Differentiation between Weissella cibaria and Weissella confusa Using Machine-Learning-Combined MALDI-TOF MS
Abstract
1. Introduction
2. Results
2.1. Molecular Identification
2.2. Analysis of Mass Spectra with BioTyper Database
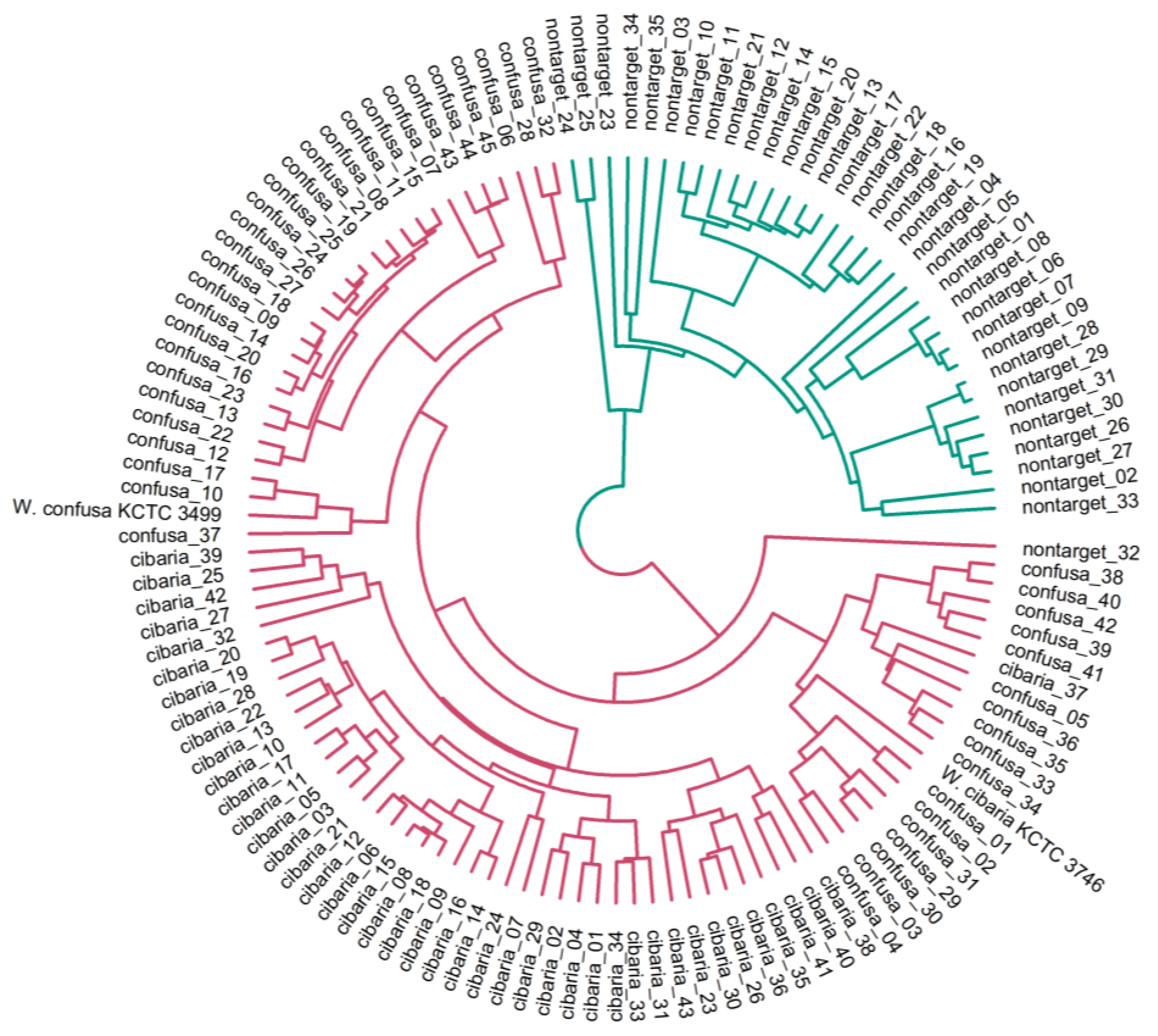
2.3. Clustering Analysis of Mass Spectra
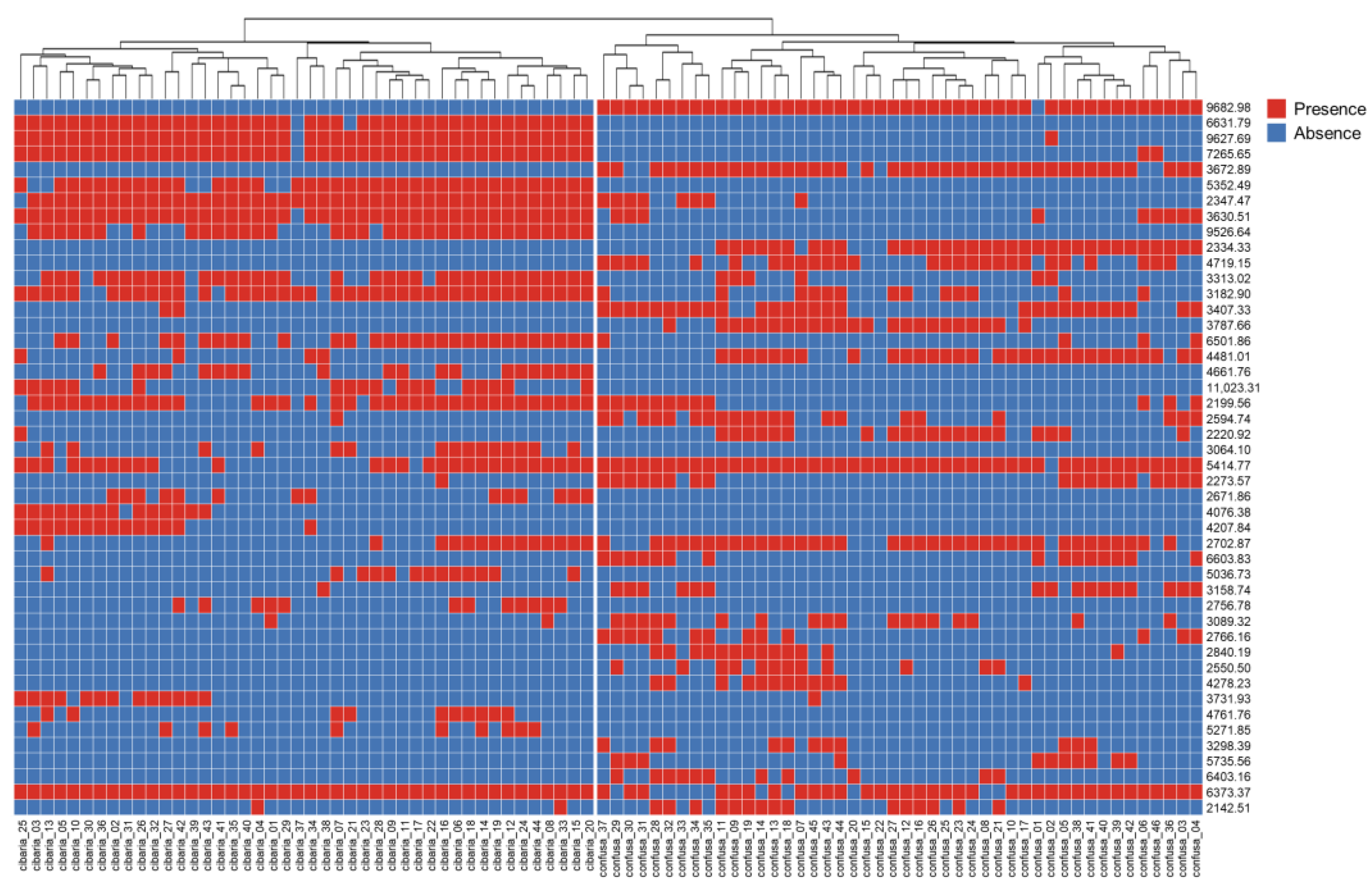
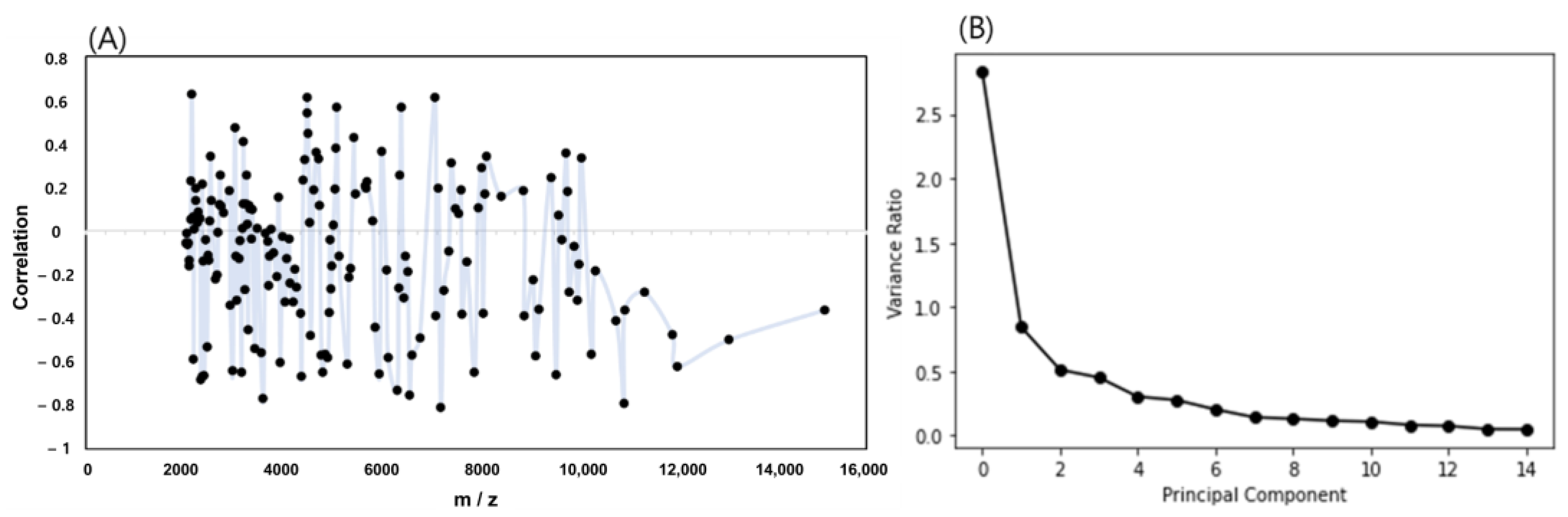
2.4. Characteristics of Mass Spectra
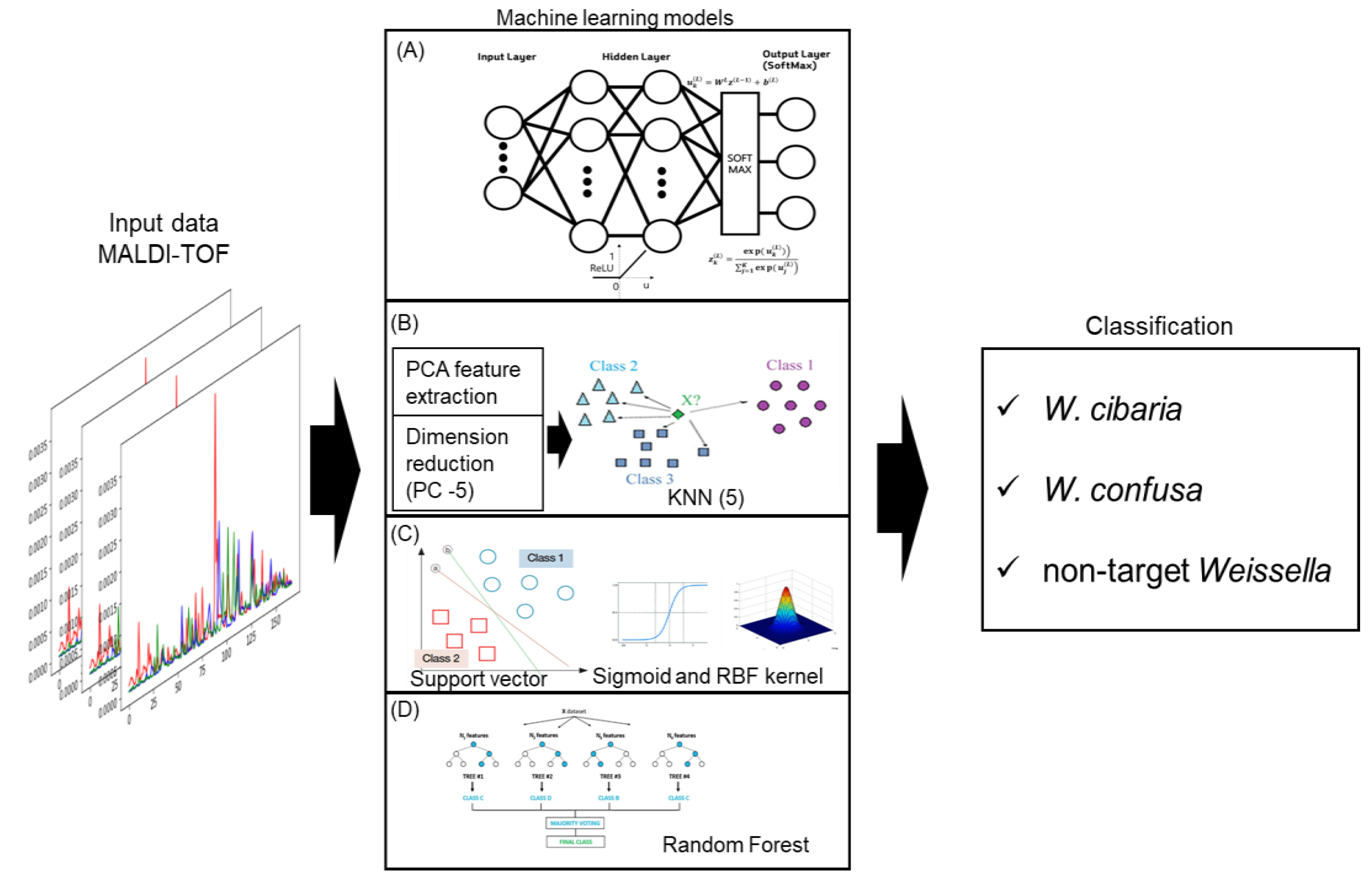
2.5. Results of the Machine Learning Classification Model
3. Discussion
4. Materials and Methods
4.1. Isolation and Identification of W. cibaria and W. confusa
4.2. MALDI-TOF MS Analysis
4.2.1. Protein Extraction
4.2.2. MALDI-TOF MS Analysis
4.3. Data Preprocessing for Machine Learning
4.3.1. Data Set
4.3.2. Data Preprocessing
4.3.3. Proteomic Analysis
4.4. Machine Learning Models for Classification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, E.; Yang, S.M.; Kim, H.Y. Weissella and the Two Janus Faces of the Genus. Appl. Microbiol. Biotechnol. 2023, 107, 1119–1127. [Google Scholar] [CrossRef] [PubMed]
- Kamboj, K.; Vasquez, A.; Balada-Llasat, J.M. Identification and Significance of Weissella Species Infections. Front. Microbiol. 2015, 6, 1204. [Google Scholar] [CrossRef] [PubMed]
- Lakra, A.K.; Domdi, L.; Hanjon, G.; Tilwani, Y.M.; Arul, V. Some Probiotic Potential of Weissella Confusa MD1 and Weissella Cibaria MD2 Isolated from Fermented Batter. Lwt 2020, 125, 109261. [Google Scholar] [CrossRef]
- Kang, M.S.; Lee, D.S.; Lee, S.A.; Kim, M.S.; Nam, S.H. Effects of Probiotic Bacterium Weissella Cibaria CMU on Periodontal Health and Microbiota: A Randomised, Double-Blind, Placebo-Controlled Trial. BMC Oral Health 2020, 20, 243. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.S.; Lee, N.K.; Choi, A.J.; Choe, J.S.; Bae, C.H.; Paik, H.D. Antagonistic and Antioxidant Effect of Probiotic Weissella Cibaria JW15. Food Sci. Biotechnol. 2019, 28, 851–855. [Google Scholar] [CrossRef]
- Wang, W.; Li, S.; Heng, X.; Chu, W. Weissella Confusa CGMCC 19,308 Strain Protects against Oxidative Stress, Increases Lifespan, and Bacterial Disease Resistance in Caenorhabditis Elegans. Probiotics Antimicrob. Proteins 2022, 14, 121–129. [Google Scholar] [CrossRef]
- Bourdichon, F.; Patrone, V.; Fontana, A.; Milani, G.; Morelli, L. Safety Demonstration of a Microbial Species for Use in the Food Chain: Weissella Confusa. Int. J. Food Microbiol. 2021, 339, 109028. [Google Scholar] [CrossRef]
- Harlan, N.P.; Kempker, R.R.; Parekh, S.M.; Burd, E.M.; Kuhar, D.T. Weissella Confusa Bacteremia in a Liver Transplant Patient with Hepatic Artery Thrombosis. Transpl. Infect. Dis. 2011, 13, 290–293. [Google Scholar] [CrossRef]
- Kim, E.; Yang, S.M.; Kim, I.S.; Kim, H.Y. Identification of Novel Molecular Targets for Weissella Species-Specific Real-Time PCR Based on Pangenome Analysis. Appl. Microbiol. Biotechnol. 2022, 106, 4157–4168. [Google Scholar] [CrossRef]
- Kim, E.; Cho, Y.; Lee, Y.; Han, S.K.; Kim, C.G.; Choo, D.W.; Kim, Y.R.; Kim, H.Y. A Proteomic Approach for Rapid Identification of Weissella Species Isolated from Korean Fermented Foods on MALDI-TOF MS Supplemented with an in-House Database. Int. J. Food Microbiol. 2017, 243, 9–15. [Google Scholar] [CrossRef]
- Nel, S.; Davis, S.B.; Endo, A.; Dicks, L.M.T. Phylogenetic Analyses of PheS, DnaA and AtpA Genes for Identification of Weissella Confusa and Weissella Cibaria Isolated from a South African Sugarcane Processing Factory. Curr. Microbiol. 2019, 76, 1138–1146. [Google Scholar] [CrossRef]
- Kim, J.M.; Kim, I.; Chung, S.H.; Chung, Y.; Han, M.; Kim, J.S. Rapid Discrimination of Methicillin-Resistant Staphylococcus Aureus by MALDI-TOF MS. Pathogens 2019, 8, 214. [Google Scholar] [CrossRef]
- Sogawa, K.; Watanabe, M.; Sato, K.; Segawa, S.; Ishii, C.; Miyabe, A.; Murata, S.; Saito, T.; Nomura, F. Use of the MALDI BioTyper System with MALDI-TOF Mass Spectrometry for Rapid Identification of Microorganisms. Anal. Bioanal. Chem. 2011, 400, 1905–1911. [Google Scholar] [CrossRef]
- Rychert, J. Benefits and Limitations of MALDI-TOF Mass Spectrometry for the Identification of Microorganisms. J. Infect. 2019, 2, 1–5. [Google Scholar] [CrossRef]
- De Bruyne, K.; Slabbinck, B.; Waegeman, W.; Vauterin, P.; De Baets, B.; Vandamme, P. Bacterial Species Identification from MALDI-TOF Mass Spectra through Data Analysis and Machine Learning. Syst. Appl. Microbiol. 2011, 34, 20–29. [Google Scholar] [CrossRef]
- Kim, E.; Yang, S.M.; Cho, E.J.; Kim, H.Y. Evaluation of Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry for the Discrimination of Lacticaseibacillus Species. Food Microbiol. 2022, 107, 104094. [Google Scholar] [CrossRef]
- Weis, C.; Cuénod, A.; Rieck, B.; Dubuis, O.; Graf, S.; Lang, C.; Oberle, M.; Brackmann, M.; Søgaard, K.K.; Osthoff, M.; et al. Direct Antimicrobial Resistance Prediction from Clinical MALDI-TOF Mass Spectra Using Machine Learning. Nat. Med. 2022, 28, 164–174. [Google Scholar] [CrossRef]
- Uysal Ciloglu, F.; Saridag, A.M.; Kilic, I.H.; Tokmakci, M.; Kahraman, M.; Aydin, O. Identification of Methicillin-Resistant: Staphylococcus Aureus Bacteria Using Surface-Enhanced Raman Spectroscopy and Machine Learning Techniques. Analyst 2020, 145, 7559–7570. [Google Scholar] [CrossRef]
- Abu-Aqil, G.; Suleiman, M.; Sharaha, U.; Lapidot, I.; Huleihel, M.; Salman, A. Instant Detection of Extended-Spectrum β-Lactamase-Producing Bacteria from the Urine of Patients Using Infrared Spectroscopy Combined with Machine Learning. Analyst 2023, 148, 1130–1140. [Google Scholar] [CrossRef]
- Weis, C.V.; Jutzeler, C.R.; Borgwardt, K. Machine Learning for Microbial Identification and Antimicrobial Susceptibility Testing on MALDI-TOF Mass Spectra: A Systematic Review. Clin. Microbiol. Infect. 2020, 26, 1310–1317. [Google Scholar] [CrossRef]
- Suarez, S.; Ferroni, A.; Lotz, A.; Jolley, K.A.; Guérin, P.; Leto, J.; Dauphin, B.; Jamet, A.; Maiden, M.C.J.; Nassif, X.; et al. Ribosomal Proteins as Biomarkers for Bacterial Identification by Mass Spectrometry in the Clinical Microbiology Laboratory. J. Microbiol. Methods 2013, 94, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Pauker, V.I.; Thoma, B.R.; Grass, G.; Bleichert, P.; Hanczaruk, M.; Zöller, L.; Zange, S. Improved Discrimination of Bacillus Anthracis from Closely Related Species in the Bacillus Cereus Sensu Lato Group Based on Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry. J. Clin. Microbiol. 2018, 56, e01900-17. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.H.; Huang, L. Rapid Species- and Subspecies-Specific Level Classification and Identification of Lactobacillus Casei Group Members Using MALDI Biotyper Combined with ClinProTools. J. Dairy Sci. 2018, 101, 979–991. [Google Scholar] [CrossRef] [PubMed]
- Manzulli, V.; Rondinone, V.; Buchicchio, A.; Serrecchia, L.; Cipolletta, D.; Fasanella, A.; Parisi, A.; Difato, L.; Iatarola, M.; Aceti, A.; et al. Discrimination of Bacillus Cereus Group Members by MALDI-TOF Mass Spectrometry. Microorganisms 2021, 9, 1202. [Google Scholar] [CrossRef]
- Haider, A.; Ringer, M.; Kotroczó, Z.; Mohácsi-Farkas, C.; Kocsis, T. The Current Level of MALDI-TOF MS Applications in the Detection of Microorganisms: A Short Review of Benefits and Limitations. Microbiol. Res. 2023, 14, 80–90. [Google Scholar] [CrossRef]
- Kim, E.; Cho, E.J.; Yang, S.M.; Kim, M.J.; Kim, H.Y. Novel Approaches for the Identification of Microbial Communities in Kimchi: MALDI-TOF MS Analysis and High-Throughput Sequencing. Food Microbiol. 2021, 94, 103641. [Google Scholar] [CrossRef]
- Kim, E.; Yang, S.M.; Kim, H.J.; Kim, H.Y. Differentiating between Enterococcus Faecium and Enterococcus Lactis by Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry. Foods 2022, 11, 1046. [Google Scholar] [CrossRef]
- Zintgraff, J.; Irazu, L.; Lara, C.S.; Rodriguez, M.; Santos, M. The Classical Bordetella Species and MALDI-TOF Technology: A Brief Experience. J. Med. Microbiol. 2018, 67, 1737–1742. [Google Scholar] [CrossRef]
- Shannon, S.; Kronemann, D.; Patel, R.; Schuetz, A.N. Routine Use of MALDI-TOF MS for Anaerobic Bacterial Identification in Clinical Microbiology. Anaerobe 2018, 54, 191–196. [Google Scholar] [CrossRef]
- Khot, P.D.; Couturier, M.R.; Wilson, A.; Croft, A.; Fisher, M.A. Optimization of Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry Analysis for Bacterial Identification. J. Clin. Microbiol. 2012, 50, 3845–3852. [Google Scholar] [CrossRef]
- Jahan, N.A.; Godden, S.M.; Royster, E.; Schoenfuss, T.C.; Gebhart, C.; Timmerman, J.; Fink, R.C. Evaluation of the Matrix-Assisted Laser Desorption Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS) System in the Detection of Mastitis Pathogens from Bovine Milk Samples. J. Microbiol. Methods 2021, 182, 106168. [Google Scholar] [CrossRef]
- Yahiaoui, R.Y.; Goessens, W.H.; Stobberingh, E.E.; Verbon, A. Differentiation between Streptococcus Pneumoniae and Other Viridans Group Streptococci by Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry. Clin. Microbiol. Infect. 2020, 26, 1088.e1–1088.e5. [Google Scholar] [CrossRef]
- Yoon, E.J.; Jeong, S.H. Maldi-Tof Mass Spectrometry Technology as a Tool for the Rapid Diagnosis of Antimicrobial Resistance in Bacteria. Antibiotics 2021, 10, 982. [Google Scholar] [CrossRef]
- Pizzato, J.; Tang, W.; Bernabeu, S.; Bonnin, R.A.; Bille, E.; Farfour, E.; Guillard, T.; Barraud, O.; Cattoir, V.; Plouzeau, C.; et al. Discrimination of Escherichia Coli, Shigella Flexneri, and Shigella Sonnei Using Lipid Profiling by MALDI-TOF Mass Spectrometry Paired with Machine Learning. Microbiologyopen 2022, 11, e1313. [Google Scholar] [CrossRef]
- Feucherolles, M.; Nennig, M.; Becker, S.L.; Martiny, D.; Losch, S.; Penny, C.; Cauchie, H.M.; Ragimbeau, C. Combination of MALDI-TOF Mass Spectrometry and Machine Learning for Rapid Antimicrobial Resistance Screening: The Case of Campylobacter Spp. Front. Microbiol. 2022, 12, 804484. [Google Scholar] [CrossRef]
- Wang, J.; Xia, C.; Wu, Y.; Tian, X.; Zhang, K.; Wang, Z. Rapid Detection of Carbapenem-Resistant Klebsiella Pneumoniae Using Machine Learning and MALDI-TOF MS Platform. Infect. Drug Resist. 2022, 15, 3703–3710. [Google Scholar] [CrossRef]
- Ho, P.L.; Yau, C.Y.; Ho, L.Y.; Chen, J.H.K.; Lai, E.L.Y.; Lo, S.W.U.; Tse, C.W.S.; Chow, K.H. Rapid Detection of CfiA Metallo-β-Lactamaseproducing Bacteroides Fragilis by the Combination of MALDI-TOF MS and CarbaNP. J. Clin. Pathol. 2017, 70, 868–873. [Google Scholar] [CrossRef]
- Mather, C.A.; Werth, B.J.; Sivagnanam, S.; SenGupta, D.J.; Butler-Wu, S.M. Rapid Detection of Vancomycin-Intermediate Staphylococcus Aureus by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. J. Clin. Microbiol. 2016, 54, 883–890. [Google Scholar] [CrossRef]
- Schulthess, B.; Bloemberg, G.V.; Zbinden, R.; Böttger, E.C.; Hombach, M. Evaluation of the Bruker MALDI Biotyper for Identification of Gram-Positive Rods: Development of a Diagnostic Algorithm for the Clinical Laboratory. J. Clin. Microbiol. 2014, 52, 1089–1097. [Google Scholar] [CrossRef]
- Gibb, S.; Strimmer, K. Maldiquant: A Versatile R Package for the Analysis of Mass Spectrometry Data. Bioinformatics 2012, 28, 2270–2271. [Google Scholar] [CrossRef]
- López-Fernández, H.; Santos, H.M.; Capelo, J.L.; Fdez-Riverola, F.; Glez-Peña, D.; Reboiro-Jato, M. Mass-Up: An All-in-One Open Software Application for MALDI-TOF Mass Spectrometry Knowledge Discovery. BMC Bioinform. 2015, 16, 318. [Google Scholar] [CrossRef] [PubMed]


| Machine Learning Models | Training Set Accuracy | Test Set Accuracy |
|---|---|---|
| ANN | 0.997 | 0.978 |
| PCA-KNN | 0.954 | 0.941 |
| SVM-sigmoid | 0.973 | 0.904 |
| SVM-RBF | 1 | 1 |
| Random Forest | 0.987 | 0.975 |
| Bacterial Strains | Origins |
|---|---|
| Reference strains | |
| W. cibaria KCTC 1 12,413 | - |
| W. confusa KCTC 3109 | - |
| W. ceti KACC 2 18,534 | - |
| W. cryptocerc KACC 18,423 | - |
| W. distrammennae KACC 16,890 | - |
| W. halotolerans KACC 11,843 | - |
| W. hellenica KACC 11,842 | - |
| W. koreensis KACC 11,853 | - |
| W. minor KCTC 3604 | - |
| W. paramesenteroides KCTC 3531 | - |
| W. soli KCTC 3789 | - |
| W. thailandensis KCTC 3751 | - |
| W. uvarum KACC 18,587 | - |
| W. viridescens KCTC 3504 | - |
| Isolates (n 3) | |
| W. cibaria (43) | Kimchi, sikhae, jeotgal |
| W. confusa (45) | Kimchi, jeotgal, tempe |
| W. hellenica (4) | Kimchi |
| W. koreensis (12) | Kimchi |
| W. paramesenteroides (7) | Tempe |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, E.; Yang, S.-M.; Jung, D.-H.; Kim, H.-Y. Differentiation between Weissella cibaria and Weissella confusa Using Machine-Learning-Combined MALDI-TOF MS. Int. J. Mol. Sci. 2023, 24, 11009. https://doi.org/10.3390/ijms241311009
Kim E, Yang S-M, Jung D-H, Kim H-Y. Differentiation between Weissella cibaria and Weissella confusa Using Machine-Learning-Combined MALDI-TOF MS. International Journal of Molecular Sciences. 2023; 24(13):11009. https://doi.org/10.3390/ijms241311009
Chicago/Turabian StyleKim, Eiseul, Seung-Min Yang, Dae-Hyun Jung, and Hae-Yeong Kim. 2023. "Differentiation between Weissella cibaria and Weissella confusa Using Machine-Learning-Combined MALDI-TOF MS" International Journal of Molecular Sciences 24, no. 13: 11009. https://doi.org/10.3390/ijms241311009
APA StyleKim, E., Yang, S.-M., Jung, D.-H., & Kim, H.-Y. (2023). Differentiation between Weissella cibaria and Weissella confusa Using Machine-Learning-Combined MALDI-TOF MS. International Journal of Molecular Sciences, 24(13), 11009. https://doi.org/10.3390/ijms241311009







