Transfer of Stress Resilient QTLs and Panicle Traits into the Rice Variety, Reeta through Classical and Marker-Assisted Breeding Approaches
Abstract
1. Introduction
2. Result
2.1. Validation of the Donor and Recipient Parents for the Target Traits
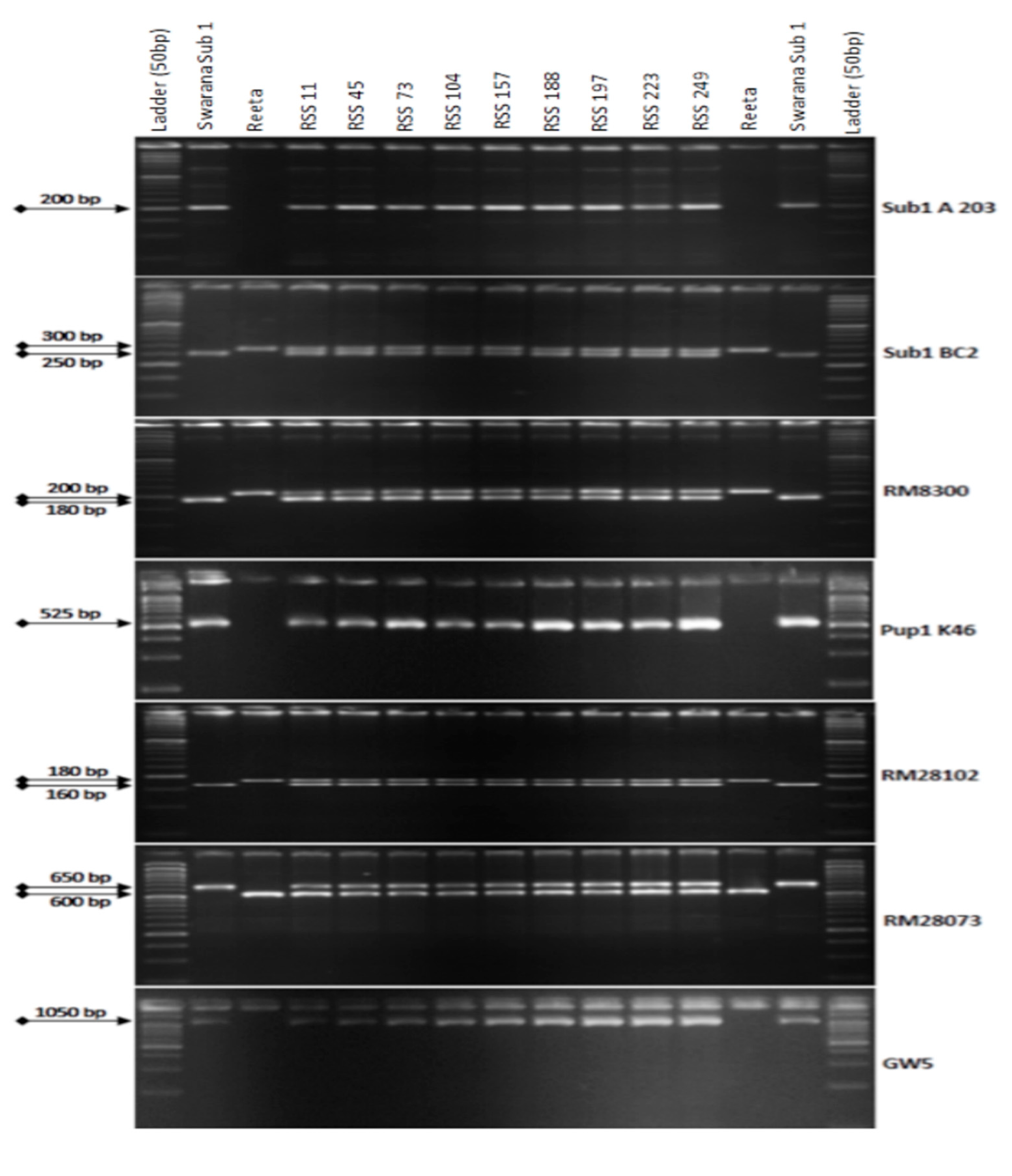
2.2. Marker-Assisted Selection in BC1F1 Progenies
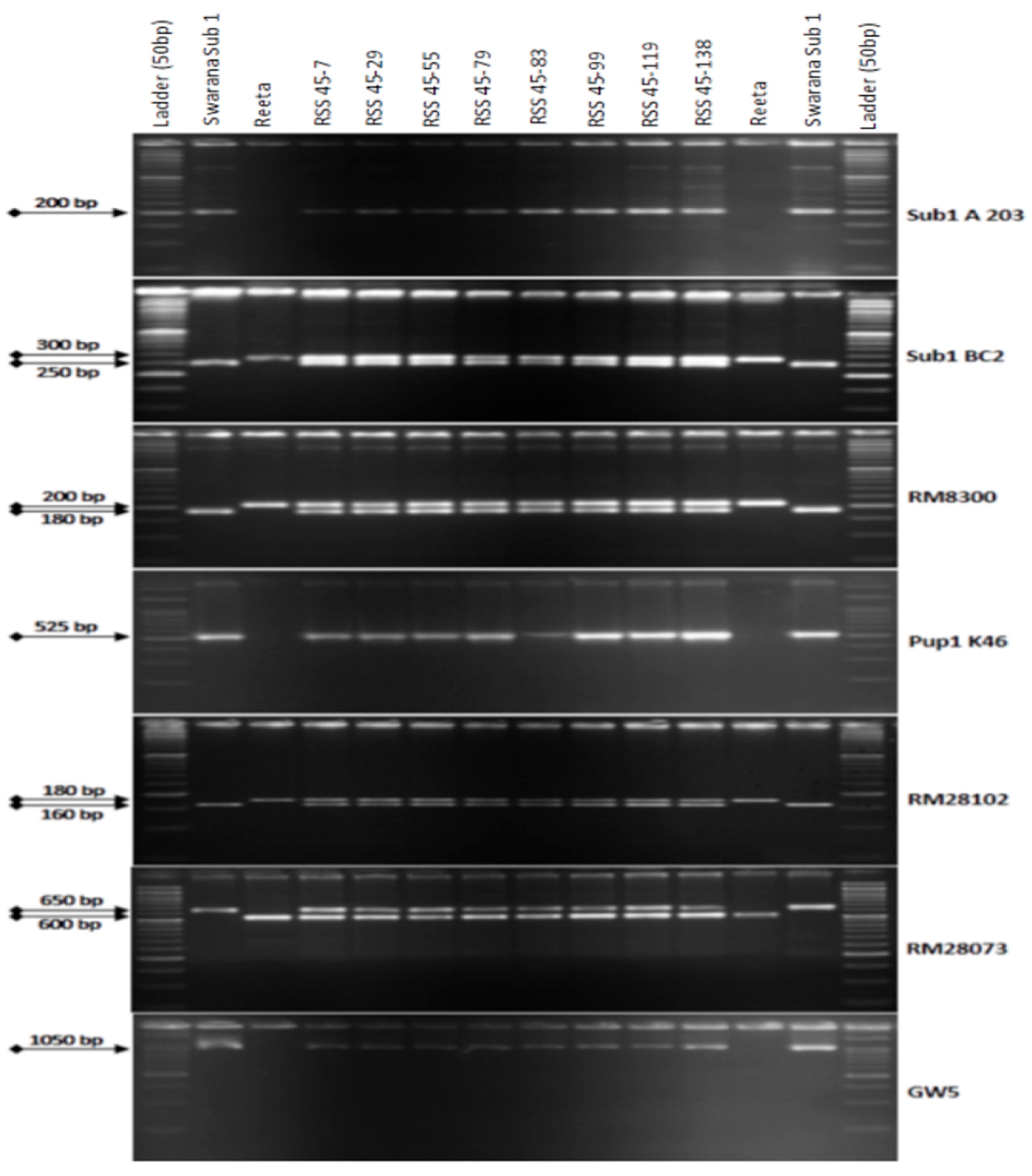
2.3. Marker-Assisted Selection in BC2F1 Generation
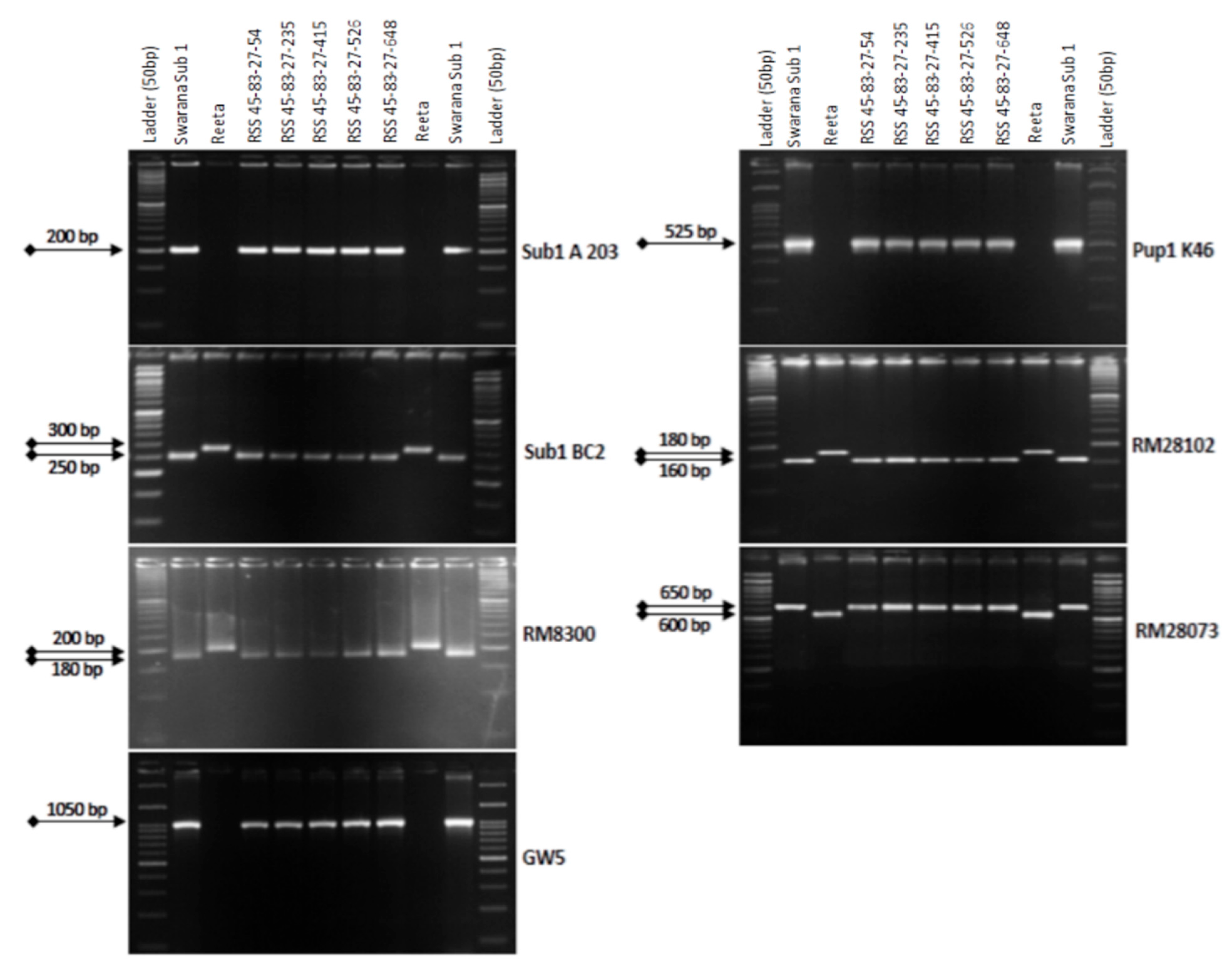
2.4. Marker-Assisted Selection in BC3F1 and BC3F2 Generations
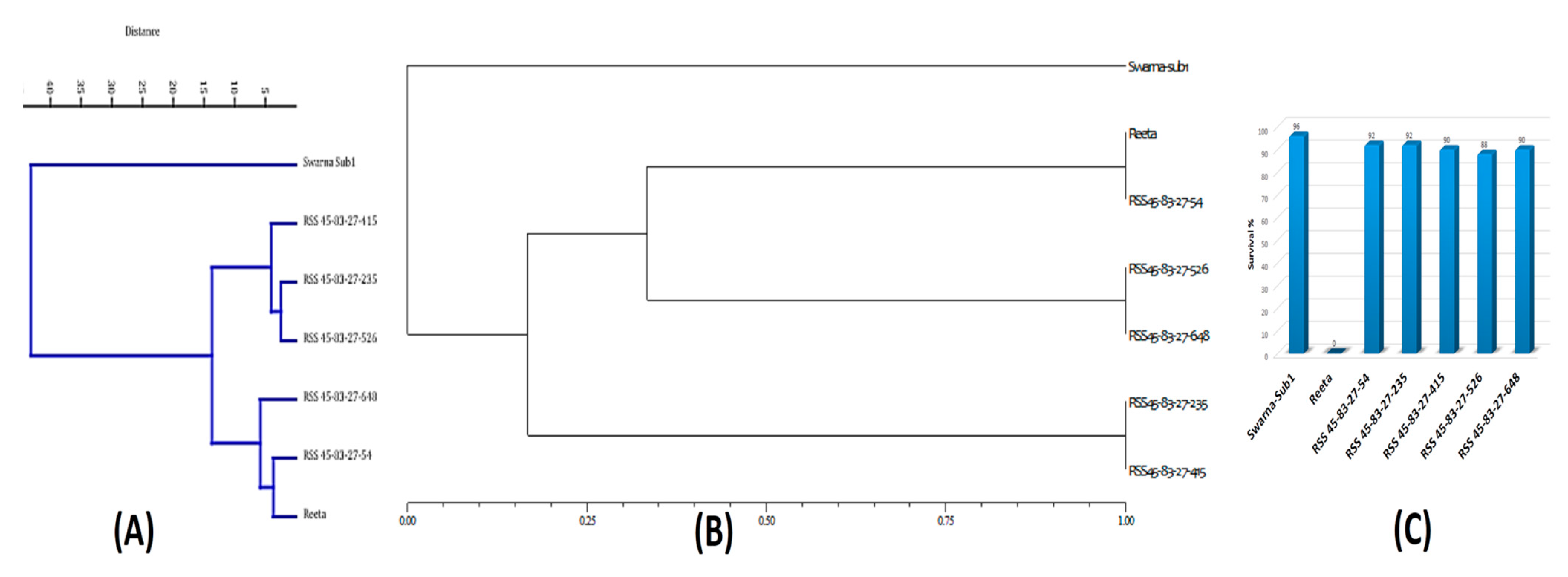
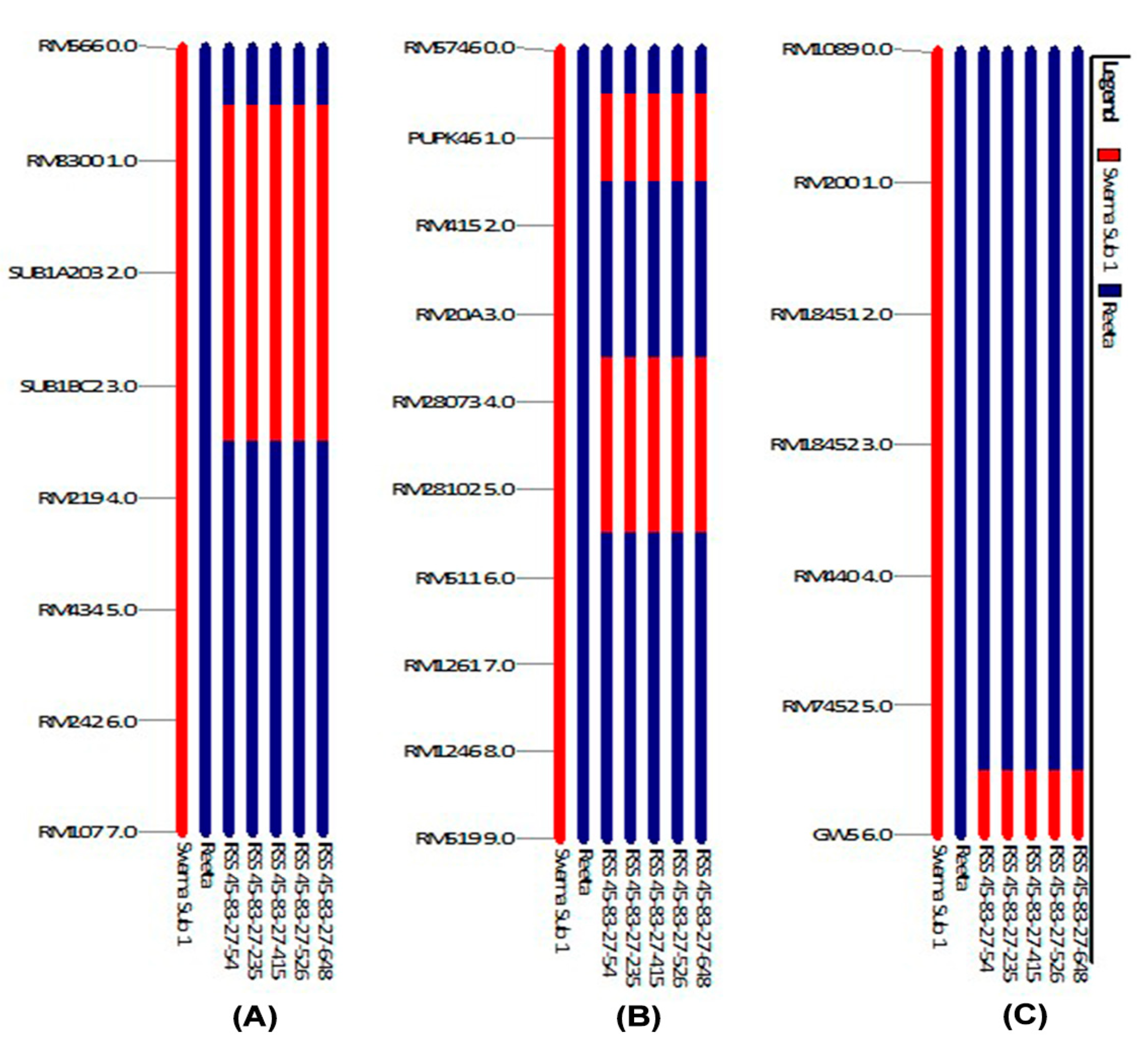
2.5. Analysis of Genome Introgression on the Carrier Chromosomes of the Pyramided Lines
2.6. Evaluation of the Pyramided Lines for Submergence Tolerance
2.7. Evaluation of the Pyramided Lines under Low Phosphorus Stress
2.8. Evaluation of the Pyramided Lines for Agro-Morphologic, Yield and Grain Quality Traits
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Breeding Program
4.2. Genomic DNA Isolation, Polymerase Chain Reaction, and Marker Analysis
4.3. Screening for Submergence Tolerance
4.4. Phenotyping for Phosphorus Uptake
4.5. Evaluation of the Pyramided Lines
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pradhan, S.K.; Pandit, E.; Pawar, S.; Bharati, B.; Chatopadhyay, K.; Singh, S.; Dash, P.; Reddy, J.N. Association mapping reveals multiple QTLs for grain protein content in rice useful for biofortification. Mol. Genet. Genom. 2019, 294, 963–983. [Google Scholar] [CrossRef] [PubMed]
- Sanghamitra, P.; Nanda, N.; Barik, S.R.; Sahoo, S.; Pandit, E.; Bastia, R.; Bagchi, T.B.; Pradhan, S.K. Genetic structure and molecular markers-trait association for physiological traits related to seed vigour in rice. Plant Gene 2021, 28, 100338. [Google Scholar] [CrossRef]
- Bastia, R.; Pandit, E.; Sanghamitra, P.; Barik, S.; Nayak, D.K.; Sahoo, A.; Moharana, A.; Meher, J.; Dash, P.K.; Raj, R. Association Mapping for Quantitative Trait Loci Controlling Superoxide Dismutase, Flavonoids, Anthocyanins, Carotenoids, γ-Oryzanol and Antioxidant Activity in Rice. Agronomy 2022, 12, 3036. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. Rice Market Monitor; Food and Agriculture Organization of the United Nations: Rome, Italy, 2017; Volume 20, pp. 1–38. [Google Scholar]
- Dar, M.H.; Waza, S.A.; Shukla, S.; Zaidi, N.W.; Nayak, S.; Hossain, M.; Singh, U.S. Drought tolerant rice for ensuring food security in Eastern India. Sustainability 2020, 12, 2214. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Barik, S.R.; Sahoo, J.; Pandit, E.; Nayak, D.K.; Pani, D.R.; Anandan, A. Comparison of Sub1 markers and their combinations for submergence tolerance and analysis of adaptation strategies of rice in rainfed lowland ecology. C. R. Biol. 2015, 338, 650–659. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Chakraborti, M.; Chakraborty, K.; Behera, L.; Meher, J.; Subudhi, H.N.; Mishra, S.K.; Pandit, E.; Reddy, J.N. Genetic Improvement of Rainfed Shallow-Lowland Rice for Higher Yield and Climate Resilience. In Rice Research for Enhancing Productivity, Profitability and Climate Resilience; Pathak, H., Nayak, A.K., Jena, M., Singh, O.N., Samal, P., Sharma, S.G., Eds.; ICAR-National Rice Research Institute: Cuttack, India, 2018; pp. 107–121. Available online: https://icar-nrri.in/wp-content/uploads/2019/02/Rice_Research_book_nrri.pdf (accessed on 7 February 2018).
- Mohapatra, S.; Panda, A.K.; Bastia, A.K.; Mukherjee, A.K.; Sanghamitra, P.; Meher, J.; Mohanty, S.P.; Pradhan, S.K. Development of Submergence-Tolerant, Bacterial Blight-Resistant, and High-Yielding Near Isogenic Lines of Popular Variety, ‘Swarna’ Through Marker-Assisted Breeding Approach. Front. Plant Sci. 2021, 12, 672618. [Google Scholar] [CrossRef]
- Pandit, E.; Pawar, S.; Barik, S.R.; Mohanty, S.P.; Meher, J.; Pradhan, S.K. Marker-Assisted Backcross Breeding for Improvement of Submergence Tolerance and Grain Yield in the Popular Rice Variety ‘Maudamani’. Agronomy 2021, 11, 1263. [Google Scholar] [CrossRef]
- Xu, K.; Mackill, D.J. A major locus for submergence tolerance mapped on rice chromosome 9. Mol. Breed. 1996, 2, 219–224. [Google Scholar] [CrossRef]
- Iftekharuddaula, K.M.; Newaz, M.A.; Salam, M.A.; Ahmed, H.U.; Mahbub, M.A.A.; Septiningsih, E.M.; Collard, B.C.Y.; Sanchez, D.L.; Pamplona, A.M.; Mackill, D.J. Rapid and high-precision marker assisted backcrossing to introgress the SUB1 QTL into BR11, the rainfed lowland rice mega variety of Bangladesh. Euphytica 2011, 178, 83–97. [Google Scholar] [CrossRef]
- Manivong, P.; Korinsak, S.; Korinsak, S.; Siangliw, J.L.; Vanavichit, A.; Toojinda, T. Marker-assisted selection to improve submergence tolerance, blast resistance and strong fragrance in glutinous rice. Genom. Genet. 2014, 7, 110–122. [Google Scholar]
- Khush, G.S.; Mackill, D.J.; Sidhu, G.S. Breeding Rice for Resistance to Bacterial Leaf Blight; IRRI: Manila, Philippines, 1989; pp. 207–217. [Google Scholar]
- Pradhan, S.K.; Barik, S.R.; Nayak, D.K.; Pradhan, A.; Pandit, E.; Nayak, P.; Das, S.R.; Pathak, H. Genetics, Molecular Mechanisms and Deployment of Bacterial Blight Resistance Genes in Rice. Crit. Rev. Plant Sci. 2020, 39, 360–385. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Pandit, E.; Barik, S.R.; Mohanty, S.P.; Nayak, D.K.; Sah, R.P.; Behera, L.; Sanghamitra, P.; Bose, L.K.; Das, S.R. Climate-Smart Rice Breeding: Progress and Challenges for the Rain-fed ecologies in India. In Advances in Rice Breeding: Stress Tolerance, Climate Resilience, Quality and High Yield; ICAR-NRRI: Cuttack, India, 2021; pp. 144–162. [Google Scholar]
- Das, S.R.; Collard, B.C.Y.; Pradhan, S.K. Breeding for climate resilient varieties suitable for rainfed rice ecologies. In Recent Innovations and Emerging Technologies for Transforming Rice Farming; ICAR-NRRI: Cuttack, India, 2023. [Google Scholar]
- Pradhan, S.K.; Pandit, E.; Pawar, S.; Baksh, S.Y.; Mukherjee, A.K.; Mohanty, S.P. Development of flash-flood tolerant and durable bacterial blight resistant versions of mega rice variety ‘Swarna’ through marker-assisted backcross breeding. Sci. Rep. 2019, 9, 12810. [Google Scholar] [CrossRef] [PubMed]
- Batjes, N.H. A world dataset of derived soil properties by FAO–UNESCO soil unit for global modelling. Soil Use Manag. 1997, 13, 9–16. [Google Scholar] [CrossRef]
- Motsara, M.R. Available nitrogen, phosphorus and potassium status of Indian soils as depicted by soil fertility maps. Ferilizer News 2002, 47, 15–21. [Google Scholar]
- Pandit, E.; Panda, R.K.; Pani, D.R.; Chandra, R.; Singh, S.; Pradhan, S.K. Molecular marker and phenotypic analyses for low phosphorus stress tolerance in cultivars and landraces of upland rice under irrigated and drought situations. Indian J. Genet. Plant Breed. 2018, 78, 59–68. [Google Scholar] [CrossRef]
- Pandit, E.; Sahoo, A.; Panda, R.K.; Mohanty, D.P.; Pani, D.R.; Anandan, A.; Pradhan, S.K. Survey of rice cultivars and landraces of upland ecology for phosphorus uptake 1 (Pup1) QTL using linked and gene specific molecular markers. Oryza 2016, 53, 1–9. [Google Scholar]
- Wissuwa, M.; Wegner, J.; Ae, N.; Yano, M. Substitution mapping of Pup1: A major QTL increasing phosphorus uptake of rice from a phosphorus deficient soil. Theor. Appl. Genet. 2002, 105, 890–897. [Google Scholar] [CrossRef]
- Gamuyao, R.; Chin, J.; Tanaka, J.; Pesaresi, P.; Catausan, S.; Dalid, C.; Loedin, I.; Mendoza, E.; Wissuwa, M.; Heuer, S. The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 2012, 488, 535–539. [Google Scholar] [CrossRef]
- Heuer, S.; Lu, X.; Chin, J.H.; Tanaka, J.P.; Kanamori, H.; Matsumoto, T.; De Leon, T.; Ulat, V.J.; Ismail, A.M.; Yano, M. Comparative sequence analyses of the major quantitative trait locus phosphorus uptake 1 (Pup1) reveal a complex genetic structure. Plant Biotechnol. J. 2009, 7, 456–457. [Google Scholar] [CrossRef]
- Chin, J.H.; Gamuyao, R.; Dalid, C.; Bustamam, M.; Prasetiyono, J.; Moeljopawiro, S.; Wissuwa, M.; Heuer, S. Developing Rice with High Yield under Phosphorus Deficiency: Pup1 Sequence to Application. Plant Physiol. 2011, 156, 1202–1216. [Google Scholar] [CrossRef]
- Weng, J.; Gu, S.; Wan, X.; Gao, H.; Guo, T.; Su, N.; Lei, C.; Zhang, X.; Cheng, Z.; Guo, X.; et al. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008, 18, 1199–1209. [Google Scholar] [CrossRef] [PubMed]
- Behera, L.; Sekhar, S.; Mohanty, S.; Devanna, B.N.; Parameswaran, C.; Pradhan, S.K. Genomics and other omics approaches for rice improvement. In Advances in Rice Breeding: Stress Tolerance, Climate Resilience, Quality and High Yield; ICAR-NRRI: Cuttack, India, 2021; pp. 369–426. [Google Scholar]
- Pradhan, S.K.; Nayak, D.K.; Mohanty, S.; Behera, L.; Barik, S.R.; Pandit, E.; Lenka, S. Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deepwater rice variety, Jalmagna. Rice 2015, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, K.C.; Pandit, E.; Mohanty, S.P.; Moharana, A.; Sanghamitra, P.; Meher, J.; Jena, B.K.; Dash, P.K.; Behera, L.; Mohapatra, P.M.; et al. Development of Broad Spectrum and Durable Bacterial Blight Resistant Variety through Pyramiding of Four Resistance Genes in Rice. Agronomy 2022, 12, 1903. [Google Scholar] [CrossRef]
- Sundaram, R.M.; Vishnupriya, M.R.; Biradar, S.K.; Laha, G.S.; Reddy, G.A.; Rani, N.S.; Sarma, N.P.; Sonti, R.V. Marker assisted introgression of bacterial blight resistance in Samba Mahsuri, an elite indica rice variety. Euphytica 2018, 160, 411–422. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Nayak, D.K.; Pandit, E.; Behera, L.; Anandan, A.; Mukherjee, A.K.; Lenka, S.; Barik, D.P. Incorporation of bacterial blight resistance genes into lowland rice cultivar through marker-assisted backcross breeding. Phytopathology 2016, 106, 710–718. [Google Scholar] [CrossRef]
- Singh, S.; Sidhu, J.S.; Huang, N.; Vikal, Y.; Li, Z.; Brar, D.S.; Dhaliwal, H.S.; Khush, G.S. Pyramiding three bacterial blight resistance genes(xa-5, xa-13 and Xa-21) using marker-assisted selection into indica rice cultivar PR-106. Theor. Appl. Genet. 2001, 102, 1011–1015. [Google Scholar] [CrossRef]
- Angeles-Shim, R.B.; Reyes, V.P.; del Valle, M.M.; Lapis, R.S.; Shim, J.; Sunohara, H.; Doi, K. Marker-assisted introgression of quantitative resistance gene pi21 confers broad spectrum resistance to rice blast. Rice Sci. 2020, 27, 113–123. [Google Scholar] [CrossRef]
- Nayak, D.K.; Pandit, E.; Mohanty, S.; Barik, D.P.; Pradhan, S.K. Marker-assisted selection in back cross progenies for transfer of bacterial leaf blight resistance genes into a popular lowland rice cultivar. Oryza 2015, 52, 163–172. [Google Scholar]
- Das, G.; Rao, G.J.N. Molecular marker assisted gene stacking for biotic and abiotic stress resistance genes in an elite rice cultivar. Front. Plant Sci. 2015, 6, 698. [Google Scholar] [CrossRef]
- Das, G.; Rao, G.J.N.; Varier, V.; Prakash, A.; Dokku, P. Improved Tapaswini having four BB resistance genes pyramided with six genes/QTLs, resistance/tolerance to biotic and abiotic stresses in rice. Sci. Rep. 2018, 8, 2413. [Google Scholar] [CrossRef]
- Pradhan, K.C.; Barik, S.R.; Mohapatra, S.; Nayak, D.K.; Pandit, E.; Jena, B.K.; Sangeeta, S.; Pradhan, A.; Samal, A.; Meher, J.; et al. Incorporation of Two Bacterial Blight Resistance Genes into the Popular Rice Variety, Ranidhan through Marker-Assisted Breeding. Agriculture 2022, 12, 1287. [Google Scholar] [CrossRef]
- Mohapatra, S.; Barik, S.R.; Dash, P.K.; Lenka, D.; Pradhan, K.C.; Reshmi Raj, K.R.; Mohanty, S.P.; Mohanty, M.R.; Sahoo, A.; Jena, B.K.; et al. Molecular Breeding for Incorporation of Submergence Tolerance and Durable Bacterial Blight Resistance into the Popular Rice Variety ‘Ranidhan’. Biomolecules 2023, 13, 198. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, S.K.; Nayak, D.K.; Pandit, E.; Barik, S.R.; Mohanty, S.P.; Anandan, A.; Reddy, J.N. Characterization of morpho-quality traits and validation of bacterial blight resistance in pyramided rice genotypes under various hot spots of India. Aust. J. Crop Sci. 2015, 9, 127–134. [Google Scholar]
- Behera, L.; Parameswaran, C.; Anandan, A.; Sanghamitra, P.; Pradhan, S.K.; Jena, M.; Umakanta, N.; Dash, S.K.; Swain, P.; Sahu, R.K.; et al. Development of genomic resources for rice improvement. In Rice Research for Enhancing Productivity, Profitability and Climate Resilience; Pathak, H., Nayak, A.K., Jena, M., Singh, O.N., Samal, P., Sharma, S.G., Eds.; ICAR-National Rice Research Institute: Cuttack, India, 2018; pp. 197–223. Available online: https://icar-nrri.in/wp-content/uploads/2019/02/Rice_Research_book_nrri.pdf (accessed on 24 March 2023).
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA mini preparation: Version II. Plant Mol. Biol. Report. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Barik, S.R.; Pandit, E.; Mohanty, S.P.; Nayak, D.K.; Pradhan, S.K. Genetic mapping of physiological traits associated with terminal stage drought tolerance in rice. BMC Genet. 2020, 21, 76. [Google Scholar] [CrossRef]
- Pandit, E.; Panda, R.K.; Sahoo, A.; Pani, D.R.; Pradhan, S.K. Genetic relationship and structure analysis of root growth angle for improvement of drought avoidance in early and mid-early maturing rice genotypes. Rice Sci. 2020, 27, 124–132. [Google Scholar] [CrossRef]
- Sanghamitra, P.; Barik, S.R.; Bastia, R.; Mohanty, S.P.; Pandit, E.; Behera, A.; Mishra, J.; Kumar, G.; Pradhan, S.K. Detection of Genomic Regions Controlling the Antioxidant Enzymes, Phenolic Content, and Antioxidant Activities in Rice Grain through Association Mapping. Plants 2022, 11, 1463. [Google Scholar] [CrossRef]
- Barik, S.R.; Pandit, E.; Pradhan, S.K.; Singh, S.; Swain, P.; Mohapatra, T. QTL mapping for relative water content trait at reproductive stage drought stress in rice. Indian J. Genet. 2018, 78, 401–408. [Google Scholar]
- Pawar, S.; Pandit, E.; Mohanty, I.C.; Saha, D.; Pradhan, S.K. Population genetic structure and association mapping for iron toxicity tolerance in rice. PLoS ONE 2021, 16, e0246232. [Google Scholar] [CrossRef]
- Van Berloo, R. GGT: Software for display of graphical genotypes. J. Hered. 1999, 90, 328–330. [Google Scholar] [CrossRef]
- Cagampang, G.B.; Perez, C.M.; Juliano, B.O. A gel consistency test for eating quality in rice. J. Sci. Food Agric. 1973, 24, 1589–1594. [Google Scholar] [CrossRef] [PubMed]
- Juliano, B.O. Rice quality screening with the Rapid ViscoAnalyser. Appl. Rapid Visco Anal. 1996, 19–24. [Google Scholar]
- SAS Institute Inc. SAS® 9.2 Language Reference: Concepts, 2nd ed.; SAS Institute Inc.: Cary, NC, USA, 2010; pp. 626–978. [Google Scholar]
- Pandit, E.; Tasleem, S.; Nayak, D.K.; Barik, S.R.; Mohanty, D.P.; Das, S.; Pradhan, S.K. Genome-wide association mapping reveals multiple QTLs governing tolerance response for seedling stage chilling stress in indica rice. Front. Plant Sci. 2017, 8, 552. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, S.K.; Pandit, E.; Pawar, S.; Naveenkumar, R.; Barik, S.R.; Mohanty, S.P.; Nayak, D.K.; Ghritlahre, S.K.; Rao, D.S.; Reddy, J.N.; et al. Linkage disequilibrium mapping for grain Fe and Zn enhancing QTLs useful for nutrient dense rice breeding. BMC Plant Biol. 2020, 20, 57. [Google Scholar] [CrossRef] [PubMed]
- Nayak, D.K.; Sahoo, S.; Barik, S.R.; Sanghamitra, P.; Sangeeta, S.; Pandit, E.; Reshmi Raj, K.R.; Basak, N.; Pradhan, S.K. Association mapping for protein, total soluble sugars, starch, amylose and chlorophyll content in rice. BMC Plant Biol. 2022, 22, 620. [Google Scholar] [CrossRef]
- Mohanty, S.P.; Nayak, D.K.; Sanghamitra, P.; Barik, S.R.; Pandit, E.; Behera, A.; Pani, D.R.; Mohapatra, S.; Reshmi Raj, K.R.; Pradhan, K.C.; et al. Mapping the Genomic Regions Controlling Germination Rate and Early Seedling Growth Parameters in Rice. Genes 2023, 14, 902. [Google Scholar] [CrossRef]




| Generation | No. of Plants Scored | No. of Progeny Heterozygotes for the 3 Target QTL | Expected % of Recurrent Parent Genome to Selected Backcross Plants | Average Recipient Parent Genome Content (%) in the Backcross Progenies | Maximum Genome Recovery (%) of Recipient Parent in the Selected Progenies | Chi-Square Value (χ2) |
|---|---|---|---|---|---|---|
| BC1F1 | 138 | 9 | 75.0 | 76.91 | 80.95 | 0.049 |
| BC2F1 | 135 | 8 | 87.5 | 89.06 | 92.26 | 0.028 |
| BC3F1 | 156 | 7 | 93.25 | 94.30 | 95.24 | 0.012 |
| Scheme | Pyramided and Parental Lines | PUptake (mg/g Tissue Dry Weight) | Grain Yield (Average Single Plant in Gram) | ||
|---|---|---|---|---|---|
| Normal | Deficient | Normal | Deficient | ||
| 1 | RSS 45-83-27-54 | 0.224 | 0.205 | 25.67 | 22.54 |
| 2 | RSS 45-83-27-235 | 0.218 | 0.212 | 24.25 | 21.55 |
| 3 | RSS 45-83-27-415 | 0.208 | 0.194 | 24.28 | 22.16 |
| 4 | RSS 45-83-27-526 | 0.232 | 0.216 | 22.56 | 20.68 |
| 5 | RSS 45-83-27-648 | 0.228 | 0.202 | 22.32 | 20.16 |
| 6 | Reeta (–ve check) | 0.156 | 0.138 | 21.56 | 15.24 |
| 7 | Swarna-Sub1 (+ve check) | 0.238 | 0.216 | 22.21 | 20.14 |
| CV% | 9.43 | 10.25 | 8.26 | 9.14 | |
| CD5% | 0.042 | 0.048 | 2.182 | 2.534 | |
| Sl. No. | Pyramided and Parental Lines | PH | DFF | NP | GN | PL | SW | SF | NPB | PW | GL | GB | H | M | HRR | AC | ASV | GC | PY |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | RSS 45-83-27-54 | 117 | 120 | 12 | 218 | 28.3 | 21.7 | 83.5 | 14.3 | 5.64 | 5.12 | 2.28 | 77 | 68.5 | 64.3 | 20.7 | 5.0 | 48 | 6.75 |
| 2 | RSS 45-83-27-235 | 115 | 119 | 11 | 212 | 28.1 | 21.4 | 83.2 | 14.1 | 5.21 | 5.18 | 2.30 | 76 | 67.8 | 65.1 | 21.2 | 5.0 | 47 | 6.65 |
| 3 | RSS 45-83-27-415 | 116 | 120 | 10 | 215 | 27.8 | 21.8 | 82.6 | 13.4 | 5.25 | 5.03 | 2.27 | 75 | 69.4 | 64.5 | 21.4 | 4.5 | 48 | 6.48 |
| 4 | RSS 45-83-27-526 | 115 | 118 | 10 | 208 | 27.5 | 21.5 | 82.3 | 13.6 | 5.56 | 5.04 | 2.28 | 76 | 66.5 | 63.6 | 21.3 | 4.0 | 49 | 6.40 |
| 5 | RSS 45-83-27-648 | 117 | 120 | 9 | 210 | 27.4 | 21.6 | 82.7 | 13.7 | 5.08 | 5.08 | 2.28 | 78 | 67.2 | 63.8 | 21.5 | 5.0 | 51 | 6.25 |
| 6 | Swarna-Sub1 | 110 | 118 | 13.25 | 164 | 26.2 | 20.32 | 83.8 | 14.2 | 5.16 | 5.175 | 2.22 | 77 | 68.45 | 61.3 | 24.25 | 4.0 | 55.5 | 6.15 |
| 7 | Reeta (Recipient) | 116 | 120 | 9.25 | 191 | 28.4 | 24.58 | 77.6 | 12.4 | 4.28 | 4.97 | 2.32 | 79 | 70.9 | 65.3 | 20.5 | 5.0 | 47 | 4.38 |
| LSD5% | 8.74 | 4.35 | 2.94 | 18.3 | 1.72 | 1.07 | 0.515 | 0.71 | 1.86 | 0.13 | 6.58 | 6.85 | 7.64 | 1.883 | 2.45 | 0.275 | |||
| CV% | 3.08 | 0.85 | 10.24 | 9.82 | 5.85 | 7.82 | 10.13 | 7.24 | 3.26 | 7.68 | 5.24 | 6.76 | 8.21 | 6.731 | 4.35 | 10.72 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barik, S.R.; Moharana, A.; Pandit, E.; Behera, A.; Mishra, A.; Mohanty, S.P.; Mohapatra, S.; Sanghamitra, P.; Meher, J.; Pani, D.R.; et al. Transfer of Stress Resilient QTLs and Panicle Traits into the Rice Variety, Reeta through Classical and Marker-Assisted Breeding Approaches. Int. J. Mol. Sci. 2023, 24, 10708. https://doi.org/10.3390/ijms241310708
Barik SR, Moharana A, Pandit E, Behera A, Mishra A, Mohanty SP, Mohapatra S, Sanghamitra P, Meher J, Pani DR, et al. Transfer of Stress Resilient QTLs and Panicle Traits into the Rice Variety, Reeta through Classical and Marker-Assisted Breeding Approaches. International Journal of Molecular Sciences. 2023; 24(13):10708. https://doi.org/10.3390/ijms241310708
Chicago/Turabian StyleBarik, Saumya Ranjan, Arpita Moharana, Elssa Pandit, Abhisarika Behera, Ankita Mishra, Shakti Prakash Mohanty, Shibani Mohapatra, Priyadarsini Sanghamitra, Jitendriya Meher, Dipti Ranjan Pani, and et al. 2023. "Transfer of Stress Resilient QTLs and Panicle Traits into the Rice Variety, Reeta through Classical and Marker-Assisted Breeding Approaches" International Journal of Molecular Sciences 24, no. 13: 10708. https://doi.org/10.3390/ijms241310708
APA StyleBarik, S. R., Moharana, A., Pandit, E., Behera, A., Mishra, A., Mohanty, S. P., Mohapatra, S., Sanghamitra, P., Meher, J., Pani, D. R., Bhadana, V. P., Datt, S., Sahoo, C. R., Raj K. R., R., & Pradhan, S. K. (2023). Transfer of Stress Resilient QTLs and Panicle Traits into the Rice Variety, Reeta through Classical and Marker-Assisted Breeding Approaches. International Journal of Molecular Sciences, 24(13), 10708. https://doi.org/10.3390/ijms241310708






