Therapeutic Targeting of the GSK3β-CUGBP1 Pathway in Myotonic Dystrophy
Abstract
1. Introduction
2. Results
2.1. RNA-Seq Approach Revealed That TG Partially Corrects Biological Processes in HSALR Muscle Associated with Cell Transport, Development and Differentiation
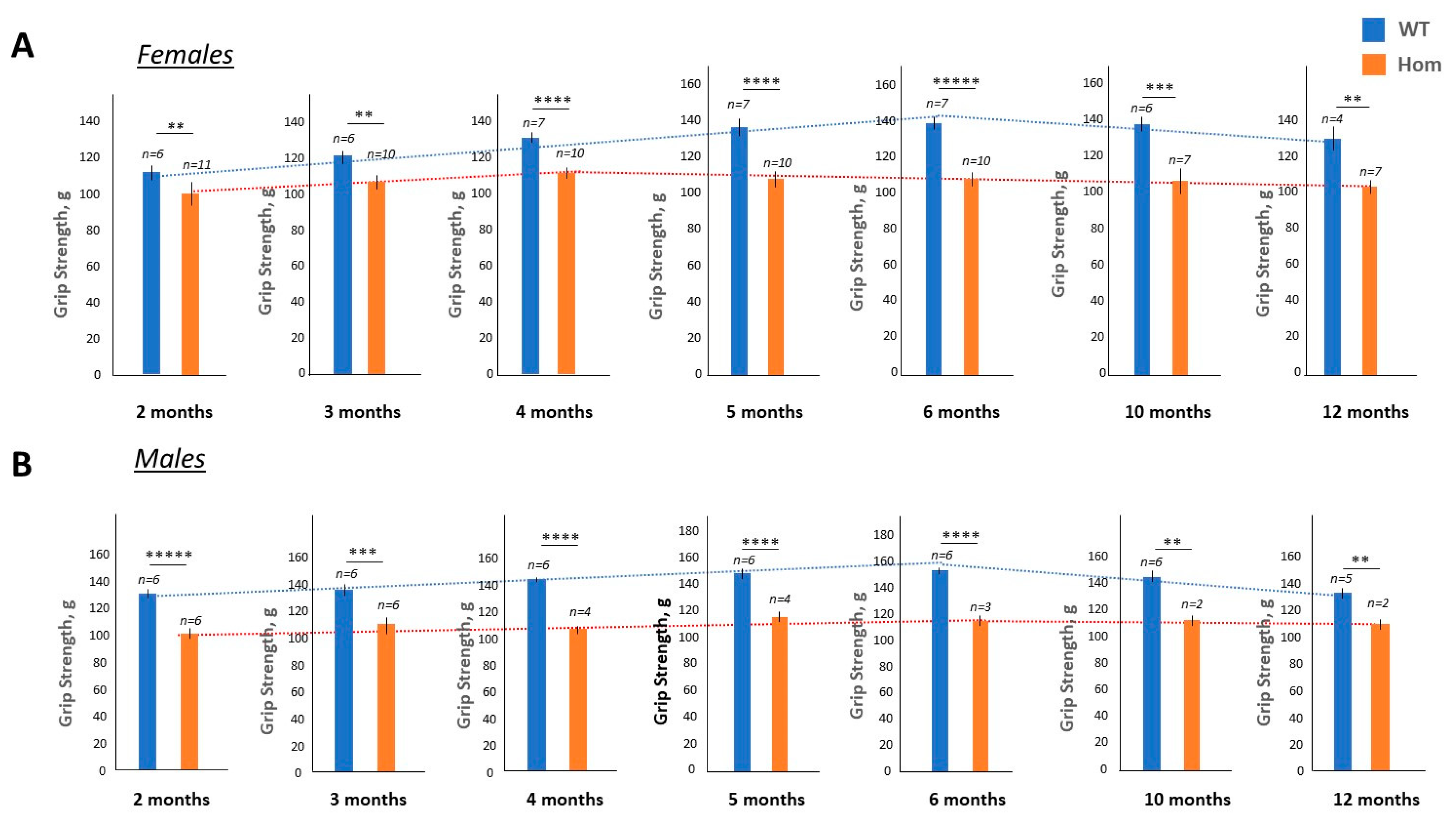
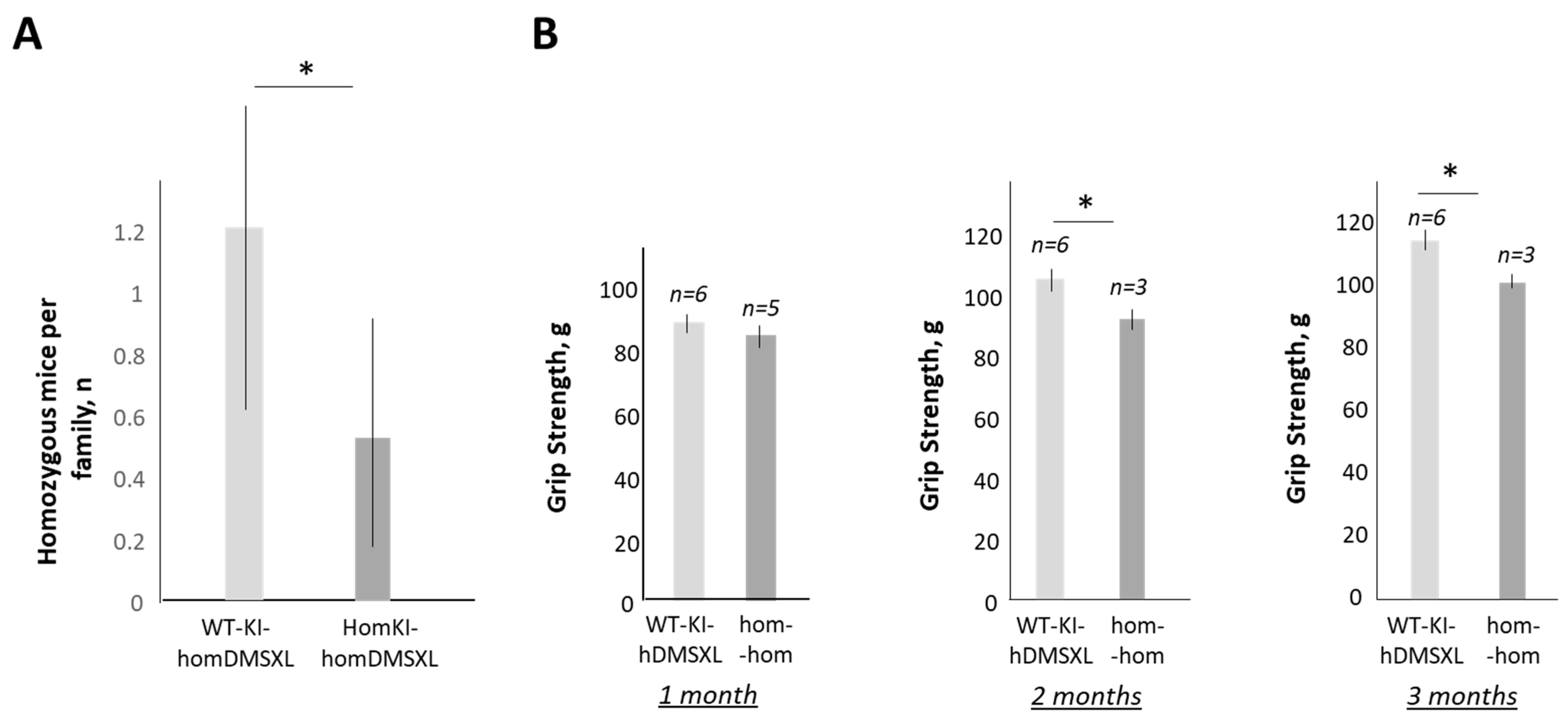
2.2. Grip weakness Is the Main Feature of Surviving Homozygous DMSXL Mice up to 1 Year of Age
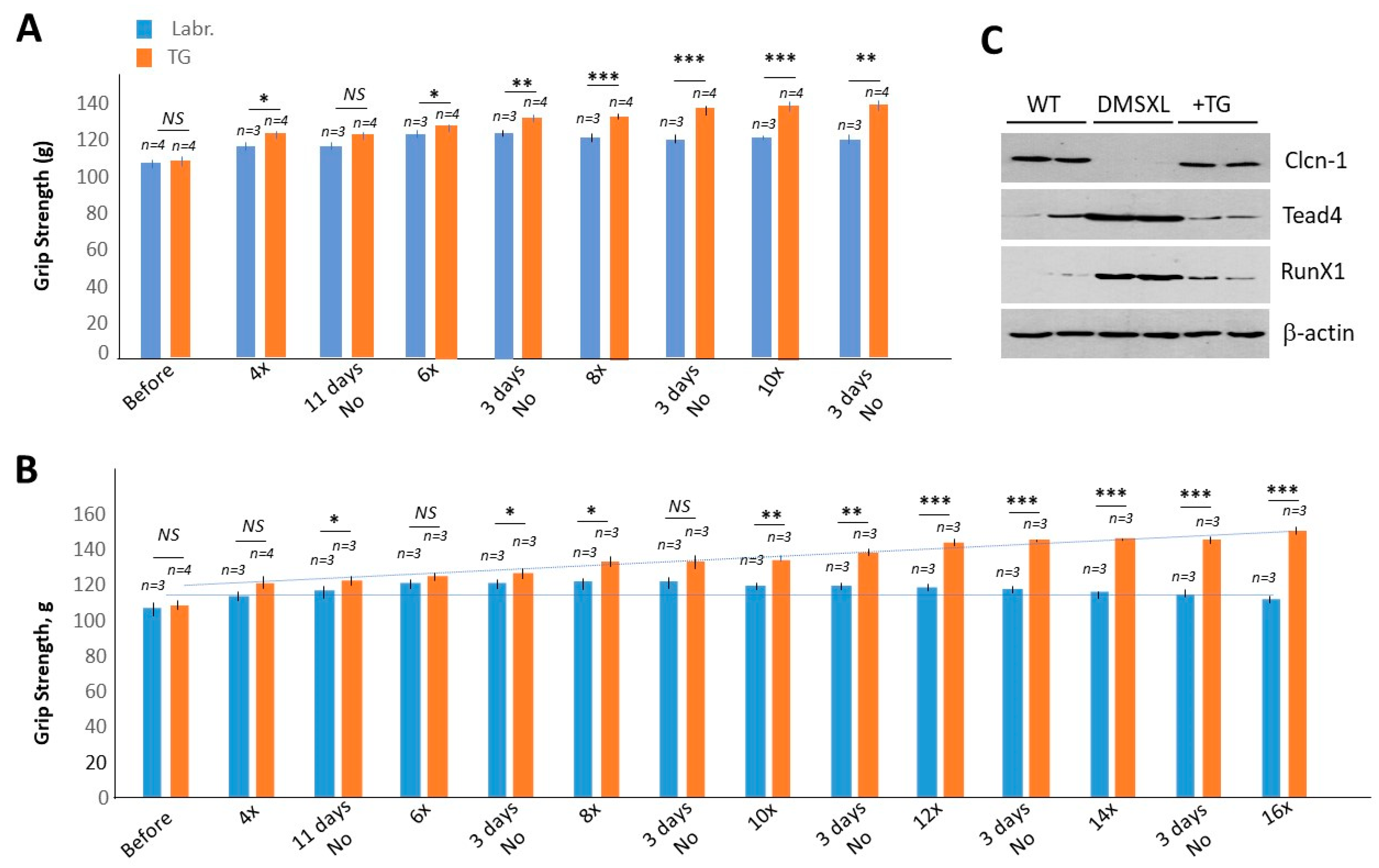
2.3. Chronic Treatments with TG Restore Muscle Strength in Adult Homozygous DMSXL Mice
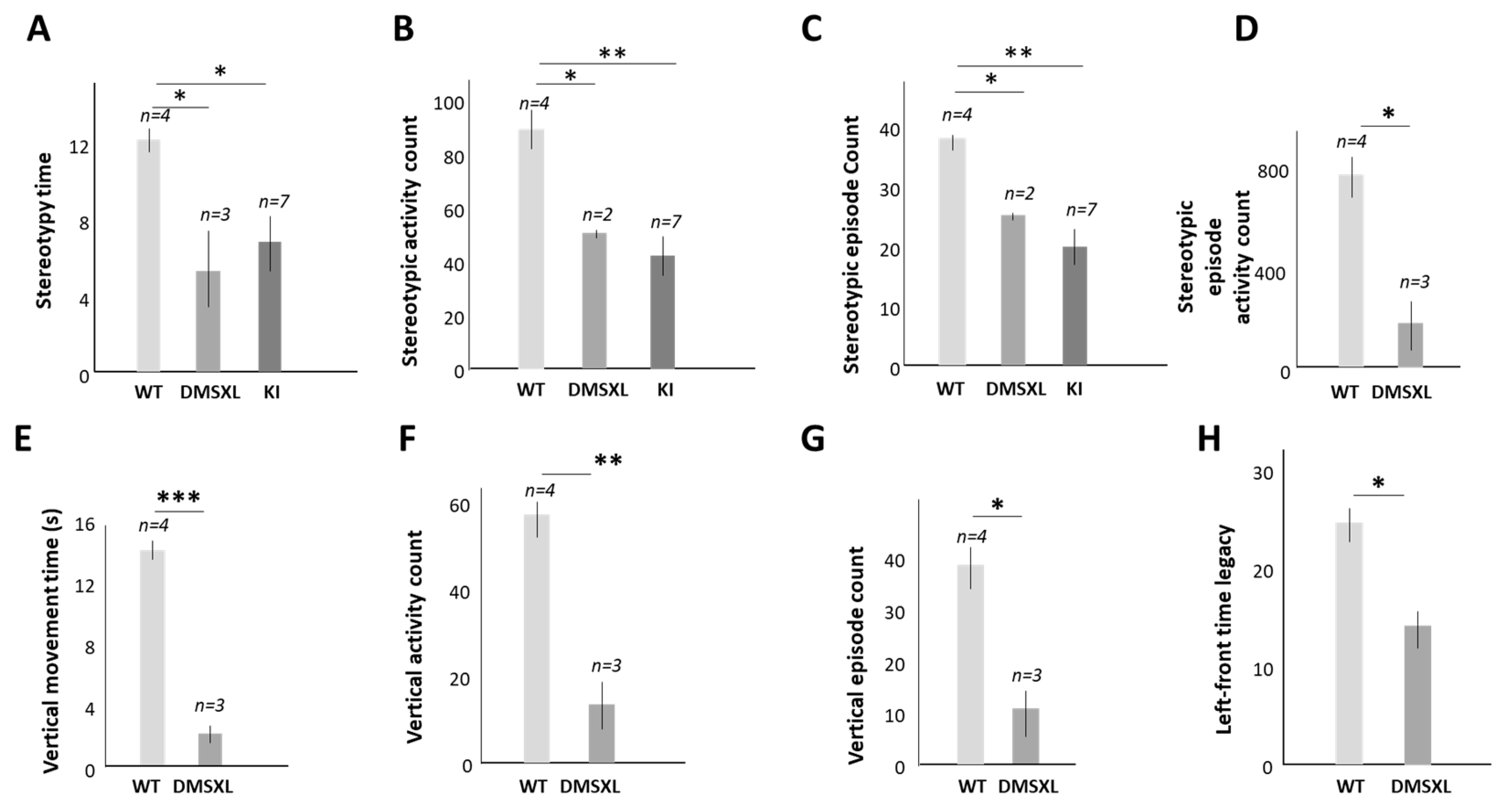
2.4. The Contribution of the Abnormal Activity of CUGBP1 to Cognitive Dysfunction in DM1
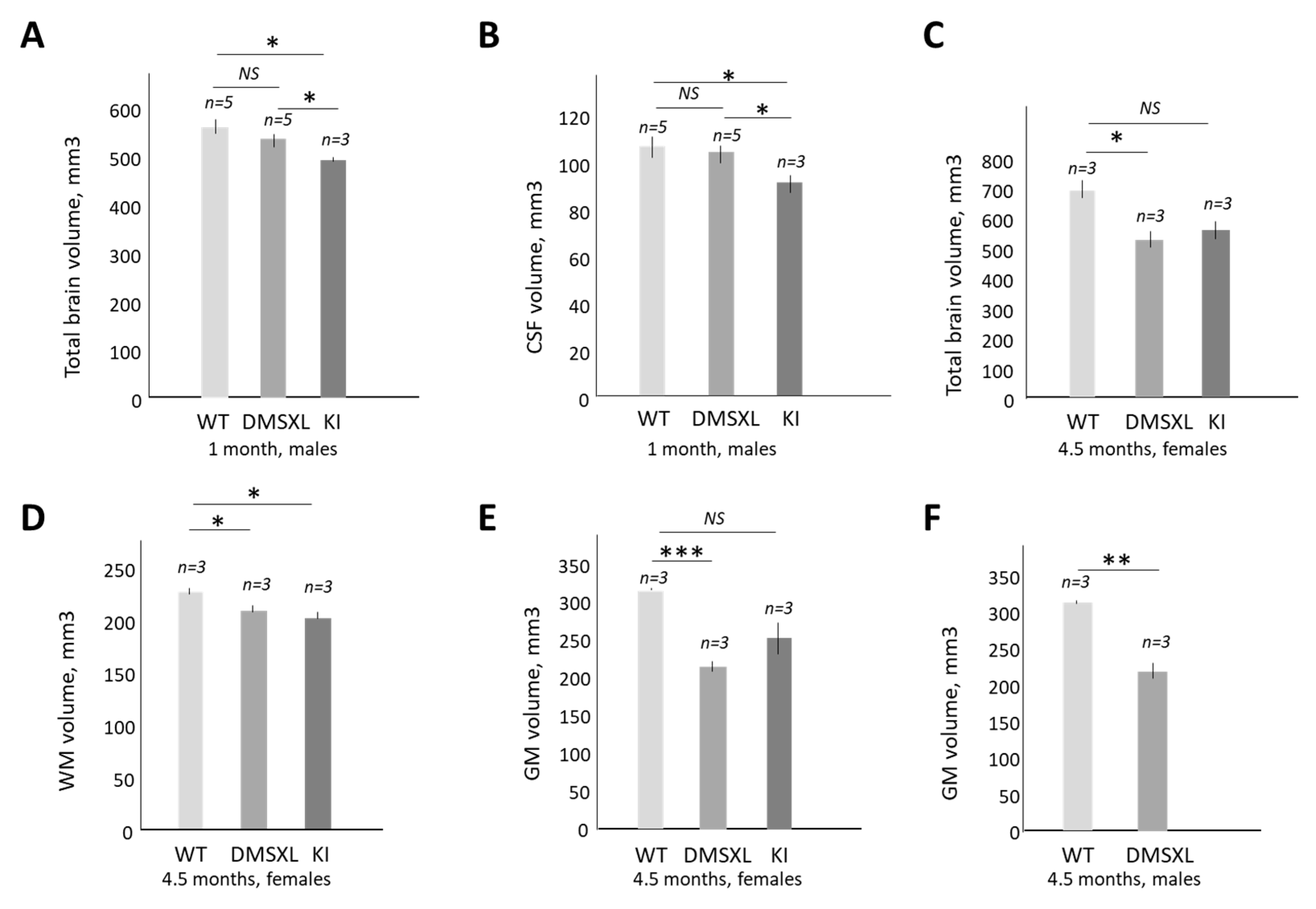
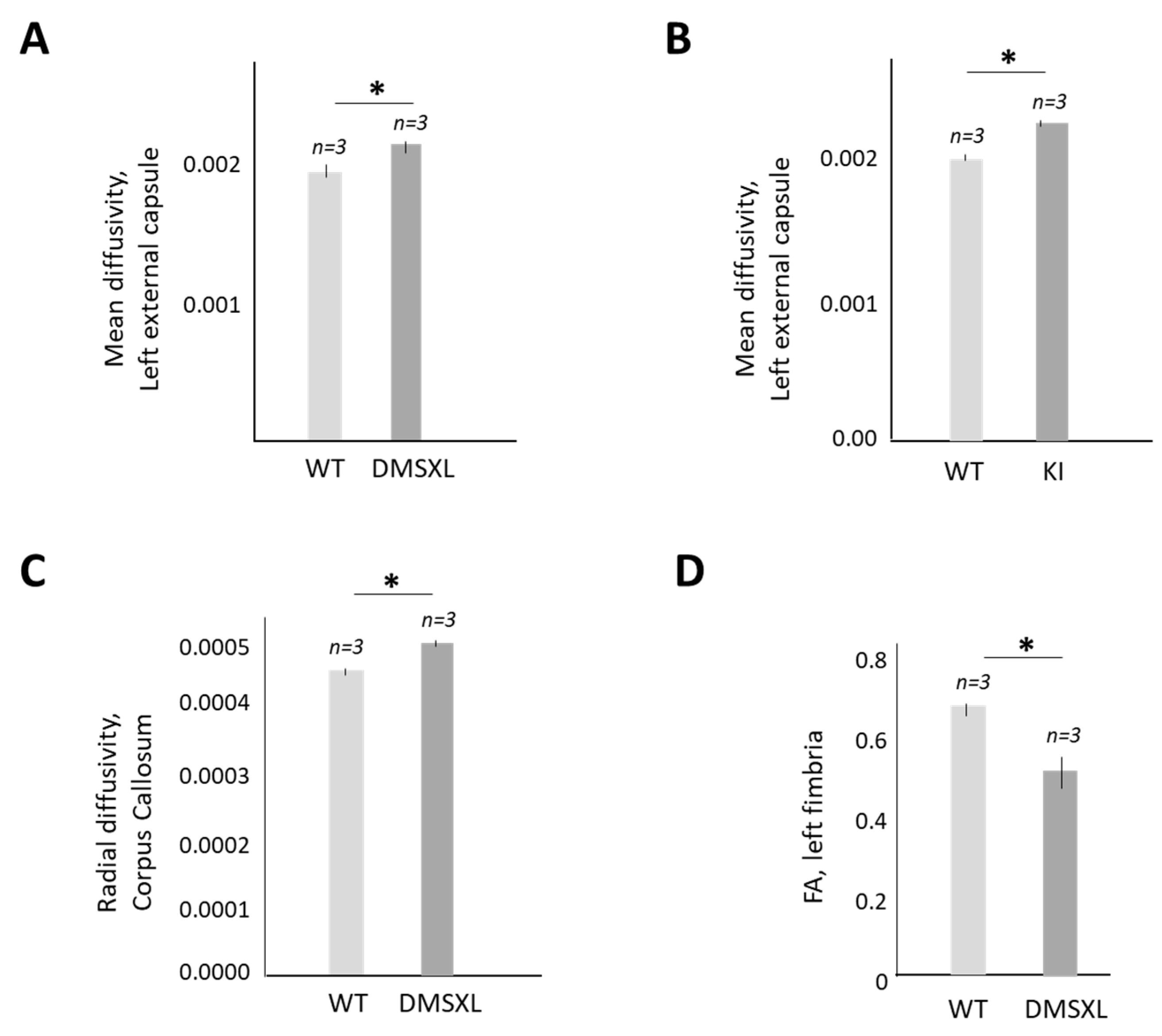
2.5. Magnetic Resonance (MRI) and Diffusion Tensor Imaging (DTI) Studies of CUGBP1 KI and DMSXL Mouse Brains
2.6. Increase in CUGBP1REP in DMSXL Mice Further Worsens the Phenotype of DMSXL Mice
3. Discussion
4. Materials and Methods
4.1. Mice and Treatments
4.2. RNAseq Analysis
4.3. Western Blot Analysis
4.4. Open Field Test (OFT)
4.5. Grip Strength Analysis
4.6. Magnetic Resonance Imaging and Diffusion Tensor Imaging
4.7. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Harper, P.S. Myotonic Dystrophy; WB Saunders: London, UK, 2001. [Google Scholar]
- Schoser, B.; Timchenko, L. Myotonic Dystrophies 1 and 2: Complex Diseases with Complex Mechanisms. Curr. Genom. 2010, 11, 79–90. [Google Scholar] [CrossRef]
- Thornton, C.A. Myotonic Dystrophy. Neurol. Clin. 2014, 32, 705–719. [Google Scholar]
- van Agtmaal, E.L.; André, L.M.; Willemse, M.; Cumming, S.A.; Kessel, I.D.G.; van den Broek, W.J.A.A.; Gourdon, G.; Furling, D.; Mouly, V.; Monckton, D.G.; et al. CRISPR/Cas9-Induced (CTG⋅CAG)n Repeat Instability in the Myotonic Dystrophy Type 1 Locus: Implications for Therapeutic Genome Editing. Mol. Ther. 2017, 25, 24–43. [Google Scholar]
- Dastidar, S.; Ardui, S.; Singh, K.; Majumdar, D.; Nair, N.; Fu, Y.; Reyon, D.; Samara, E.; Gerli, M.F.M.; Klein, A.F.; et al. Efficient CRISPR/Cas9-mediated editing of trinucleotide repeat expansion in myotonic dystrophy patient-derived iPS and myogenic cells. Nucl. Acids Res. 2018, 46, 8275–8298. [Google Scholar] [PubMed]
- Lo Scrudato, M.; Poulard, K.; Sourd, C.; Tomé, S.; Klein, A.F.; Corre, G.; Huguet, A.; Furling, D.; Gourdon, G.; Buj-Bello, A. Genome Editing of Expanded CTG Repeats within the Human DMPK Gene Reduces Nuclear RNA Foci in the Muscle of DM1 Mice. Mol. Ther. 2019, 27, 1372–1388. [Google Scholar]
- Wang, Y.; Hao, L.; Wang, H.; Santostefano, K.; Thapa, A.; Cleary, J.; Li, H.; Guo, X.; Terada, N.; Ashizawa, T.; et al. Therapeutic Genome Editing for Myotonic Dystrophy Type 1 Using CRISPR/Cas9. Mol. Ther. 2018, 26, 2617–2630. [Google Scholar] [CrossRef]
- Raaijmakers, R.H.L.; Ripken, L.; Ausems, C.R.M.; Wansink, D.G. CRISPR/Cas Applications in Myotonic Dystrophy: Expanding Opportunities. Int. J. Mol. Sci. 2019, 20, 3689–3705. [Google Scholar]
- Mulders, S.A.; van den Broek, W.J.; Wheeler, T.M.; Croes, H.J.; van Kuik-Romeijn, P.; de Kimpe, S.J.; Furling, D.; Platenburg, G.J.; Gourdon, G.; Thornton, C.A.; et al. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc. Natl. Acad. Sci. USA 2009, 106, 13915–13920. [Google Scholar] [PubMed]
- Wheeler, T.M.; Leger, A.J.; Pandey, S.K.; MacLeod, A.R.; Nakamori, M.; Cheng, S.H.; Wentworth, B.M.; Bennett, C.F.; Thornton, C.A. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature 2012, 488, 111–115. [Google Scholar] [PubMed]
- Benichou, A.S.; Jauvi, D.; De-Serres-Berard, T.; Bennett, F.; Rigo, F.; Gourdon, G.; Boutjdir, M.; Chahine, M.; Puymirat, J. Enhanced Delivery of Ligand-Conjugated Antisense Oligonucleotides (C16-HA-ASO) Targeting DMPK Transcripts for the Treatment of Myotonic Dystrophy Type 1. Hum. Gene Ther. 2022, 33, 810–820. [Google Scholar] [CrossRef]
- Benichou, A.S.; Jauvin, D.; De Serres-Bérard, T.; Pierre, M.; Ling, K.K.; Bennett, C.F.; Rigo, F.; Gourdon, G.; Chahine, M.; Puymirat, J. Antisense oligonucleotides as a potential treatment for brain deficits observed in myotonic dystrophy type 1. Gene Ther. 2022, 29, 698–709. [Google Scholar] [CrossRef]
- López-Morató, M.; Brook, J.D.; Wojciechowska, M. Small Molecules Which Improve Pathogenesis of Myotonic Dystrophy Type 1. Front Neurol. 2018, 9, 349. [Google Scholar] [CrossRef] [PubMed]
- Castel, A.L.; Overby, S.J.; Artero, R. MicroRNA-Based Therapeutic Perspectives in Myotonic Dystrophy. Int. J. Mol. Sci. 2019, 20, 5600–5619. [Google Scholar]
- Timchenko, L. Development of Therapeutic Approaches for Myotonic Dystrophies Type 1 and Type 2. Int. J. Mol. Sci. 2022, 23, 10491. [Google Scholar] [CrossRef]
- Thornton, C.A.; Moxley, R.T., 3rd; Eichinger, K.; Heatwole, C.; Mignon, L.; Arnold, W.D.; Ashizawa, T.; Day, J.W.; Dent, G.; Tanner, M.K.; et al. Antisense oligonucleotide targeting DMPK in patients with myotonic dystrophy type 1: A multicentre, randomised, dose-escalation, placebo-controlled, phase 1/2a trial. Lancet Neurol. 2023, 22, 218–228. [Google Scholar] [CrossRef]
- Huichalaf, C.; Sakai, K.; Jin, B.; Jones, K.; Wang, G.L.; Schoser, B.; Schneider-Gold, C.; Sarkar, P.; Pereira-Smith, O.M.; Timchenko, N.; et al. Expansion of CUG RNA repeats causes stress and inhibition of translation in myotonic dystrophy 1 (DM1) cells. FASEB J. 2010, 24, 3706–3719. [Google Scholar] [CrossRef]
- Salisbury, E.; Sakai, K.; Schoser, B.; Huichalaf, C.; Schneider-Gold, C.; Nguyen, H.; Wang, G.L.; Albrecht, J.H.; Timchenko, L.T. Ectopic expression of cyclin D3 corrects differentiation of DM1 myoblasts through activation of RNA CUG-binding protein, CUGBP1. Exp. Cell Res. 2008, 314, 2266–2278. [Google Scholar] [CrossRef]
- Timchenko, L. Correction of RNA-Binding Protein CUGBP1 and GSK3β Signaling as Therapeutic Approach for Congenital and Adult Myotonic Dystrophy Type 1. Int. J. Mol. Sci. 2019, 21, 94. [Google Scholar] [CrossRef]
- Jones, K.; Wei, C.; Iakova, P.; Bugiardini, E.; Schneider-Gold, C.; Meola, G.; Woodgett, J.; Killian, J.; Timchenko, N.A.; Timchenko, L.T. GSK3β mediates muscle pathology in myotonic dystrophy. J. Clin. Investig. 2012, 122, 4461–4472. [Google Scholar] [CrossRef]
- Wei, C.; Stock, L.; Valanejad, L.; Zalewski, Z.A.; Karns, R.; Puymirat, J.; Nelson, D.; Witte, D.; Woodgett, J.; Timchenko, N.A.; et al. Correction of GSK3β at young age prevents muscle pathology in mice with myotonic dystrophy type 1. FASEB J. 2018, 32, 2073–2085. [Google Scholar]
- Wang, M.; Weng, W.C.; Stock, L.; Lindquist, D.; Martinez, A.; Gourdon, G.; Timchenko, N.; Snape, M.; Timchenko, L. Correction of Glycogen Synthase Kinase 3β in Myotonic Dystrophy 1 Reduces the Mutant RNA and Improves Postnatal Survival of DMSXL Mice. Mol. Cell. Biol. 2019, 39, e00155-19. [Google Scholar] [CrossRef]
- Horrigan, J.; Gomes, T.B.; Snape, M.; Nikolenko, N.; McMorn, A.; Evans, S.; Yaroshinsky, A.; Della Pasqua, O.; Oosterholt, S.; Lochmüller, H. A Phase 2 Study of AMO-02 (Tideglusib) in Congenital and Childhood-Onset Myotonic Dystrophy Type 1 (DM1). Pediatr. Neurol. 2020, 112, 84–93. [Google Scholar]
- Mankodi, A.; Logigian, E.; Callahan, L.; McClain, C.; White, R.; Henderson, D.; Krym, M.; Thornton, C.A. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science 2000, 289, 1769–1772. [Google Scholar]
- Huguet, A.; Medja, F.; Nicole, A.; Vignaud, A.; Guiraud-Dogan, C.; Ferry, A.; Decostre, V.; Hogrel, J.Y.; Metzger, F.; Hoeflich, A.; et al. Molecular, physiological, and motor performance defects in DMSXL mice carrying >1000 CTG repeats from the human DM1 locus. PLoS Genet. 2012, 8, e1003043. [Google Scholar]
- Lewis, K.; Valanejad, L.; Cast, A.; Wright, M.; Wei, C.; Iakova, P.; Stock, L.; Karns, R.; Timchenko, L.; Timchenko, N. RNA Binding Protein CUGBP1 inhibits liver cancer in a phosphorylation-dependent Manner. Mol. Cell. Biol. 2017, 37, e00128-17. [Google Scholar] [CrossRef]
- Charlet-B, N.; Savkur, R.S.; Singh, G.; Philips, A.V.; Grice, E.A.; Cooper, T.A. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell 2002, 10, 45–53. [Google Scholar] [CrossRef]
- Mankodi, A.; Takahashi, M.P.; Jiang, H.; Beck, C.L.; Bowers, W.J.; Moxley, R.T.; Cannon, S.C.; Thornton, C.A. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell 2002, 10, 35–44. [Google Scholar] [CrossRef]
- Wang, X.; Blagden, C.; Fan, J.; Nowak, S.J.; Taniuchi, I.; Littman, D.R.; Burden, S.J. Runx1 prevents wasting, myofibrillar disorganization, and autophagy of skeletal muscle. Genes Dev. 2005, 19, 1715–1722. [Google Scholar] [CrossRef]
- Yang, X.; Li, M.; Ji, Y.; Lin, Y.; Xu, L.; Gu, X.; Sun, H.; Wang, W.; Shen, Y.; Liu, H.; et al. Changes of Gene Expression Patterns of Muscle Pathophysiology-Related Transcription Factors During Denervated Muscle Atrophy. Front. Physiol. 2022, 13, 923190. [Google Scholar] [CrossRef]
- Joshi, S.; Davidson, G.; Le Gras, S.; Watanabe, S.; Braun, T.; Mengus, G.; Davidson, I. TEAD transcription factors are required for normal primary myoblast differentiation in vitro and muscle regeneration in vivo. PLoS Genet. 2017, 13, e1006600. [Google Scholar] [CrossRef]
- Chargé, S.B.; Rudnicki, M.A. Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 2004, 84, 209–238. [Google Scholar] [CrossRef]
- Nagakura, R.; Yamamoto, M.; Jeong, J.; Hinata, N.; Katori, Y.; Chang, W.J.; Abe, S. Switching of Sox9 expression during musculoskeletal system development. Sci. Rep. 2020, 10, 8425. [Google Scholar] [CrossRef]
- von Maltzahn, J.; Bentzinger, C.F.; Rudnicki, M.A. Wnt7a-Fzd7 signalling directly activates the Akt/mTOR anabolic growth pathway in skeletal muscle. Nat. Cell Biol. 2011, 14, 186–191. [Google Scholar] [CrossRef]
- Kruegel, J.; Miosge, N. Basement membrane components are key players in specialized extracellular matrices. Cell. Mol. Life Sci. 2010, 67, 2879–2895. [Google Scholar] [CrossRef]
- Kumar, A.; Bhatnagar, S.; Paul, P.K. TWEAK and TRAF6 regulate skeletal muscle atrophy. Curr. Opin. Clin. Nutr. Metab. Care 2012, 15, 233–239. [Google Scholar] [CrossRef]
- Adams, J.C.; Lawler, J. The thrombospondins. Cold Spring Harb. Perspect. Biol. 2011, 3, a009712. [Google Scholar] [CrossRef]
- Inoue, M.; Jiang, Y.; Barnes, R.H., 2nd; Tokunaga, M.; Martinez-Santibañez, G.; Geletka, L.; Lumeng, C.N.; Buchner, D.A.; Chun, T.H. Thrombospondin 1 mediates high-fat diet-induced muscle fibrosis and insulin resistance in male mice. Endocrinology 2013, 154, 4548–4559. [Google Scholar] [CrossRef]
- Vlachopapadopoulou, E.; Zachwieja, J.J.; Gertner, J.M.; Manzione, D.; Bier, D.M.; Matthews, D.E.; Slonim, A.E. Metabolic and clinical response to recombinant human insulin-like growth factor I in myotonic dystrophy—A clinical research center study. J. Clin. Endocrinol. Metab. 1995, 80, 3715–3723. [Google Scholar] [CrossRef]
- Heatwole, C.R.; Eichinger, K.J.; Friedman, D.I.; Hilbert, J.E.; Jackson, C.E.; Logigian, E.L.; Martens, W.B.; McDermott, M.P.; Pandya, S.K.; Quinn, C.; et al. Open-label trial of recombinant human insulin-like growth factor 1/recombinant human insulin-like growth factor binding protein 3 in myotonic dystrophy type 1. Arch. Neurol. 2011, 68, 37–44. [Google Scholar] [CrossRef]
- Bolós, M.; Fernandez, S.; Torres-Aleman, I. Oral administration of a GSK3 inhibitor increases brain insulin-like growth factor I levels. J. Biol. Chem. 2010, 285, 17693–17700. [Google Scholar] [CrossRef]
- Douniol, M.; Jacquette, A.; Cohen, D.; Bodeau, N.; Rachidi, L.; Angeard, N.; Cuisset, J.-M.; Vallee, L.; Eymard, B.; Plaza, M.; et al. Psychiatric and cognitive phenotype of childhood myotonic dystrophy type 1. Dev. Med. Child Neurol. 2012, 54, 905–911. [Google Scholar] [CrossRef]
- Ekstrom, A.-B.; Hakenas-Plate, L.; Tulinius, M.; Wentz, E. Cognition and adaptive skills in myotonic dystrophy type 1: A study of 55 individuals with congenital and childhood forms. Dev. Med. Child Neurol. 2009, 51, 982–990. [Google Scholar] [CrossRef]
- Bosco, G.; Diamanti, S.; Meola, G.; the DM-CNS Group. Workshop Report: Consensus on biomarkers of cerebral involvement in myotonic dystrophy, 2-3 December 2014, Milan, Italy. Neuromuscul. Disord. 2015, 25, 813–823. [Google Scholar] [CrossRef]
- van der Plas, E.; Hamilton, M.J.; Miller, J.N.; Koscik, T.R.; Long, J.D.; Cumming, S.; Povilaikaite, J.; Farrugia, M.E.; McLean, J.; Jampana, R.; et al. Brain Structural Features of Myotonic Dystrophy Type 1 and their Relationship with CTG Repeats. J. Neuromuscul. Dis. 2019, 6, 321–332. [Google Scholar] [CrossRef]
- Smith, S.M. Fast robust automated brain extraction. Hum. Brain Mapp. 2002, 17, 143–155. [Google Scholar] [CrossRef]
- Jiang, H.; van Zijl, P.C.M.; Jinsuh Kim, J.; Pearlson, G.D.; Mori, S. DtiStudio: Resource program for diffusion tensor computation and fiber bundle tracking. Comput. Methods Programs Biomed. 2006, 81, 106–116. [Google Scholar] [CrossRef]

| Number of Altered Genes | Number of Altered Genes, Improved by 2-Week Treatment by TG 115 | Percent of Genes, Normalized by TG | |
|---|---|---|---|
| Total altered genes (>1.5 fold) in HSALR muscle (gastroc) vs WT | 665 | 117 | ~17% |
| Total altered genes (>1.5 fold) in TG-treated HSALR muscle (gastroc) vs vehicle-treated HSALR muscle | 675 |
| Pathway | Altered Genes (up and down) in HSALR vs WT Muscle (n) | p Value | Genes, Improved by TG in HSALR Muscle (n) | Genes Improved in TG-Treated HSALR Muscle |
|---|---|---|---|---|
| Regulation of hormone levels | 35 | 1.4 × 10−7 (down); 0.1365 (up) | 7 | Snap25, Fgb, Acvr2b, Crhr2, Nrxn1,Stxbp4, Runx1 a,b |
| Regulation of transport | 93 | 1 × 10−6 (down); 0.001689 (up) | 10 | Slc1a2, Stxbp4, Pop4, Clcn1 a, Snap25, Dpp6, Fgb, Rgcc, Runx1 a,b, Rrad |
| Secretion | 30 | 1.26 × 10−6 (down); 0.368332 (up) | 14 | Snap25, Acvr2b, Sptbn2, Pla2g12a, Myc, Pak1, Vmp1, Syt14, Syt9, Syt12, Park7, Xbp1, Cd27, Trp73 |
| Synaptic vesicle localization | 12 | 1.5 × 10−6 (down); 0.328787 (up) | 2 | Snap25, Sptbn2 |
| Synaptic vesicle cycle | 14 | 1.75 × 10−6 (down); 0.0558 (up) | 3 | Sptbn2, Snap25, Nrxn1 |
| System development | 185 | 1.8 × 10−6 (down); 6.74 × 10−6 (up) | 34 | Snap25, Avil, Nrxn1, Pcp4, Crhr2, Acvr2b, Kdr; Fzd7, Cdnf, Sptbn2, Akap5, Slcla2, Angptl4, Sh3pxd2b, Tnfrsf12, Sox9 a,b, Atf3, Chodl, Hbegf, Tmem100, Esr2, Klhl40, Fscn2, Xirp1, Plk2, Baiap2, Thbs1 a,b, Hspb1, Dlg5, Tead4 a, Lamc2 a,b, Peg10, Runx1, Myf6 a,b |
| Secretion by cell | 24 | 3.88 × 10−6 (down); 0.4887 (up) | 6 | Akap5, Nrxn1, Acvr2b, Sptbn2, Snap25, Rab3c |
| Developmental Process | 223 | 6.39 × 10−6 (down); 1.35 × 10−5 (up) | 32 | Aldoc, Sptbn2, Mug1, Slc1a2, 20Akap5, Avil, Snap25, Pcp4, Nrxn1, Acvr2b, Crhr2, Lrrc38, Fzd7, Kdr, Hspb1, Baiap2, Runx1 a,b, Peg10, Tead4, Dlg5, Myf6 a,b,Tnfrsf12, Sh3pxd2 b, Chodl, Atf3, Sox9 a,b, Nes, Xirp1, Klhl40, Fscn2, Esr2, Tmem100 |
| Cell differentiation | 164 | 3.89 × 10−6 (down); 2.36 × 10−6 (up) | 20 | Pcp4, Cdnf, Avil, Snap25, Fzd7, Kdr, Crhr2, Acvr2b; Myf6 a,b, Peg10, Runx1 a,b, Dlg5, Tead4 a, Baiap2, Tmem120b, Klh140, Tmem100, Esr2, Atf3, Sox9 a,b |
| Genotype | Born Mice Number (23 crosses, 232 pups) | % | Survival Rate, % |
|---|---|---|---|
| hetKI-hetDMSXL | 56 | 24.1 | 94.6 |
| WTKI-homDMSXL | 28 | 12.1 | 71.4 |
| homKI-homDMSXL | 12 | 5.15 | 41.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lutz, M.; Levanti, M.; Karns, R.; Gourdon, G.; Lindquist, D.; Timchenko, N.A.; Timchenko, L. Therapeutic Targeting of the GSK3β-CUGBP1 Pathway in Myotonic Dystrophy. Int. J. Mol. Sci. 2023, 24, 10650. https://doi.org/10.3390/ijms241310650
Lutz M, Levanti M, Karns R, Gourdon G, Lindquist D, Timchenko NA, Timchenko L. Therapeutic Targeting of the GSK3β-CUGBP1 Pathway in Myotonic Dystrophy. International Journal of Molecular Sciences. 2023; 24(13):10650. https://doi.org/10.3390/ijms241310650
Chicago/Turabian StyleLutz, Maggie, Miranda Levanti, Rebekah Karns, Genevieve Gourdon, Diana Lindquist, Nikolai A. Timchenko, and Lubov Timchenko. 2023. "Therapeutic Targeting of the GSK3β-CUGBP1 Pathway in Myotonic Dystrophy" International Journal of Molecular Sciences 24, no. 13: 10650. https://doi.org/10.3390/ijms241310650
APA StyleLutz, M., Levanti, M., Karns, R., Gourdon, G., Lindquist, D., Timchenko, N. A., & Timchenko, L. (2023). Therapeutic Targeting of the GSK3β-CUGBP1 Pathway in Myotonic Dystrophy. International Journal of Molecular Sciences, 24(13), 10650. https://doi.org/10.3390/ijms241310650







