developments in Applied Phycology; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar] [CrossRef]
- Imlay, J.A. Pathways of Oxidative Damage. Annu. Rev. Microbiol. 2003, 57, 395–418. [Google Scholar] [CrossRef] [PubMed]
- Fufezan, C.; Gross, C.M.; Sjödin, M.; Rutherford, A.W.; Krieger-Liszkay, A.; Kirilovsky, D. Influence of the Redox Potential of the Primary Quinone Electron Acceptor on Photoinhibition in Photosystem II. J. Biol. Chem. 2007, 282, 12492–12502. [Google Scholar] [CrossRef] [PubMed]
- Mehler, A.H. Studies on reactions of illuminated chloroplasts. I. Mechanisms of the reduction of oxygen and other hill reagents. Arch. Biochem. Biophys. 1951, 33, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Delwiche, C.; Kuhsel, M.; Palmer, J. Phylogenetic Analysis of tufA Sequences Indicates a Cyanobacterial Origin of All Plastids. Mol. Phylogenet. Evol. 1995, 4, 110–128. [Google Scholar] [CrossRef]
- Ahmad, P.; Sarwat, M.; Sharma, S. Reactive oxygen species, antioxidants and signaling in plants. J. Plant Biol. 2008, 51, 167–173. [Google Scholar] [CrossRef]
- St-Pierre, J.; Buckingham, J.A.; Roebuck, S.J.; Brand, M.D. Topology of Superoxide Production from Different Sites in the Mitochondrial Electron Transport Chain. J. Biol. Chem. 2002, 277, 44784–44790. [Google Scholar] [CrossRef]
- Han, D.; Antunes, F.; Canali, R.; Rettori, D.; Cadenas, E. Voltage-dependent Anion Channels Control the Release of the Superoxide Anion from Mitochondria to Cytosol. J. Biol. Chem. 2003, 278, 5557–5563. [Google Scholar] [CrossRef]
- Han, L.-M.; Hua, W.-P.; Cao, X.-Y.; Yan, J.-A.; Chen, C.; Wang, Z.-Z. Genome-wide identification and expression analysis of the superoxide dismutase (SOD) gene family in Salvia miltiorrhiza. Gene 2020, 742, 144603. [Google Scholar] [CrossRef]
- Abouzari A, Fakheri BA Reactive oxygen species: Generation, oxidative damage, and signal transduction. Int. J. Life Sci. 2015, 9, 3–17. [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.B.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Al Mahmud, J.; Fujita, M.; Fotopoulos, V. Reactive Oxygen Species and Antioxidant Defense in Plants under Abiotic Stress: Revisiting the Crucial Role of a Universal Defense Regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef]
- Czarnocka, W.; Karpiński, S. Friend or foe? Reactive oxygen species production, scavenging and signaling in plant response to environmental stresses. Free Radic. Biol. Med. 2018, 122, 4–20. [Google Scholar] [CrossRef]
- Miao, R.; Ma, X.; Deng, X.; Huang, K. High level of reactive oxygen species inhibits triacylglycerols accumulation in Chlamydomonas reinhardtii. Algal Res. 2019, 38, 101400. [Google Scholar] [CrossRef]
- Perkins, A.; Nelson, K.J.; Parsonage, D.; Poole, L.B.; Karplus, P.A. Peroxiredoxins: Guardians against oxidative stress and modulators of peroxide signaling. Trends Biochem. Sci. 2015, 40, 435–445. [Google Scholar] [CrossRef]
- Weids, A.J.; Grant, C.M. The yeast peroxiredoxin Tsa1 protects against proteinaggregate-induced oxidative stress. J. Cell Sci. 2014, 127, 1327–1335. [Google Scholar] [CrossRef]
- Karplus, P.A. A primer on peroxiredoxin biochemistry. Free Radic. Biol. Med. 2015, 80, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Karuppanapandian, T.; Moon, J.C.; Kim, C.; Manoharan, K.; Kim, W. Reactive oxygen species in plants: Their generation, signal transduction, and scavenging mechanisms. Aust. J. Crop Sci. 2011, 5, 709–725. [Google Scholar]
- Pancha, I.; Chokshi, K.; Maurya, R.; Trivedi, K.; Patidar, S.K.; Ghosh, A.; Mishra, S. Salinity induced oxidative stress enhanced biofuel production potential of microalgae Scenedesmus sp. CCNM 1077. Bioresour. Technol. 2015, 189, 341–348. [Google Scholar] [CrossRef]
- Goiris, K.; Van Colen, W.; Wilches, I.; León-Tamariz, F.; De Cooman, L.; Muylaert, K. Impact of nutrient stress on antioxidant production in three species of microalgae. Algal Res. 2015, 7, 51–57. [Google Scholar] [CrossRef]
- Foo, S.C.; Yusoff, F.M.; Ismail, M.; Basri, M.; Yau, S.K.; Khong, N.M.; Chan, K.W.; Ebrahimi, M. Antioxidant capacities of fucoxanthin-producing algae as influenced by their carotenoid and phenolic contents. J. Biotechnol. 2017, 241, 175–183. [Google Scholar] [CrossRef]
- Safafar, H.; van Wagenen, J.; Møller, P.; Jacobsen, C. Carotenoids, phenolic compounds and tocopherols contribute to the antioxidative properties of some microalgae species grown on industrial wastewater. Mar. Drugs 2015, 13, 7069. [Google Scholar] [CrossRef]
- Norbert, H.; Peter, S.; Kristiina, V.; Pauline, S. Abiotic stress modifies the synthesis of alpha-tocopherol and beta-carotene in phytoplankton species. J. Psychol. 2014, 50, 753–759. [Google Scholar] [CrossRef]
- Huang, X.; Huang, Z.; Wen, W.; Yan, J. Effects of nitrogen supplementation of the culture medium on the growth, total lipid content and fatty acid profiles of three microalgae (Tetraselmis subcordiformis, Nannochloropsis oculata and Pavlova viridis). J. Appl. Phycol. 2012, 25, 129–137. [Google Scholar] [CrossRef]
- Jacob, J.P.; Mathew, S. Effect of lipases from candida cylinderacea on enrichment of pufa in marine microalgae. J. Food Process. Preserv. 2016, 41, e12928. [Google Scholar] [CrossRef]
- Slocombe, S.P.; Cornah, J.; Pinfield-Wells, H.; Soady, K.; Zhang, Q.; Gilday, A.; Dyer, J.M.; Graham, I.A. Oil accumulation in leaves directed by modification of fatty acid breakdown and lipid synthesis pathways. Plant Biotechnol. J. 2010, 7, 694–703. [Google Scholar] [CrossRef] [PubMed]
- Durrett, T.P.; Benning, C.; Ohlrogge, J. Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 2010, 54, 593–607. [Google Scholar] [CrossRef]
- Liufu, W.; Di, M.; Yingying, P.; Liqiu, S.; Wenzhi, W. Nitrogen limitation and hydrogen peroxide act synergistically to enhancelipids accumulation via ROS/Ca2+ dependent mechanism in Chlorella sorokiniana. Algal Res. 2023, 70, 102974. [Google Scholar] [CrossRef]
- Cock, J.; Vanoosthuyse, V.; Gaude, T. Receptor kinase signalling in plants and animals: Distinct molecular systems with mechanistic similarities. Curr. Opin. Cell Biol. 2002, 14, 230–236. [Google Scholar] [CrossRef]
- Lehti-Shiu, M.D.; Shiu, S.-H.; Talevich, E.; Tobin, A.B.; Kannan, N.; Doerig, C. Diversity, classification and function of the plant protein kinase superfamily. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 2619–2639. [Google Scholar] [CrossRef]
- Stone, J.M.; Walker, J. Plant Protein Kinase Families and Signal Transduction. Plant Physiol. 1995, 108, 451–457. [Google Scholar] [CrossRef]
- Tichtinsky, G.; Vanoosthuyse, V.; Cock, J.M.; Gaude, T. Making inroads into plant receptor kinase signalling pathways. Trends Plant Sci. 2003, 8, 231–237. [Google Scholar] [CrossRef]
- Nongpiur, R.; Soni, P.; Karan, R.; Singla-Pareek, S.L.; Pareek, A. Histidine kinases in plants. Cross talk between hormone and stress responses. Plant Signal. Behav. 2012, 7, 1230–1237. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.-H.; Bleecker, A.B. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 2001, 98, 10763–10768. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.H.; Bleecker, A.B. Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci. Signal Transduct. Knowl. Environ. 2001, 113, RE22. [Google Scholar] [CrossRef]
- Shiu, S.-H.; Bleecker, A.B. Expansion of the Receptor-Like Kinase/Pelle Gene Family and Receptor-Like Proteins in Arabidopsis. Plant Physiol. 2003, 132, 530–543. [Google Scholar] [CrossRef]
- West, A.H.; Stock, A.M. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 2001, 26, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Ding, Y.; Jiang, Q.; Wang, F.; Sun, J.; Zhu, C. The role of receptor-like protein kinases (RLKs) in abiotic stress response in plants. Plant Cell Rep. 2017, 36, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Idnheimo, N. The Role of Cysteine-Rich Receptor-like Protein Kinases in ROS Signaling in Arabidopsis thaliana. Ph.D. Dissertation, University of Helsinki Finland, Helsinki, Finland, 2015. [Google Scholar]
- Wrzaczek, M.; Brosché, M.; Salojärvi, J.; Kangasjärvi, S.; Idänheimo, N.; Mersmann, S.; Robatzek, S.; Karpiński, S.; Karpińska, B.; Kangasjärvi, J. Transcriptional regulation of the CRK/DUF26 group of Receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol. 2010, 10, 95. [Google Scholar] [CrossRef]
- Pham, J.; Liu, J.; Bennett, M.H.; Mansfield, J.W.; Desikan, R. Arabidopsis histidine kinase 5 regulates salt sensitivity and resistance against bacterial and fungal infection. New Phytol. 2012, 194, 168–180. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Widmann, C.; Gibson, S.; Jarpe, M.B.; Johnson, G.L. Mitogen-activated protein kinase: Conservation of a three-kinase module from yeast to human. Physiol. Rev. 1999, 79, 143–180. [Google Scholar] [CrossRef]
- Johnson, G.L.; Lapadat, R. Mitogen-Activated Protein Kinase Pathways Mediated by ERK, JNK, and p38 Protein Kinases. Science 2002, 298, 1911–1912. [Google Scholar] [CrossRef] [PubMed]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.-E.; Rhee, J.-K.; Kim, H.-S.; Ahn, J.-W.; Hwang, H.; Yang, J.-W. Chemical genetics approach reveals importance of camp and map kinase signaling to lipid and carotenoid biosynthesis in microalgae. J. Microbiol. Biotechnol. 2015, 25, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Jalmi, S.K.; Sinha, A.K. ROS mediated MAPK signaling in abiotic and biotic stress- striking similarities and differences. Front. Plant Sci. 2015, 6, 769. [Google Scholar] [CrossRef] [PubMed]
- Barford, D.; Das, A.K.; Egloff, M.-P. The Structure and Mechanism of Protein Phosphatases: Insights into Catalysis and Regulation. Annu. Rev. Biophys. Biomol. Struct. 1998, 27, 133–164. [Google Scholar] [CrossRef]
- Keyse, S.M. Protein phosphatases and the regulation of mitogen-activated protein kinase signalling. Curr. Opin. Cell Biol. 2000, 12, 186–192. [Google Scholar] [CrossRef]
- Bheri, M.; Pandey, G.K. Protein phosphatases meet reactive oxygen species in plant signaling networks. Environ. Exp. Bot. 2019, 161, 26–40. [Google Scholar] [CrossRef]
- Gupta, R.; Luan, S. Redox Control of Protein Tyrosine Phosphatases and Mitogen-Activated Protein Kinases in Plants. Plant Physiol. 2003, 132, 1149–1152. [Google Scholar] [CrossRef]
- Hu, J.; Wang, D.; Li, J.; Jing, G.; Ning, K.; Xu, J. Genome-wide identification of transcription factors and transcription-factor binding sites in oleaginous microalgae Nannochloropsis. Sci. Rep. 2014, 4, 5454. [Google Scholar] [CrossRef]
- Hong, S.-Y.; Roze, L.V.; Linz, J.E. Oxidative Stress-Related Transcription Factors in the Regulation of Secondary Metabolism. Toxins 2013, 5, 683–702. [Google Scholar] [CrossRef]
- Kwon, S.; Kang, N.K.; Koh, H.G.; Shin, S.-E.; Lee, B.; Jeong, B.-R.; Chang, Y.K. Enhancement of biomass and lipid productivity by overexpression of a bZIP transcription factor in Nannochloropsis salina. Biotechnol. Bioeng. 2018, 115, 331–340. [Google Scholar] [CrossRef]
- Han, X.; Yin, L.; Xue, H. Co-expression Analysis Identifies CRC and AP1 the Regulator of Arabidopsis Fatty Acid Biosynthesis. J. Integr. Plant Biol. 2012, 54, 486–499. [Google Scholar] [CrossRef]
- Kang, N.K.; Jeon, S.; Kwon, S.; Koh, H.G.; Shin, S.-E.; Lee, B.; Choi, G.-G.; Yang, J.-W.; Jeong, B.-R.; Chang, Y.K. Effects of overexpression of a bHLH transcription factor on biomass and lipid production in Nannochloropsis salina. Biotechnol. Biofuels 2015, 8, 200. [Google Scholar] [CrossRef]
- Kalia, R.; Sareen, S.; Nagpal, A.; Katnoria, J.; Bhardwaj, R. ROS-induced transcription factors during oxidative stress in plants: A tabulated review. In Reactive Oxygen Species and Antioxidant Systems in Plants: Role and Regulation under Abiotic Stress; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar] [CrossRef]
- Ding, J.; Li, X.; Hu, H. Systematic Prediction of cis-Regulatory Elements in the Chlamydomonas reinhardtii Genome Using Comparative Genomics. Plant Physiol. 2012, 160, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, P.; Park, S.; Melis, A. Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metab. Eng. 2010, 12, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Chokshi, K.; Pancha, I.; Trivedi, K.; George, B.; Maurya, R.; Ghosh, A.; Mishra, S. Biofuel potential of the newly isolated microalgae Acutodesmus dimorphus under temperature induced oxidative stress conditions. Bioresour. Technol. 2015, 180, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Morgan, B.; Van Laer, K.; Owusu, T.N.E.; Ezeriņa, D.; Pastor-Flores, D.; Amponsah, P.S.; Tursch, A.; Dick, B.M. Real-time monitoring of basal H2O2 levels with peroxiredoxin-based probes. Nat. Chem. Biol. 2016, 12, 437–443. [Google Scholar] [CrossRef]
- Behera, B.; Unpaprom, Y.; Ramaraj, R.; Maniam, G.P.; Govindan, N.; Paramasivan, B. Integrated biomolecular and bioprocess engineering strategies for enhancing the lipid yield from microalgae. Renew. Sustain. Energy Rev. 2021, 148, 111270. [Google Scholar] [CrossRef]
- Niemeyer, J.; Scheuring, D.; Oestreicher, J.; Morgan, B.; Schroda, M. Real-time monitoring of subcellular H2O2 distribution in Chlamydomonas reinhardtii. Plant Cell 2021, 33, 2935–2949. [Google Scholar] [CrossRef]
- Battah, M.; El-Ayoty, Y.; Abomohra, A.E.-F.; El-Ghany, S.A.; Esmael, A. Effect of Mn2+, Co2+ and H2O2 on biomass and lipids of the green microalga Chlorella vulgaris as a potential candidate for biodiesel production. Ann. Microbiol. 2015, 65, 155–162. [Google Scholar] [CrossRef]
- Harris, E.H. Chlamydomonasas amodelorganism. Annu. Rev. Plant Biol. 2001, 52, 363–406. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Hu, X.; Zhao, X.; Chen, S.; Wu, Y.; Li, D.; Yu, Y.; Geng, L.; Ji, X.; Huang, H. Transcriptomic Analysis of the Regulation of Lipid Fraction Migration and Fatty Acid Biosynthesis in Schizochytrium sp. Sci. Rep. 2017, 7, 3562. [Google Scholar] [CrossRef]
- Cirulis, J.T.; Scott, J.A.; Ross, G.M. Management of oxidative stress by microalgae. Can. J. Physiol. Pharmacol. 2013, 91, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Waszczak, C.; Hunter, K.; Wrzaczek, M. Bound by fate: Reactive oxygen species in receptor-like kinase signaling. Plant Cell 2017, 29, 638–654. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Sommerfeld, M.; Jarvis, E.; Ghirardi, M.; Posewitz, M.; Seibert, M.; Darzins, A. Microalgal triacylglycerols as feedstocks for biofuel production: Perspectives and advances. Plant J. 2008, 54, 621–639. [Google Scholar] [CrossRef]
- Lupette, J.; Benning, C. Human health benefits of very-long-chain polyunsaturated fatty acids from microalgae. Biochimie 2020, 178, 15–25. [Google Scholar] [CrossRef]
- Capell, T.; Christou, P. Progress in plant metabolic engineering. Curr. Opin. Biotechnol. 2004, 15, 148–154. [Google Scholar] [CrossRef]
- Courchesne, N.M.D.; Parisien, A.; Wang, B.; Lan, C.Q. Enhancement of lipid production using biochemical, genetic and transcription factor engineering approaches. J. Biotechnol. 2009, 141, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, N.; Zhao, J.; Ge, F.; Xu, Y.; Chen, Y. Disturbance of photosystem II-oxygen evolution complex induced the oxidative damage in Chlorella vulgaris under the stress of cetyltrimethylammonium chloride. Chemosphere 2019, 223, 659–667. [Google Scholar] [CrossRef]
- Che, X.; Ding, R.; Li, Y.; Zhang, Z.; Gao, H.; Wang, W. Mechanism of long-term toxicity of CuO NPs to microalgae. Nanotoxicology 2018, 12, 923–939. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, L.; Chen, Y.; Wang, S.; Fang, Y.; Zhang, X.; Wu, Y.; Xue, D. Genome-wide identification of the SOD gene family and expression analysis under drought and salt stress in barley. Plant Growth Regul. 2021, 94, 49–60. [Google Scholar] [CrossRef]




| Microalgae | Treatment | Changes of ROS Content | Changes of Lipid Content/Lipid Productivity | Ref. |
|---|---|---|---|---|
| Monoraphidium sp. | Salt stress and 1 mmol·L−1 H2O2 | Total ROS content increased | Lipid productivity was 107.25 mg·L−1·d−1 | [12] |
| Acutodesmus dimorphus | NaCl | H2O2 content increased | Lipid content increased by 43% | [13] |
| Chromochloris zofingiensis | DPI and nitrogen starvation | Total ROS content decreased | Total fatty acids and neutral lipids decreased | [14] |
| Haematococcus pluvialis | Nitrogen limitation, highlight and BHT | Total ROS content decreased | Lipid content increased by 10.71% | [15] |
| Monoraphidium sp. | Cadmium stress | Total ROS content increased | Lipid content was 52.78% | [16] |
| Scenedesmus sp. | UV and 2 mmol·L−1 H2O2 | — | Lipid production increased by 3 times | [17] |
| Scenedesmus sp. | 0, 17.64, 35.29 mmol·L−1 NaNO3 and 10 mmol·L−1 H2O2 | — | The lipid production was 1.3, 1.2 and 1.3 times that of the group without H2O2, respectively | [18] |
| Chlorella pyrenoidosa | ·OH | — | Total fatty acids and neutral lipids increased | [19] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Yang, T.; Pan, Y.; Shi, L.; Jin, Y.; Huang, X. The Metabolism of Reactive Oxygen Species and Their Effects on Lipid Biosynthesis of Microalgae. Int. J. Mol. Sci. 2023, 24, 11041. https://doi.org/10.3390/ijms241311041
Wang L, Yang T, Pan Y, Shi L, Jin Y, Huang X. The Metabolism of Reactive Oxygen Species and Their Effects on Lipid Biosynthesis of Microalgae. International Journal of Molecular Sciences. 2023; 24(13):11041. https://doi.org/10.3390/ijms241311041
Chicago/Turabian StyleWang, Liufu, Tian Yang, Yingying Pan, Liqiu Shi, Yaqi Jin, and Xuxiong Huang. 2023. "The Metabolism of Reactive Oxygen Species and Their Effects on Lipid Biosynthesis of Microalgae" International Journal of Molecular Sciences 24, no. 13: 11041. https://doi.org/10.3390/ijms241311041
APA StyleWang, L., Yang, T., Pan, Y., Shi, L., Jin, Y., & Huang, X. (2023). The Metabolism of Reactive Oxygen Species and Their Effects on Lipid Biosynthesis of Microalgae. International Journal of Molecular Sciences, 24(13), 11041. https://doi.org/10.3390/ijms241311041





