Creation of a Prognostic Model Using Cuproptosis-Associated Long Noncoding RNAs in Hepatocellular Carcinoma
Abstract
1. Introduction
2. Results
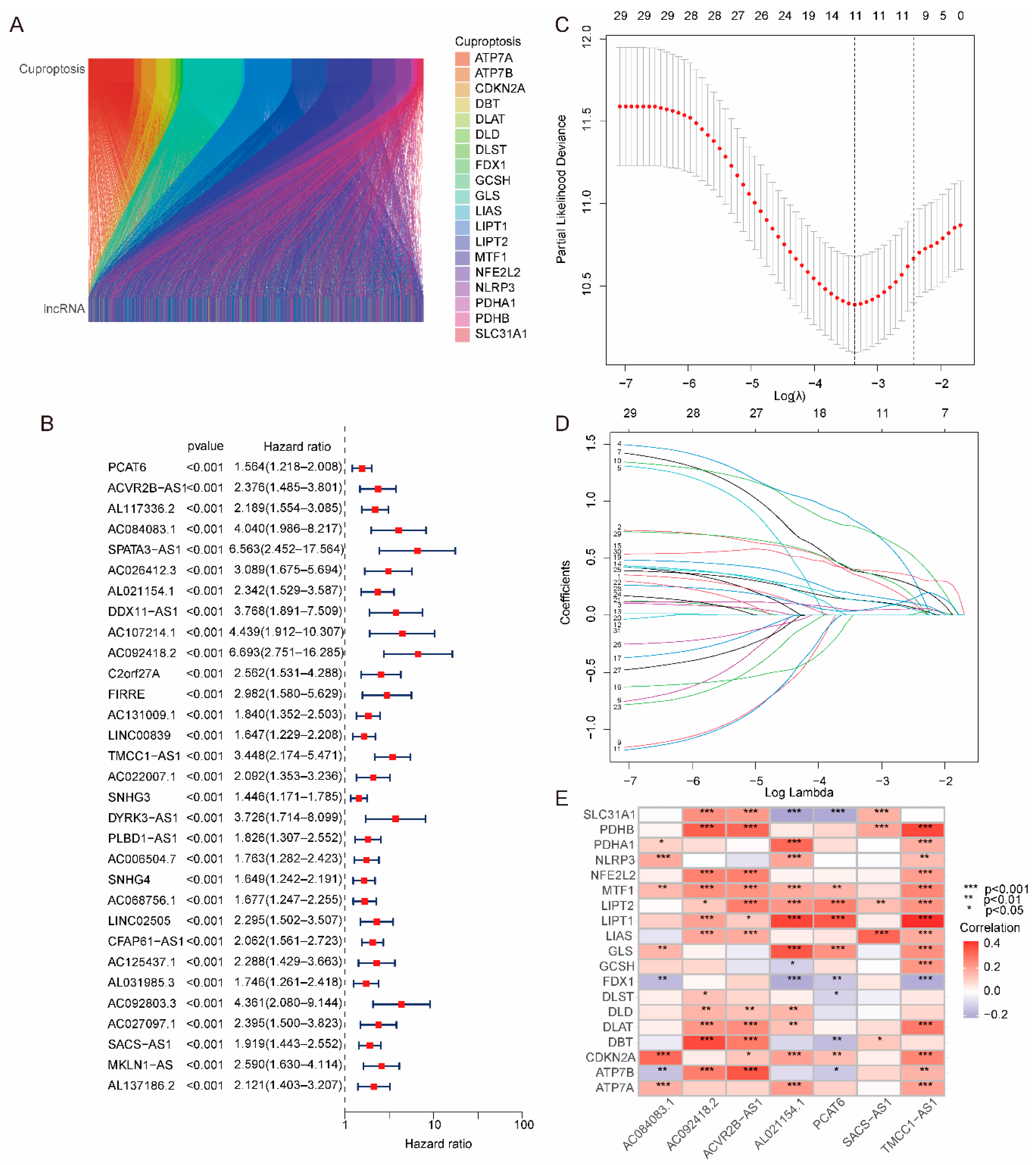
2.1. Construction and Validation of the Risk Model
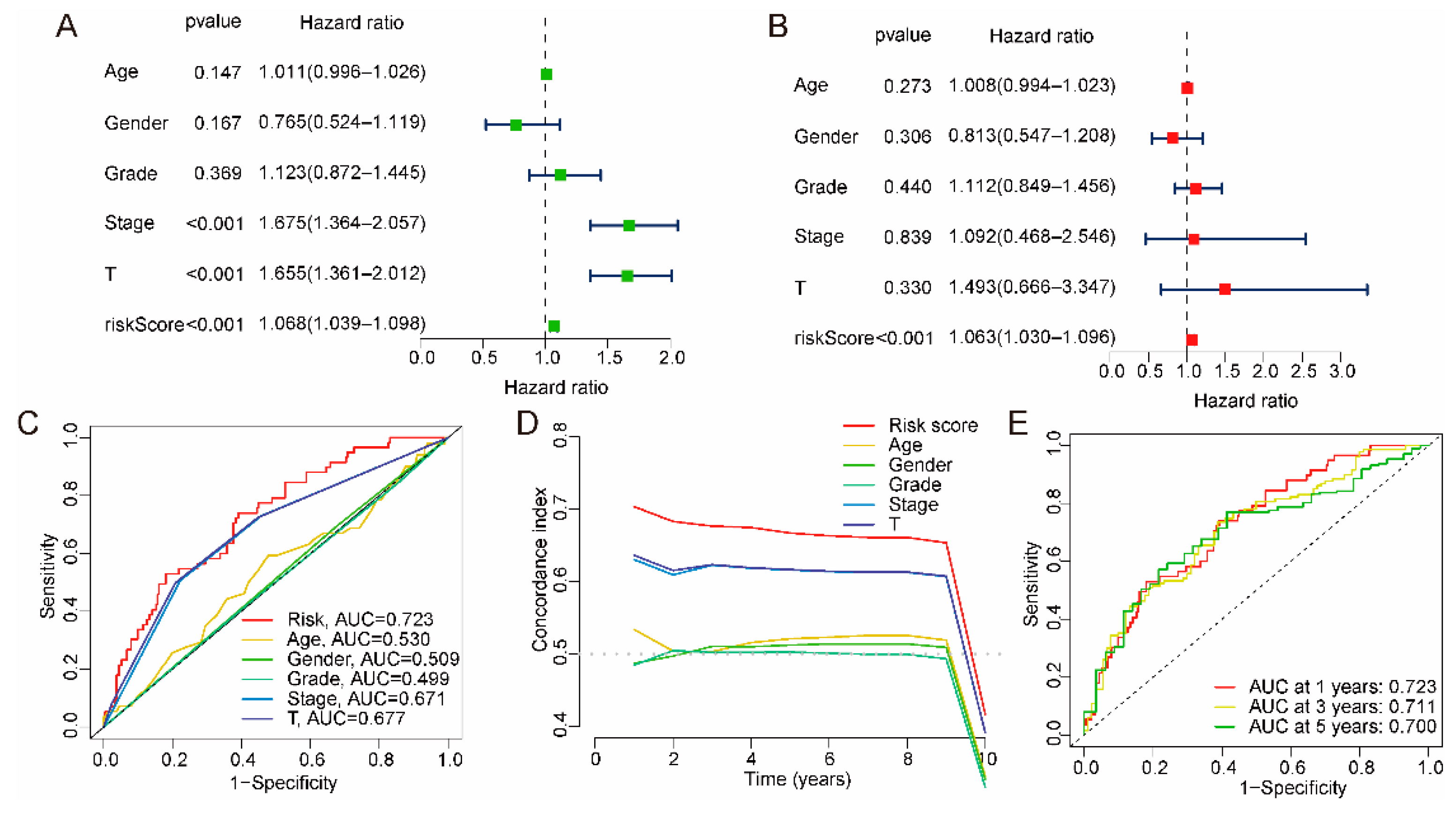
2.2. Independent Prognostic Validation of the Risk Model
2.3. Association between Risk Model and Clinical Characteristics
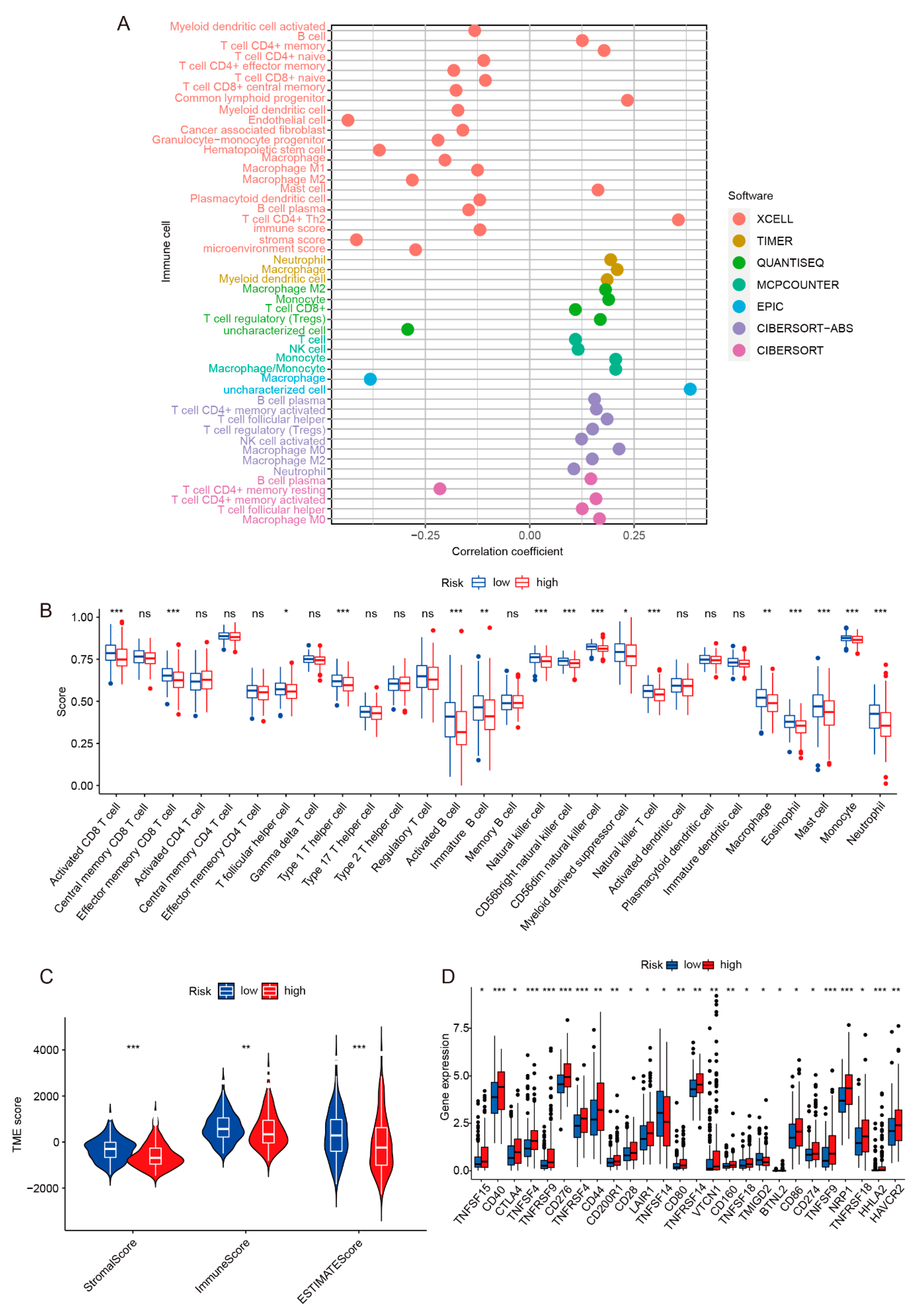
2.4. Immune Infiltration and Immunotherapy Response Analysis of Risk Model
2.5. Sensitivity of Risk Models to Anticancer Drugs
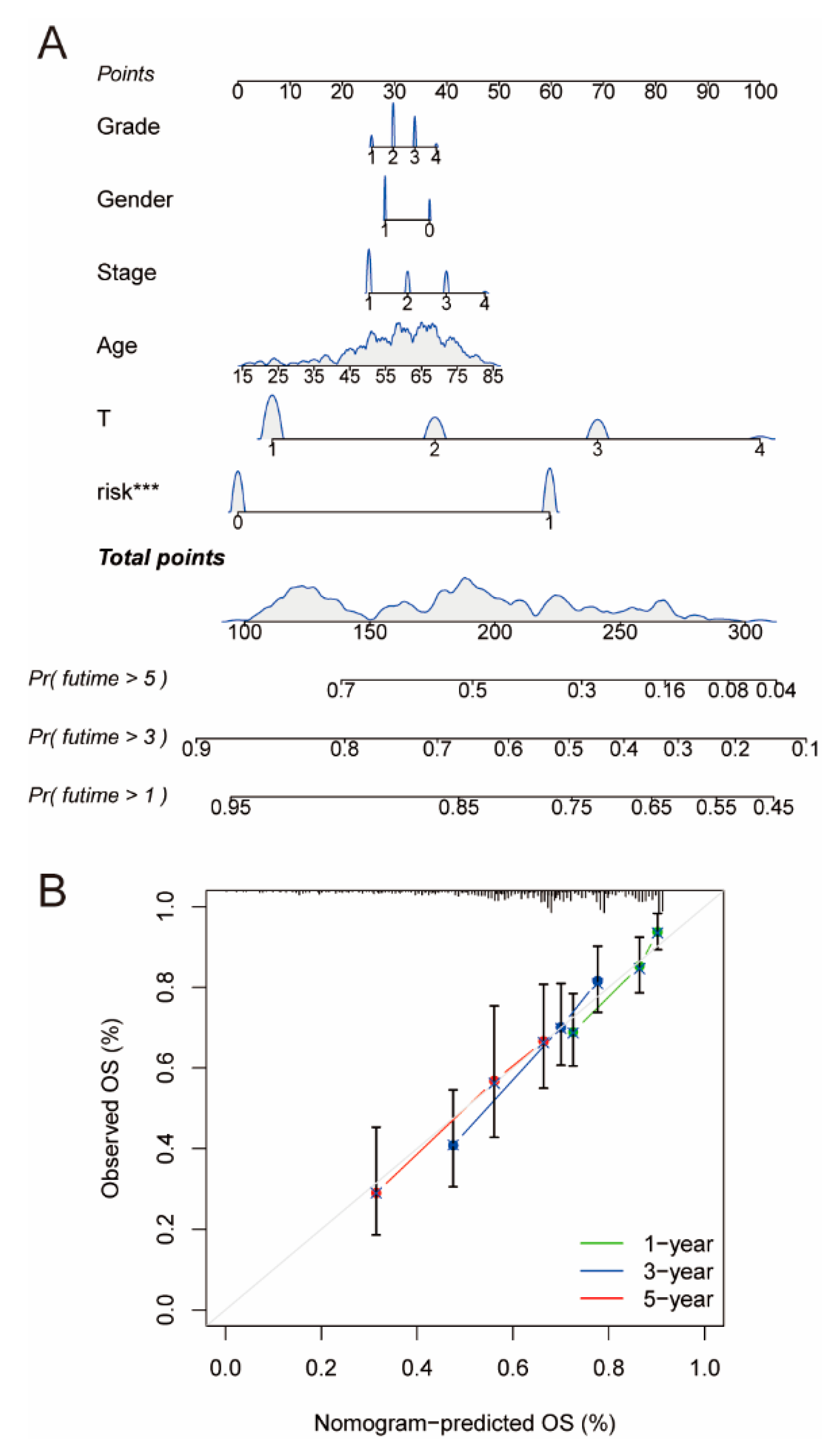
2.6. Construction and Assessment of Nomogram
2.7. Potential lncRNA Biomarkers and Targets for HCC
2.8. Inhibition of PCAT6 Inhibits the Proliferation and Migratory Capacity of HCC Cells
3. Discussion
4. Materials and Methods
4.1. Preparation of Transcriptomic, Mutation Data, and Clinical Information
4.2. Screening of Cuproptosis-Related lncRNAs
4.3. Establishment of the Risk Model Based on Cuproptosis-Related lncRNAs
4.4. Validation of the Risk Model
4.5. Function Analysis
4.6. Immune Response and Analysis
4.7. Exploration of Potential Antitumor Drugs
4.8. Quantitative Real-Time PCR (RT-qPCR)
4.9. Cell Culture and Transfection
4.10. Cell Counting Kit-8 Experiment
4.11. Wound Healing
4.12. CFSE Assay
4.13. Cell Cycle Assay
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, C.; Zhang, H.; Zhang, L.; Zhu, A.X.; Bernards, R.; Qin, W.; Wang, C. Evolving therapeutic landscape of advanced hepatocellular carcinoma. Nat. Rev. Gastro. Hepat. 2023, 20, 203–222. [Google Scholar] [CrossRef]
- Bosch, F.X.; Ribes, J.; Díaz, M.; Cléries, R. Primary liver cancer: Worldwide incidence and trends. Gastroenterology 2004, 127, S5–S16. [Google Scholar] [CrossRef]
- Li, X.; Ramadori, P.; Pfister, D.; Seehawer, M.; Zender, L.; Heikenwalder, M. The immunological and metabolic landscape in primary and metastatic liver cancer. Nat. Rev. Cancer 2021, 21, 541–557. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Schwartz, M.; Mazzaferro, V. Resection and liver transplantation for hepatocellular carcinoma. Semin. Liver Dis. 2005, 25, 181–200. [Google Scholar] [CrossRef]
- Liu, C.Y.; Chen, K.F.; Chen, P.J. Treatment of Liver Cancer. CSH Perspect. Med. 2015, 5, a021535. [Google Scholar] [CrossRef]
- Verslype, C.; Van Cutsem, E.; Dicato, M.; Arber, N.; Berlin, J.D.; Cunningham, D.; De Gramont, A.; Diaz-Rubio, E.; Ducreux, M.; Gruenberger, T.; et al. The management of hepatocellular carcinoma. Current expert opinion and recommendations derived from the 10th World Congress on Gastrointestinal Cancer, Barcelona, 2008. Ann. Oncol. 2009, 20, vii1–vii6. [Google Scholar] [CrossRef] [PubMed]
- Oliveri, V. Selective targeting of cancer cells by copper ionophores: An overview. Front. Mol. Biosci. 2022, 9, 841814. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, L.M.; Libedinsky, A.; Elorza, A.A. Role of copper on mitochondrial function and metabolism. Front. Mol. Biosci. 2021, 8, 711227. [Google Scholar] [CrossRef] [PubMed]
- Pavithra, V.; Sathisha, T.G.; Kasturi, K.; Mallika, D.S.; Amos, S.J.; Ragunatha, S. Serum levels of metal ions in female patients with breast cancer biochemistry section MAterIAls And MethOds. J. Clin. Diagn. Res. 2015, 9, BC25–BC27. [Google Scholar] [CrossRef]
- Baltaci, A.K.; Dundar, T.K.; Aksoy, F.; Mogulkoc, R. Changes in the serum levels of trace elements before and after the operation in thyroid cancer patients. Biol. Trace. Elem. Res. 2017, 175, 57–64. [Google Scholar] [CrossRef]
- Ge, E.J.; Bush, A.I.; Casini, A.; Cobine, P.A.; Cross, J.R.; DeNicola, G.M.; Dou, Q.P.; Franz, K.J.; Gohil, V.M.; Gupta, S.; et al. Connecting copper and cancer: From transition metal signalling to metalloplasia. Nat. Rev. Cancer 2022, 22, 102–113. [Google Scholar] [CrossRef]
- Chen, L.; Min, J.; Wang, F. Copper homeostasis and cuproptosis in health and disease. Signal Transduct. Tar. 2022, 7, 378. [Google Scholar] [CrossRef]
- Tsvetkov, P.; Detappe, A.; Cai, K.; Keys, H.R.; Brune, Z.; Ying, W.; Thiru, P.; Reidy, M.; Kugener, G.; Rossen, J.; et al. Mitochondrial metabolism promotes adaptation to proteotoxic stress. Nat. Chem. Biol. 2019, 15, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Corsello, S.M.; Nagari, R.T.; Spangler, R.D.; Rossen, J.; Kocak, M.; Bryan, J.G.; Humeidi, R.; Peck, D.; Wu, X.; Tang, A.A.; et al. Discovering the anti-cancer potential of non-oncology drugs by systematic viability profiling. Nat. Cancer 2020, 1, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Tsvetkov, P.; Coy, S.; Petrova, B.; Dreishpoon, M.; Verma, A.; Abdusamad, M.; Rossen, J.; Joesch-Cohen, L.; Humeidi, R.; Spangler, R.D.; et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022, 375, 1254–1261. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, M.; Fujii, J.; Suzuki, K.; Inoue, T.; Inoue, T.; Gutteridge, J.M.; Taniguchi, N. A marked increase in free copper levels in the plasma and liver of LEC rats: An animal model for Wilson disease and liver cancer. Free Radic. Res. 1998, 28, 441–450. [Google Scholar] [CrossRef]
- Siddiqui, M.A.; Alhadlaq, H.A.; Ahmad, J.; Al-Khedhairy, A.A.; Musarrat, J.; Ahamed, M. Copper oxide nanoparticles induced mitochondria mediated apoptosis in human hepatocarcinoma cells. PLoS ONE 2013, 8, e69534. [Google Scholar] [CrossRef]
- Polishchuk, E.V.; Merolla, A.; Lichtmannegger, J.; Romano, A.; Indrieri, A.; Ilyechova, E.Y.; Concilli, M.; De Cegli, R.; Crispino, R.; Mariniello, M.; et al. Activation of autophagy, observed in liver tissues from patients with wilson disease and from ATP7B-deficient animals, protects hepatocytes from copper-induced apoptosis. Gastroenterology 2019, 156, 1173–1189.e5. [Google Scholar] [CrossRef]
- Tadini-Buoninsegni, F.; Smeazzetto, S. Mechanisms of charge transfer in human copper ATPases ATP7A and ATP7B. IUBMB Life 2017, 69, 218–225. [Google Scholar] [CrossRef]
- Dong, J.; Wang, X.; Xu, C.; Gao, M.; Wang, S.; Zhang, J.; Tong, H.; Wang, L.; Han, Y.; Cheng, N.; et al. Inhibiting NLRP3 inflammasome activation prevents copper-induced neuropathology in a murine model of Wilson’s disease. Cell Death Dis. 2021, 12, 87. [Google Scholar] [CrossRef]
- Zou, Y.; Zhong, Y.; Wu, J.; Xiao, H.; Zhang, X.; Liao, X.; Li, J.; Mao, X.; Liu, Y.; Zhang, F. Long non-coding PANDAR as a novel biomarker in human cancer: A systematic review. Cell Prolif. 2018, 51, e12422. [Google Scholar] [CrossRef]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Zhou, J.K.; Peng, Y.; He, W.; Huang, C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol. Cancer 2020, 19, 77. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Lv, B.; Liu, R.; Dai, Z.; Zhang, F.; Liang, Y.; Yu, B.; Zeng, D.; Lv, X.B.; Zhang, Z. Role of LncRNAs in regulating cancer amino acid metabolism. Cancer Cell Int. 2021, 21, 209. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chen, L.; Gu, J.; Zhang, H.; Yuan, J.; Lian, Q.; Lv, G.; Wang, S.; Wu, Y.; Yang, Y.T.; et al. Recurrently deregulated lncRNAs in hepatocellular carcinoma. Nat. Commun. 2017, 8, 14421. [Google Scholar] [CrossRef]
- Li, D.; Wang, R.; Wu, N.; Yu, Y. LncRNA HULC as a potential predictor of prognosis and clinicopathological features in patients with digestive system tumors: A meta-analysis. Aging 2022, 14, 1797–1811. [Google Scholar] [CrossRef]
- Bai, Y.; Zhang, Q.; Liu, F.; Quan, J. A novel cuproptosis-related lncRNA signature predicts the prognosis and immune landscape in bladder cancer. Front. Immunol. 2022, 13, 1027449. [Google Scholar] [CrossRef]
- Xie, T.; Liu, B.; Liu, D.; Zhou, Y.; Yang, Q.; Wang, D.; Tang, M.; Liu, W. Cuproptosis-related lncRNA signatures predict prognosis and immune relevance of kidney renal papillary cell carcinoma. Front. Pharmacol. 2022, 13, 1103986. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. B 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Laumont, C.M.; Banville, A.C.; Gilardi, M.; Hollern, D.P.; Nelson, B.H. Tumour-infiltrating B cells: Immunological mechanisms, clinical impact and therapeutic opportunities. Nat. Rev. Cancer 2022, 22, 414–430. [Google Scholar] [CrossRef]
- Xiang, S.; Li, J.; Shen, J.; Zhao, Y.; Wu, X.; Li, M.; Yang, X.; Kaboli, P.J.; Du, F.; Zheng, Y.; et al. Identification of prognostic genes in the tumor microenvironment of hepatocellular carcinoma. Front. Immunol. 2021, 12, 653836. [Google Scholar] [CrossRef]
- Jiang, P.; Gu, S.; Pan, D.; Fu, J.; Sahu, A.; Hu, X.; Li, Z.; Traugh, N.; Bu, X.; Li, B.; et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018, 24, 1550–1558. [Google Scholar] [CrossRef]
- Allgäuer, M.; Budczies, J.; Christopoulos, P.; Endris, V.; Lier, A.; Rempel, E.; Volckmar, A.L.; Kirchner, M.; von Winterfeld, M.; Leichsenring, J.; et al. Implementing tumor mutational burden (TMB) analysis in routine diagnostics-a primer for molecular pathologists and clinicians. Transl. Lung Cancer Res. 2018, 7, 703–715. [Google Scholar] [CrossRef]
- Chen, C.; Su, N.; Li, G.; Shen, Y.; Duan, X. Long non-coding RNA TMCC1-AS1 predicts poor prognosis and accelerates epithelial-mesenchymal transition in liver cancer. Oncol. Lett. 2021, 22, 773. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Bi, Q.; Chen, S.; Chen, X.; Li, S.; Zhong, Z.; Guo, W.; Li, X.; Deng, Y.; Yang, Y. Identification of a five-autophagy-related-lncRNA signature as a novel prognostic biomarker for hepatocellular carcinoma. Front. Mol. Biosci. 2020, 7, 611626. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Jiao, Y.; Li, Y.; Li, W. Investigation of the clinical significance and prognostic value of the lncRNA ACVR2B-As1 in liver cancer. Biomed. Res. Int. 2019, 2019, 4602371. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zheng, J.; Hao, W.; Zeng, H.; Zhang, Z.; Shao, G. lncRNA PCAT6 facilitates cell proliferation and invasion via regulating the miR-326/hnRNPA2B1 axis in liver cancer. Oncol. Lett. 2021, 21, 471. [Google Scholar] [CrossRef]
- Zhu, N.; Hou, J.; Wu, Y.; Liu, J.; Li, G.; Zhao, W.; Ma, G.; Chen, B.; Song, Y. Integrated analysis of a competing endogenous RNA network reveals key lncRNAs as potential prognostic biomarkers for human bladder cancer. Medicine 2018, 97, e11887. [Google Scholar] [CrossRef]
- Mahoney, K.M.; Rennert, P.D.; Freeman, G.J. Combination cancer immunotherapy and new immunomodulatory targets. Nat. Rev. Drug Discov. 2015, 14, 561–584. [Google Scholar] [CrossRef]
- Wang, S.; Chen, Z.; Gu, J.; Chen, X.; Wang, Z. The role of lncRNA PCAT6 in cancers. Front. Oncol. 2021, 11, 701495. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Lv, Q.; Hu, C.; Li, Z.; Wu, H.; Gao, S.; Wang, H.; Zhao, Y.; Shao, Q. PCAT6 may be a whistler and checkpoint target for precision therapy in human cancers. Cancers 2021, 13, 6101. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Khoshbakht, T.; Taheri, M.; Ebrahimzadeh, K. A review on the role of PCAT6 lncRNA in tumorigenesis. Biomed. Pharmacother 2021, 142, 112010. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Hong, C.; Qu, J.; Chen, J.; Qin, Z. Knockdown of lncRNA PCAT6 suppresses the growth of non-small cell lung cancer cells by inhibiting macrophages M2 polarization via miR-326/KLF1 axis. Bioengineered 2022, 13, 12834–12846. [Google Scholar] [CrossRef]
- Wu, W.; Wei, H.; Hu, S.; Wang, Q.; Zhang, C.; Xu, J.; He, Z.; Li, Z.; Pan, X.; Cao, J.; et al. Long Noncoding RNA PCAT6 regulates cell proliferation and migration in human esophageal squamous cell carcinoma. J. Cancer 2022, 13, 681–690. [Google Scholar] [CrossRef]
- Lang, C.; Yin, C.; Lin, K.; Li, Y.; Yang, Q.; Wu, Z.; Du, H.; Ren, D.; Dai, Y.; Peng, X. m(6) A modification of lncRNA PCAT6 promotes bone metastasis in prostate cancer through IGF2BP2-mediated IGF1R mRNA stabilization. Clin. Transl. Med. 2021, 11, e426. [Google Scholar] [CrossRef]
- Chen, Y.; Tang, L.; Huang, W.; Abisola, F.H.; Zhang, Y.; Zhang, G.; Yao, L. Identification of a prognostic cuproptosis-related signature in hepatocellular carcinoma. Biol. Direct. 2023, 18, 4. [Google Scholar] [CrossRef]
- Ding, W.; Chen, J.; Feng, G.; Chen, G.; Wu, J.; Guo, Y.; Ni, X.; Shi, T. DNMIVD: DNA methylation interactive visualization database. Nucl. Acids Res. 2020, 48, D856–D862. [Google Scholar] [CrossRef]
- Zhou, Q.; Yan, X.; Zhu, H.; Xin, Z.; Zhao, J.; Shen, W.; Yin, W.; Guo, Y.; Xu, H.; Zhao, M.; et al. Identification of three tumor antigens and immune subtypes for mRNA vaccine development in diffuse glioma. Theranostics 2021, 11, 9775–9790. [Google Scholar] [CrossRef]
- Xu, G.; Li, C.; Di, Z.; Yang, Y.; Liang, L.; Yuan, Q.; Yang, Q.; Dong, X.; Xu, S.; Wu, G. Development of the expression and prognostic significance of m(5) C-related LncRNAs in breast cancer. Cancer Med. 2023, 12, 7667–7681. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, B.; He, M.; Cai, Y.; Ying, X.; Jiang, C.; Ji, W.; Zeng, J. N6-methylandenosine-related lncRNAs predict prognosis and immunotherapy response in bladder cancer. Front. Oncol. 2021, 11, 710767. [Google Scholar] [CrossRef]
- Xu, F.; Huang, X.; Li, Y.; Chen, Y.; Lin, L. m(6)A-related lncRNAs are potential biomarkers for predicting prognoses and immune responses in patients with LUAD. Mol. Ther. Nucl. Acids 2021, 24, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef]
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautès-Fridman, C.; Fridman, W.H.; et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016, 17, 218. [Google Scholar] [CrossRef]
- Li, B.; Severson, E.; Pignon, J.C.; Zhao, H.; Li, T.; Novak, J.; Jiang, P.; Shen, H.; Aster, J.C.; Rodig, S.; et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol. 2016, 17, 174. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Racle, J.; de Jonge, K.; Baumgaertner, P.; Speiser, D.E.; Gfeller, D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. eLife 2017, 6, e26476. [Google Scholar] [CrossRef] [PubMed]
- Finotello, F.; Mayer, C.; Plattner, C.; Laschober, G.; Rieder, D.; Hackl, H.; Krogsdam, A.; Loncova, Z.; Posch, W.; Wilflingseder, D.; et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 2019, 11, 34. [Google Scholar] [CrossRef]
- Yadav, V.K.; De, S. An assessment of computational methods for estimating purity and clonality using genomic data derived from heterogeneous tumor tissue samples. Brief. Bioinform. 2015, 16, 232–241. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Ru, B.; Wong, C.N.; Tong, Y.; Zhong, J.Y.; Zhong, S.S.W.; Wu, W.C.; Chu, K.C.; Wong, C.Y.; Lau, C.Y.; Chen, I.; et al. TISIDB: An integrated repository portal for tumor-immune system interactions. Bioinformatics 2019, 35, 4200–4202. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, C.; Wang, Z.; Shen, J.; Xiang, P.; Chen, X.; Nan, J.; Lin, Y. Lipofectamine 2000/siRNA complexes cause endoplasmic reticulum unfolded protein response in human endothelial cells. J. Cell Physiol. 2019, 234, 21166–21181. [Google Scholar] [CrossRef] [PubMed]





| Covariates | Type | Total | Testing | Training | pvalue |
|---|---|---|---|---|---|
| Age | ≤65 | 232 (62.7%) | 118 (63.78%) | 114 (61.62%) | 0.7471 |
| Age | >65 | 138 (37.3%) | 67 (36.22%) | 71 (38.38%) | |
| Gender | FEMALE | 121 (32.7%) | 59 (31.89%) | 62 (33.51%) | 0.8246 |
| Gender | MALE | 249 (67.3%) | 126 (68.11%) | 123 (66.49%) | |
| Grade | G1 | 55 (14.86%) | 26 (14.05%) | 29 (15.68%) | 0.6769 |
| Grade | G2 | 177 (47.84%) | 87 (47.03%) | 90 (48.65%) | |
| Grade | G3 | 121 (32.7%) | 66 (35.68%) | 55 (29.73%) | |
| Grade | G4 | 12 (3.24%) | 5 (2.7%) | 7 (3.78%) | |
| Grade | Unknown | 5 (1.35%) | 1 (0.54%) | 4 (2.16%) | |
| Stage | Stage I | 171 (46.22%) | 85 (45.95%) | 86 (46.49%) | 0.9642 |
| Stage | Stage II | 85 (22.97%) | 43 (23.24%) | 42 (22.7%) | |
| Stage | Stage III | 85 (22.97%) | 44 (23.78%) | 41 (22.16%) | |
| Stage | Stage IV | 5 (1.35%) | 3 (1.62%) | 2 (1.08%) | |
| Stage | Unknown | 24 (6.49%) | 10 (5.41%) | 14 (7.57%) | |
| T | T1 | 181 (48.92%) | 92 (49.73%) | 89 (48.11%) | 0.5286 |
| T | T2 | 93 (25.14%) | 44 (23.78%) | 49 (26.49%) | |
| T | T3 | 80 (21.62%) | 40 (21.62%) | 40 (21.62%) | |
| T | T4 | 13 (3.51%) | 9 (4.86%) | 4 (2.16%) | |
| T | Unknown | 3 (0.81%) | 0 (0%) | 3 (1.62%) | |
| M | M0 | 266 (71.89%) | 139 (75.14%) | 127 (68.65%) | 1 |
| M | M1 | 4 (1.08%) | 2 (1.08%) | 2 (1.08%) | |
| M | Unknown | 100 (27.03%) | 44 (23.78%) | 56 (30.27%) | |
| N | N0 | 252 (68.11%) | 136 (73.51%) | 116 (62.7%) | 0.7399 |
| N | N1 | 4 (1.08%) | 3 (1.62%) | 1 (0.54%) | |
| N | unknown | 114 (30.81%) | 46 (24.86%) | 68 (36.76%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, L.; Jia, X.; Fu, Y.; Tian, J.; Liu, Y.; Lin, J. Creation of a Prognostic Model Using Cuproptosis-Associated Long Noncoding RNAs in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2023, 24, 9987. https://doi.org/10.3390/ijms24129987
Yang L, Jia X, Fu Y, Tian J, Liu Y, Lin J. Creation of a Prognostic Model Using Cuproptosis-Associated Long Noncoding RNAs in Hepatocellular Carcinoma. International Journal of Molecular Sciences. 2023; 24(12):9987. https://doi.org/10.3390/ijms24129987
Chicago/Turabian StyleYang, Lihong, Xiao Jia, Yueyue Fu, Jiao Tian, Yijin Liu, and Jianping Lin. 2023. "Creation of a Prognostic Model Using Cuproptosis-Associated Long Noncoding RNAs in Hepatocellular Carcinoma" International Journal of Molecular Sciences 24, no. 12: 9987. https://doi.org/10.3390/ijms24129987
APA StyleYang, L., Jia, X., Fu, Y., Tian, J., Liu, Y., & Lin, J. (2023). Creation of a Prognostic Model Using Cuproptosis-Associated Long Noncoding RNAs in Hepatocellular Carcinoma. International Journal of Molecular Sciences, 24(12), 9987. https://doi.org/10.3390/ijms24129987








