Lost and Found: The Family of NF-κB Inhibitors Is Larger than Assumed in Salmonid Fish
Abstract
1. Introduction
2. Results
2.1. Iκb Proteins from Rainbow Trout Are Encoded on 14 Distinct Genes
2.2. IκBα-Encoding nfkbia-a Transcripts Are Most Strongly Expressed in Immune Tissues and Immune-Cell Fractions
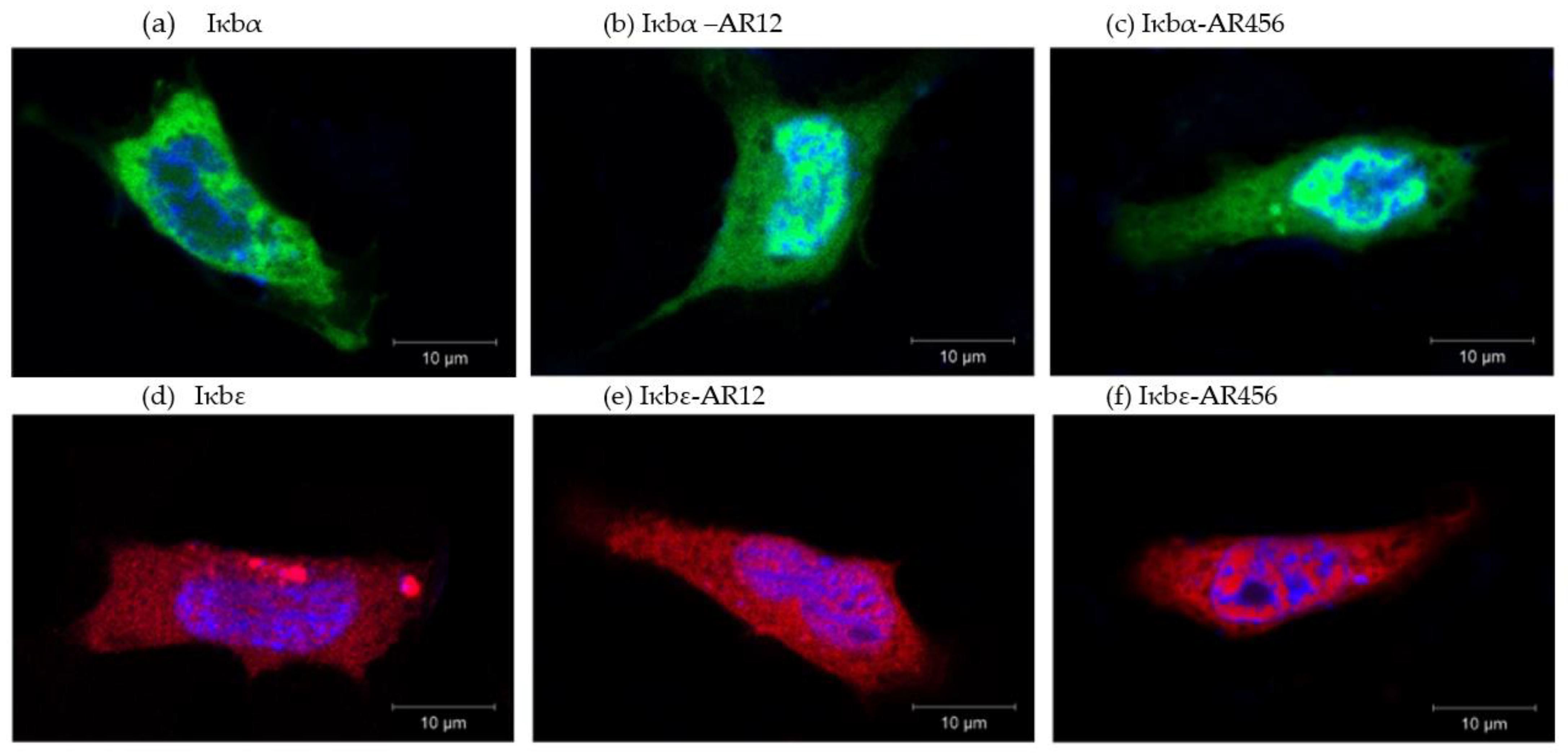
2.3. Iκbα and iκbε Localise to the Cytoplasm as well as the Nucleus of Salmonid Model CHSE-214 Cells
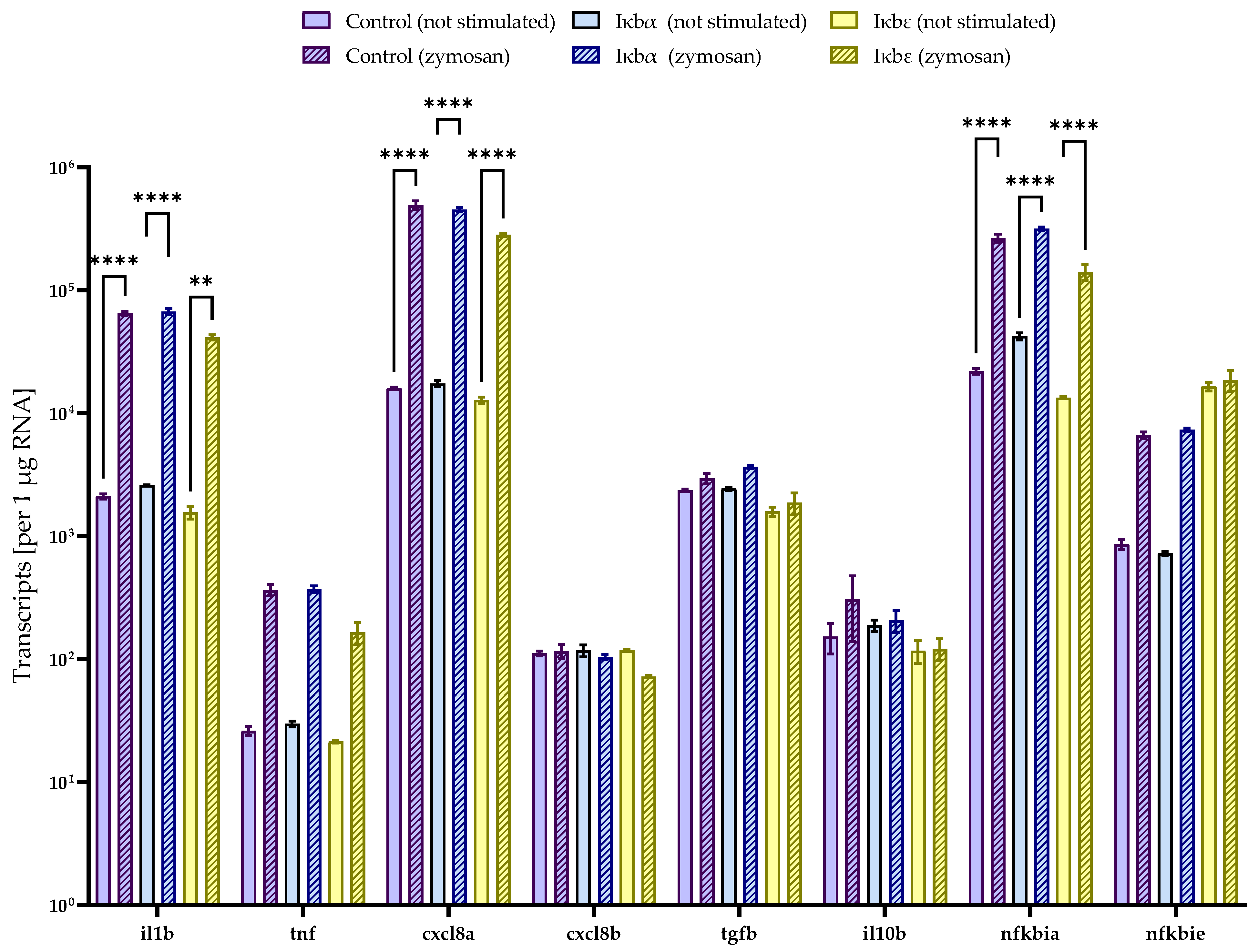
2.4. Iκbα and iκbε Reduce the Basal and Stimulated nf-κb Activity
3. Discussion
4. Materials and Methods
4.1. Quantitative PCR (qPCR) Analysis
4.2. Construction of Nfkbi-Expression Constructs
4.3. Cell Transfection, Luciferase Assay and Confocal Microscopy
4.4. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Visvanathan, K.V.; Goodbourn, S. Double-stranded RNA Activates Binding of NF-kappa B to an Inducible Element in the Human Beta-interferon Promoter. EMBO J. 1989, 8, 1129–1138. [Google Scholar] [CrossRef] [PubMed]
- Sen, R.; Baltimore, D. Inducibility of κ Immunoglobulin Enhancer-Binding Protein NF-κB by a Posttranslational Mechanism. Cell 1986, 47, 921–928. [Google Scholar] [CrossRef]
- Hayden, M.S.; Ghosh, S. NF-ΚB in Immunobiology. Cell Res. 2011, 21, 223–244. [Google Scholar] [CrossRef] [PubMed]
- Dorrington, M.G.; Fraser, I.D.C. NF-ΚB Signaling in Macrophages: Dynamics, Crosstalk, and Signal Integration. Front. Immunol. 2019, 10, 705. [Google Scholar] [CrossRef] [PubMed]
- Baeuerle, P.A.; Henkel, T. Function and Activation of NF-KappaB in the Immune System. Annu. Rev. Immunol. 1994, 12, 141–179. [Google Scholar] [CrossRef] [PubMed]
- Sha, W.C.; Liou, H.C.; Tuomanen, E.I.; Baltimore, D. Targeted Disruption of the P50 Subunit of NF-ΚB Leads to Multifocal Defects in Immune Responses. Cell 1995, 80, 321–330. [Google Scholar] [CrossRef]
- Alcamo, E.; Mizgerd, J.P.; Horwitz, B.H.; Bronson, R.; Beg, A.A.; Scott, M.; Doerschuk, C.M.; Hynes, R.O.; Baltimore, D. Targeted Mutation of TNF Receptor I Rescues the RelA-Deficient Mouse and Reveals a Critical Role for NF-ΚB in Leukocyte Recruitment. J. Immunol. 2001, 167, 1592–1600. [Google Scholar] [CrossRef]
- Franzoso, G.; Carlson, L.; Poljak, L.; Shores, E.W.; Epstein, S.; Leonardi, A.; Grinberg, A.; Tran, T.; Scharton-Kersten, T.; Anver, M.; et al. Mice Deficient in Nuclear Factor (NF)-κB/P52 Present with Defects in Humoral Responses, Germinal Center Reactions, and Splenic Microarchitecture. J. Exp. Med. 1998, 187, 147–159. [Google Scholar] [CrossRef]
- Diamond, G.; Kaiser, V.; Rhodes, J.; Russell, J.P.; Bevins, C.L. Transcriptional Regulation of β-Defensin Gene Expression in Tracheal Epithelial Cells. Infect. Immun. 2000, 68, 113–119. [Google Scholar] [CrossRef]
- Baeuerle, P.A.; Baltimore, D. I Kappa B: A Specific Inhibitor of the NF-Kappa B Transcription Factor. Science 1988, 242, 540–546. [Google Scholar] [CrossRef]
- DiDonato, J.A.; Hayakawa, M.; Rothwarf, D.M.; Zandi, E.; Karin, M. A Cytokine-Responsive IκB Kinase That Activates the Transcription Factor NF-κB. Nature 1997, 388, 548–554. [Google Scholar] [CrossRef]
- Rebl, A.; Goldammer, T. Under Control: The Innate Immunity of Fish from the Inhibitors’ Perspective. Fish Shellfish Immunol. 2018, 77, 328–349. [Google Scholar] [CrossRef] [PubMed]
- Inoue, J.-i.; Kerr, L.D.; Kakizuka, A.; Verma, I.M. IκBγ, a 70 Kd Protein Identical to the C-Terminal Half of P110 NF-ΚB: A New Member of the IκB Family. Cell 1992, 68, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Rice, N.R.; MacKichan, M.L.; Israël, A. The Precursor of NF-ΚB P50 Has IκB-like Functions. Cell 1992, 71, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Zabel, U.; Baeuerle, P.A. Purified Human IκB Can Rapidly Dissociate the Complex of the NF-κB Transcription Factor with Its Cognate DNA. Cell 1990, 61, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Kerr, L.D.; Duckett, C.S.; Wamsley, P.; Zhang, Q.; Chiao, P.; Nabel, G.; Baeuerle, P.A.; Verma, I.M. The Proto-Oncogene B CL-3 Encodes an IKB Protein. Genes Dev. 1992, 5, 2352–2363. [Google Scholar] [CrossRef]
- Li, Z.; Nabel, G.J. A New Member of the I KappaB Protein Family, I KappaB Epsilon, Inhibits RelA (P65)-Mediated NF-KappaB Transcription. Mol. Cell. Biol. 1997, 17, 6184–6190. [Google Scholar] [CrossRef]
- Yamazaki, S.; Muta, T.; Takeshige, K. A Novel IκB Protein, IκB-ζ, Induced by Proinflammatory Stimuli, Negatively Regulates Nuclear Factor-κB in the Nuclei. J. Biol. Chem. 2001, 276, 27657–27662. [Google Scholar] [CrossRef]
- Fiorini, E.; Schmitz, I.; Marissen, W.E.; Osborn, S.L.; Touma, M.; Sasada, T.; Reche, P.A.; Tibaldi, E.V.; Hussey, R.E.; Kruisbeek, A.M.; et al. Peptide-Induced Negative Selection of Thymocytes Activates Transcription of an NF-ΚB Inhibitor. Mol. Cell 2002, 9, 637–648. [Google Scholar] [CrossRef]
- Yamauchi, S.; Ito, H.; Miyajima, A. IκBη, a Nuclear IκB Protein, Positively Regulates the NF-ΚB-Mediated Expression of Proinflammatory Cytokines. Proc. Natl. Acad. Sci. USA 2010, 107, 11924–11929. [Google Scholar] [CrossRef]
- Scheinman, R.I.; Beg, A.A.; Baldwin, A.S. NF-Kappa B P100 (Lyt-10) Is a Component of H2TF1 and Can Function as an I Kappa B-like Molecule. Mol. Cell. Biol. 1993, 13, 6089–6101. [Google Scholar] [CrossRef]
- Naumann, M.; Wulczyn, F.G.; Scheidereit, C. The NF-ΚB Precursor P105 and the Proto-Oncogene Product Bcl-3 Are IκB Molecules and Control Nuclear Translocation of NF-ΚB. EMBO J. 1993, 12, 213–222. [Google Scholar] [CrossRef]
- Davis, N.; Ghosh, S.; Simmons, D.L.; Tempst, P.; Liou, H.C.; Baltimore, D.; Bose, H.R. Rel-Associated Pp40: An Inhibitor of the Rel Family of Transcription Factors. Science 1991, 253, 1268–1271. [Google Scholar] [CrossRef]
- Liou, H.C.; Nolan, G.P.; Ghosh, S.; Fujita, T.; Baltimore, D. The NF-ΚB P50 Precursor, P105, Contains an Internal IκB-like Inhibitor That Preferentially Inhibits P50. EMBO J. 1992, 11, 3003–3009. [Google Scholar] [CrossRef] [PubMed]
- Nolan, G.P.; Fujita, T.; Bhatia, K.; Huppi, C.; Liou, H.C.; Scott, M.L.; Baltimore, D. The Bcl-3 Proto-Oncogene Encodes a Nuclear I Kappa B-like Molecule That Preferentially Interacts with NF-Kappa B P50 and P52 in a Phosphorylation-Dependent Manner. Mol. Cell. Biol. 1993, 13, 3557–3566. [Google Scholar] [CrossRef] [PubMed]
- Beg, A.A.; Ruben, S.M.; Scheinman, R.I.; Haskill, S.; Rosen, C.A.; Baldwin, A.S.J. I Kappa B Interacts with the Nuclear Localization Sequences of the Subunits of NF-Kappa B: A Mechanism for Cytoplasmic Retention. Genes Dev. 1992, 6, 1899–1913. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, S.T.; Epinat, J.C.; Rice, N.R.; Israël, A. I Kappa B Epsilon, a Novel Member of the IκB Family, Controls RelA and CRel NF-ΚB Activity. EMBO J. 1997, 16, 1413–1426. [Google Scholar] [CrossRef]
- Huang, T.T.; Kudo, N.; Yoshida, M.; Miyamoto, S. A Nuclear Export Signal in the N-Terminal Regulatory Domain of IkappaBalpha Controls Cytoplasmic Localization of Inactive NF-KappaB/IkappaBalpha Complexes. Proc. Natl. Acad. Sci. USA 2000, 97, 1014–1019. [Google Scholar] [CrossRef]
- Bours, V.; Franzoso, G.; Azarenko, V.; Park, S.; Kanno, T.; Brown, K.; Siebenlist, U. The Oncoprotein Bcl-3 Directly Transactivates through ΚB Motifs via Association with DNA-Binding P50B Homodimers. Cell 1993, 72, 729–739. [Google Scholar] [CrossRef]
- Hirotani, T.; Lee, P.Y.; Kuwata, H.; Yamamoto, M.; Matsumoto, M.; Kawase, I.; Akira, S.; Takeda, K. The Nuclear IκB Protein IκBNS Selectively Inhibits Lipopolysaccharide-Induced IL-6 Production in Macrophages of the Colonic Lamina Propria. J. Immunol. 2005, 174, 3650–3657. [Google Scholar] [CrossRef]
- Kuwata, H.; Matsumoto, M.; Atarashi, K.; Morishita, H.; Hirotani, T.; Koga, R.; Takeda, K. IκBNS Inhibits Induction of a Subset of Toll-like Receptor-Dependent Genes and Limits Inflammation. Immunity 2006, 24, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Motoyama, M.; Yamazaki, S.; Eto-Kimura, A.; Takeshige, K.; Muta, T. Positive and Negative Regulation of Nuclear Factor-κB-Mediated Transcription by IκB-ζ, an Inducible Nuclear Protein. J. Biol. Chem. 2005, 280, 7444–7451. [Google Scholar] [CrossRef] [PubMed]
- Massoumi, R.; Chmielarska, K.; Hennecke, K.; Pfeifer, A.; Fässler, R. Cyld Inhibits Tumor Cell Proliferation by Blocking Bcl-3-Dependent NF-κB Signaling. Cell 2006, 125, 665–677. [Google Scholar] [CrossRef]
- Lux, S.E.; John, K.M.; Bennettt, V. Analysis of EDNA for Human Erythrocyte Ankyrin Indicates a Repeated Structure with Homology to Tissue-Differentiation and Cell-Cycle Control Proteins. Nature 1990, 344, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Huxford, T.; Huang, D.B.; Malek, S.; Ghosh, G. The Crystal Structure of the IκBα/NF-κB Complex Reveals Mechanisms of NF-ΚB Inactivation. Cell 1998, 95, 759–770. [Google Scholar] [CrossRef]
- Kieran, M.; Blank, V.; Logeat, F.; Vandekerckhove, J.; Lottspeich, F.; Le Bail, O.; Urban, M.B.; Kourilsky, P.; Baeuerle, P.A.; Israël, A. The DNA Binding Subunit of NF-κB Is Identical to Factor KBF1 and Homologous to the Rel Oncogene Product. Cell 1990, 62, 1007–1018. [Google Scholar] [CrossRef]
- Mercurio, F.; Didonato, J.; Rosette, C.; Karin, M. Molecular Cloning and Characterization of a Novel Rel/NF-ΧB Family Member Displaying Structural and Functional Homology to NF-ΧB P50/P105. DNA Cell Biol. 1992, 11, 523–537. [Google Scholar] [CrossRef]
- Michel, F.; Soler-Lopez, M.; Petosa, C.; Cramer, P.; Siebenlist, U.; Müller, C.W. Crystal Structure of the Ankyrin Repeat Domain of Bcl-3: A Unique Member of the IκB Protein Family. EMBO J. 2001, 20, 6180–6190. [Google Scholar] [CrossRef]
- Beg, A.A.; Baldwin, A.S. The I Kappa B Proteins: Multifunctional Regulators of Rel/NF-Kappa B Transcription Factors. Genes Dev. 1993, 7, 2064–2070. [Google Scholar] [CrossRef]
- Ernst, M.K.; Dunn, L.L.; Rice, N.R. The PEST-like Sequence of I Kappa B Alpha Is Responsible for Inhibition of DNA Binding but Not for Cytoplasmic Retention of c-Rel or RelA Homodimers. Mol. Cell. Biol. 1995, 15, 872–882. [Google Scholar] [CrossRef]
- Thompson, J.E.; Phillips, R.J.; Erdjument-Bromage, H.; Tempst, P.; Ghosh, S. IκB-β Regulates the Persistent Response in a Biphasic Activation of NF-κB. Cell 1995, 80, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Mathes, E.; O’Dea, E.L.; Hoffmann, A.; Ghosh, G. NF-ΚB Dictates the Degradation Pathway of IκBα. EMBO J. 2008, 27, 1357–1367. [Google Scholar] [CrossRef] [PubMed]
- McKinsey, T.A.; Chu, Z.L.; Ballard, D.W. Phosphorylation of the PEST Domain of IκBβ Regulates the Function of NF-κB/IκBβ Complexes. J. Biol. Chem. 1997, 272, 22377–22380. [Google Scholar] [CrossRef]
- McKinsey, T.A.; Brockman, J.A.; Scherer, D.C.; Al-Murrani, S.W.; Green, P.L.; Ballard, D.W. Inactivation of IkappaBbeta by the Tax Protein of Human T-Cell Leukemia Virus Type 1: A Potential Mechanism for Constitutive Induction of NF-KappaB. Mol. Cell. Biol. 1996, 16, 2083–2090. [Google Scholar] [CrossRef] [PubMed]
- DiDonato, J.; Mercurio, F.; Rosette, C.; Wu-Li, J.; Suyang, H.; Ghosh, S.; Karin, M. Mapping of the Inducible IkappaB Phosphorylation Sites That Signal Its Ubiquitination and Degradation. Mol. Cell. Biol. 1996, 16, 1295–1304. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Baltimore, D. Activation in Vitro of NF-κB” by Phosphorylation of Its Inhibitor IκB”. Nature 1990, 344, 678–682. [Google Scholar] [CrossRef]
- Chiao, P.J.; Miyamoto, S.; Verma, I.M. Autoregulation of I Kappa B Alpha Activity. Proc. Natl. Acad. Sci. USA 1994, 91, 28–32. [Google Scholar] [CrossRef]
- Kearns, J.D.; Basak, S.; Werner, S.L.; Huang, C.S.; Hoffmann, A. IκBε Provides Negative Feedback to Control NF-κB Oscillations, Signaling Dynamics, and Inflammatory Gene Expression. J. Cell Biol. 2006, 173, 659–664. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, J.; Ghosh, G. KappaB-Ras Binds to the Unique Insert within the Ankyrin Repeat Domain of IkappaBbeta and Regulates Cytoplasmic Retention of IkappaBbeta x NF-KappaB Complexes. J. Biol. Chem. 2003, 278, 23101–23106. [Google Scholar] [CrossRef]
- Totzke, G.; Essmann, F.; Pohlmann, S.; Lindenblatt, C.; Jänicke, R.U.; Schulze-Osthoff, K. A Novel Member of the IκB Family, Human IκB-ζ, Inhibits Transactivation of P65 and Its DNA Binding. J. Biol. Chem. 2006, 281, 12645–12654. [Google Scholar] [CrossRef]
- Sarais, F.; Rebl, H.; Verleih, M.; Ostermann, S.; Krasnov, A.; Köllner, B.; Goldammer, T.; Rebl, A. Characterisation of the Teleostean κB-Ras Family: The Two Members NKIRAS1 and NKIRAS2 from Rainbow Trout Influence the Activity of NF-κB in Opposite Ways. Fish Shellfish Immunol. 2020, 106, 1004–1013. [Google Scholar] [CrossRef] [PubMed]
- Andrea Oeckinghaus, A.; Post-ler, T.S.; Lienhard, G.E. ΚB-Ras Proteins Regulate Both NF-κB-Dependent Inflammation and Ral-Dependent Proliferation. CellReports 2014, 8, 1793–1807. [Google Scholar] [CrossRef] [PubMed]
- Sangrador-Vegas, A.; Smith, T.J.; Cairns, M.T. Cloning and Characterization of a Homologue of the Alpha Inhibitor of NF-κB in Rainbow Trout (Oncorhynchus mykiss). Vet. Immunol. Immunopathol. 2005, 103, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Geisler, R.; Bergmann, A.; Hiromi, Y.; Nüsslein-Volhard, C. Cactus, a Gene Involved in Dorsoventral Pattern Formation of Drosophila, Is Related to the IκB Gene Family of Vertebrates. Cell 1992, 71, 613–621. [Google Scholar] [CrossRef]
- Lee, Y.; Umasuthan, N.; Whang, I.; Revathy, K.S.; Lee, S.; De Zoysa, M.; Oh, C.; Kang, D.H.; Noh, J.K.; Lee, J. Two NF-κB Inhibitor-Alpha (IκBα) Genes from Rock Bream (Oplegnathus fasciatus): Molecular Characterization, Genomic Organization and MRNA Expression Analysis after Immune Stimulation. Fish Shellfish Immunol. 2014, 41, 633–642. [Google Scholar] [CrossRef]
- Mai, W.; Huang, F.; Liu, P. The Role of EaIκB-α, an IκB-α Homologue in Epinephelus Akaara, Involved in Innate Immune Response. Biotechnol. Lett. 2014, 36, 1421–1429. [Google Scholar] [CrossRef]
- Gao, R.; Huang, Y.; Huang, X.; Guan, L.; Wei, S.; Zhou, Y.; Qin, Q. Molecular Cloning and Characterization of Two Types of IκBα Orthologues in Orange-Spotted Grouper, Epinephelus Coioides. Fish Shellfish Immunol. 2014, 38, 101–110. [Google Scholar] [CrossRef]
- Correa, R.G.; Tergaonkar, V.; Ng, J.K.; Dubova, I.; Izpisua-Belmonte, J.C.; Verma, I.M. Characterization of NF-κΒ/IκΒ Proteins in Zebra Fish and Their Involvement in Notochord Development. Mol. Cell. Biol. 2004, 24, 5257–5268. [Google Scholar] [CrossRef]
- Jakovlić, I.; Liu, H.; Wang, W.-M. Identification, Origin and Evidence for Retained Functionality of Two IκBα Paralogs in Megalobrama Amblycephala. Dev. Comp. Immunol. 2016, 62, 89–96. [Google Scholar] [CrossRef]
- Zhang, M.; Xiao, Z.; Sun, L. Overexpression of NF-κB Inhibitor Alpha in Cynoglossus Semilaevis Impairs Pathogen-Induced Immune Response. Dev. Comp. Immunol. 2012, 36, 253–257. [Google Scholar] [CrossRef]
- Wang, Y.; Wei, H.; Wang, X.; Du, L.; Zhang, A.; Zhou, H. Cellular Activation, Expression Analysis and Functional Characterization of Grass Carp IκBα: Evidence for Its Involvement in Fish NF-κB Signaling Pathway. Fish Shellfish Immunol. 2015, 42, 408–412. [Google Scholar] [CrossRef] [PubMed]
- Yazawa, R.; Kondo, H.; Hirono, I.; Aoki, T. Cloning and Characterization of the IκBα Gene from Japanese Flounder, Paralichthys Olivaceus. Fish Shellfish Immunol. 2007, 23, 808–814. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Xu, Y.; Lin, P.; Peng, X.; Wang, Y.; Zhang, Z. Identification of IκBα in Japanese Eel Anguilla Japonica That Impairs the IKKα-Dependent Activation of NF-ΚB, AP1, and Type I IFN Signaling Pathways. Dev. Comp. Immunol. 2021, 122, 104044. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhou, Z.-C.; Guo, C.-J.; Rao, X.-Y.; Xiao, J.; Weng, S.-P.; Yin, Z.-X.; Yu, X.-Q.; He, J.-G. The Alpha Inhibitor of NF-KappaB (IkappaBalpha) from the Mandarin Fish Binds with P65 NF-KappaB. Fish Shellfish Immunol. 2009, 26, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.P.; Arora, J.; Isambert, H. Identification of Ohnolog Genes Originating from Whole Genome Duplication in Early Vertebrates, Based on Synteny Comparison across Multiple Genomes. PLoS Comput. Biol. 2015, 11, e1004394. [Google Scholar] [CrossRef]
- Morris, R.T.; Drouin, G. Ectopic Gene Conversions in the Genome of Ten Hemiascomycete Yeast Species. Int. J. Evol. Biol. 2011, 2011, 970768. [Google Scholar] [CrossRef]
- Campbell, M.A.; Hale, M.C.; McKinney, G.J.; Nichols, K.M.; Pearse, D.E. Long-Term Conservation of Ohnologs Through Partial Tetrasomy Following Whole-Genome Duplication in Salmonidae. G3 Genes Genomes Genet. 2019, 9, 2017–2028. [Google Scholar] [CrossRef]
- Reams, A.B.; Roth, J.R. Mechanisms of Gene Duplication and Amplification. Cold Spring Harb. Perspect. Biol. 2015, 7, a016592. [Google Scholar] [CrossRef]
- Okamoto, T.; Ono, T.; Hori, M.; Yang, J.P.; Tetsuka, T.; Kawabe, T.; Sonta, S. Assignment of the IkappaB-Beta Gene NFKBIB to Human Chromosome Band 19q13.1 by in Situ Hybridization. Cytogenet. Cell Genet. 1998, 82, 105–106. [Google Scholar] [CrossRef]
- Eslamloo, K.; Xue, X.; Booman, M.; Smith, N.C.; Rise, M.L. Transcriptome Profiling of the Antiviral Immune Response in Atlantic Cod Macrophages. Dev. Comp. Immunol. 2016, 63, 187–205. [Google Scholar] [CrossRef]
- Sood, N.; Verma, D.K.; Paria, A.; Yadav, S.C.; Yadav, M.K.; Bedekar, M.K.; Kumar, S.; Swaminathan, T.R.; Mohan, C.V.; Rajendran, K.V.; et al. Transcriptome Analysis of Liver Elucidates Key Immune-Related Pathways in Nile Tilapia Oreochromis Niloticus Following Infection with Tilapia Lake Virus. Fish Shellfish Immunol. 2021, 111, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Schiøtz, B.L.; Jørgensen, S.M.; Rexroad, C.; Gjøen, T.; Krasnov, A. Transcriptomic Analysis of Responses to Infectious Salmon Anemia Virus Infection in Macrophage-like Cells. Virus Res. 2008, 136, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Jiang, D.; Wang, J.; Qi, Q. The Effects of TPT and Dietary Quercetin on Growth, Hepatic Oxidative Damage and Apoptosis in Zebrafish. Ecotoxicol. Environ. Saf. 2021, 224, 112697. [Google Scholar] [CrossRef] [PubMed]
- Kaleo, I.V.; Gao, Q.; Liu, B.; Sun, C.; Zhou, Q.; Zhang, H.; Shan, F.; Xiong, Z.; Bo, L.; Song, C. Effects of Moringa Oleifera Leaf Extract on Growth Performance, Physiological and Immune Response, and Related Immune Gene Expression of Macrobrachium Rosenbergii with Vibrio Anguillarum and Ammonia Stress. Fish Shellfish Immunol. 2019, 89, 603–613. [Google Scholar] [CrossRef]
- Pacitti, D.; Lawan, M.M.; Feldmann, J.; Sweetman, J.; Wang, T.; Martin, S.A.M.; Secombes, C.J. Impact of Selenium Supplementation on Fish Antiviral Responses: A Whole Transcriptomic Analysis in Rainbow Trout (Oncorhynchus mykiss) Fed Supranutritional Levels of Sel-Plex®. BMC Genom. 2016, 17, 116. [Google Scholar] [CrossRef]
- Magray, A.R.; Ribera, J.M.; Isernhagen, L.; Galuska, S.P.; Günther, J.; Verleih, M.; Viergutz, T.; Brunner, R.M.; Ganai, B.A.; Ahmad, F.; et al. Evaluation of Blood Cell Viability Rate, Gene Expression, and O-GlcNAcylation Profiles as Indicative Signatures for Fungal Stimulation of Salmonid Cell Models. Mol. Immunol. 2022, 142, 120–129. [Google Scholar] [CrossRef]
- Lee, S.H.; Hannink, M. Characterization of the Nuclear Import and Export Functions of IκBε. J. Biol. Chem. 2002, 277, 23358–23366. [Google Scholar] [CrossRef]
- Zheng, C.; Yin, Q.; Wu, H. Structural Studies of NF-ΚB Signaling. Cell Res. 2011, 21, 183–195. [Google Scholar] [CrossRef]
- Jacobs, M.D.; Harrison, S.C. Structure of an IkappaBalpha/NF-KappaB Complex. Cell 1998, 95, 749–758. [Google Scholar] [CrossRef]
- Sivasubramanian, N.; Adhikary, G.; Sil, P.C.; Sen, S. Cardiac Myotrophin Exhibits Rel/NF-κB Interacting Activity in Vitro. J. Biol. Chem. 1996, 271, 2812–2816. [Google Scholar] [CrossRef]
- Wolff, B.; Naumann, M. INK4 Cell Cycle Inhibitors Direct Transcriptional Inactivation of NF-κB. Oncogene 1999, 18, 2663–2666. [Google Scholar] [CrossRef] [PubMed]
- Sarais, F.; Kummerow, S.; Montero, R.; Rebl, H.; Köllner, B.; Goldammer, T.; Collet, B.; Rebl, A. PIAS Factors from Rainbow Trout Control NF-κB- and STAT-Dependent Gene Expression. Int. J. Mol. Sci. 2021, 22, 12815. [Google Scholar] [CrossRef] [PubMed]
- Rebl, A.; Rebl, H.; Verleih, M.; Haupt, S.; Köbis, J.M.; Goldammer, T.; Seyfert, H.M. At Least Two Genes Encode Many Variants of Irak3 in Rainbow Trout, but Neither the Full-Length Factor Nor Its Variants Interfere Directly With the TLR-Mediated Stimulation of Inflammation. Front. Immunol. 2019, 10, 2246. [Google Scholar] [CrossRef]
- Chenna, R.; Sugawara, H.; Koike, T.; Lopez, R.; Gibson, T.J.; Higgins, D.G.; Thompson, J.D. Multiple Sequence Alignment with the Clustal Series of Programs. Nucleic Acids Res. 2003, 31, 3497–3500. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent Updates, New Developments and Status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y. Protein Structure and Function Prediction Using I-TASSER. Curr. Protoc. Bioinform. 2015, 52, 5.8.1–5.8.15. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure Visualization for Researchers, Educators, and Developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]







| Gene | Chromosome | LOC Symbol | CDS Length [nt] | UTR Length [bp] | Polyadenylation Motifs a | Instability Motifs | Prot. NCBI Acc.# | Protein Length [aa] | Ankyrin Repeats | Orthologs in Chinook Salmon | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5′ | 3′ | ||||||||||
| nfkbia-a1 | 4 | LOC110522049 | 942 | 91 | 368 | 4 | 4 | XP_021455792 | 313 | 6 | LOC112217446 |
| nfkbia-a2 | 8 | LOC100136058 | 945 | 49 | 407 | 4 | 2 | NP_001117840 | 314 | 6 | LOC112249975 |
| nfkbia-b1 | 19 | LOC110497729 | 1005 | 66 | 454 | 3 | 8 | XP_021429724 | 334 | 5 | LOC112244922 |
| nfkbia-b2 | 25 | LOC110505735 | 963 | 75 | 438 | 4 | 8 | XP_021440813 | 320 | 5 | LOC112256803 |
| nfkbia-c1 | 10 | LOC110533787 | 1176 | 403 | 1408 | 9 | 8 | XP_021473979 | 391 | 6 | LOC112250205 |
| nfkbia-c2 | 12 | LOC110537332 | 1191 | 425 | 1420 | 9 | 13 | XP_021478957 | 396 | 6 | LOC112257706 |
| nfkbie-a1 | 4 | LOC110522047 | 1059 | 392 | 885 | 8 | 16 | XP_021455790 | 352 | 6 | LOC112217444 |
| nfkbie-a2 | 8 | LOC110529432 | 1065 | 449 | 1421 | 5 | 14 | XP_021467277 | 354 | 6 | LOC112249971 |
| nfkbid-a1 | 2 | LOC110538540 | 1461 | 117 | 721 | 2 | 2 | XP_021481102 | 486 | 6 | LOC112245876 |
| nfkbid-a2 | 3 | LOC118936618 | 1707 | 536 | 797 | 1 | 2 | XP_036826041 | 568 | 6 | LOC112227108 |
| nfkbiz-a1 | 7 | LOC110527232 | 1605 | 106 | 1369 | 4 | 8 | XP_036838138 | 534 | 6 | LOC112247547 |
| nfkbiz-a2 | 18 | LOC110495544 | 1605 | 108 | 1369 | 4 | 8 | XP_036807230 | 534 | 6 | LOC112266936 |
| bcl3-a1 | 2 | LOC110537869 | 2061 | 444 | 380 | 7 | 9 | XP_021479984 | 686 | 6 | LOC112235280 |
| bcl3-a2 | 3 | LOC110510160 | 2010 | 1728 | 414 | 8 | 11 | XP_021447196 | 669 | 6 | LOC112226570 |
| Nfkbi Transcript Isoform | Chromosome | Nucleotide NCBI Acc.# | LOC Symbol | CDS Length [nt] | UTR Length [bp] | Polyadenylation Motifs a | Instability Motifs | Prot. NCBI Acc.# | Protein Length [aa] | Ankyrin Repeats | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5′ | 3′ | ||||||||||
| nfkbia-c2.2 | 12 | XM_021623283 | LOC110537332 | 1101 | 425 | 1420 | 9 | 13 | XP_021478958 | 366 | 6 |
| nfkbiz-a1.2 | 7 | XR_005052489 | LOC110527232 | None | 0 | No CDS | |||||
| nfkbiz-a1.3 | 7 | XM_036982244 | LOC110527232 | 1407 | 107 | 555 | None | 0 | XP_036838139 | 468 | 5 |
| nfkbiz-a1.4 | 7 | XM_036982245 | LOC110527232 | 1407 | 107 | 524 | None | 0 | XP_036838140 | 468 | 5 |
| nfkbiz-a2.2 | 18 | XR_005037322 | LOC110495544 | None | 0 | No CDS | |||||
| Gene | Primer Sequence 5′→3′ (Sense, Antisense) | NCBI-Nucleotide Accession Code | Fragment Length [bp] |
|---|---|---|---|
| nfkbia-a | GCATGTCTGATGATGAACAGATG, GAACTCCAGGTCCCAGAAGCC | XM_021600117, NM_001124368 b | 149 |
| nfkbia-b | ACCCAGCTCCCAGCCATTATG, GACATCGATGCACAGGAGCAG | XM_021574049 a, XM_021585138 | 135 |
| nfkbia-c | GGGAGCTGAGGCAGGACTGT, CAACTACTCGGGGGTGAGTGC | XM_021618304, XM_021623282 a, XM_021623283 a | 91 |
| nfkbid-a1 | AGTCAGCCGTATCATCTATGTTTT, CTTTATGTAGGCCGTTTGTGATC | XM_036970146 | 153 |
| nfkbid-a2 | AGGTTGAATCCAGACATCTGTAC, AATAATGGCTAGCTAGTAATGAGC | XM_021625427 | 191 |
| nfkbie-a1 | CTGTAGGGTTATTTATCGTTGTTG, ATTCTCTGCTAGCAAAGTGGTAC | XM_021600115 | 107 |
| nfkbie-a2 | GCAACCGCTACCTTTGGTTTCA, CGGTCAAGACTACCTGGAGTG | XM_021611602 | 140 |
| nfkbiz-a1.1, nfkbiz-a2.1 | TCGTCAATGTCAAGGCATTCAGT, AAGAACCTGGAGAATGAGCAGC | XM_036982243, XM_036951335 | 144 |
| nfkbiz-a1.3, nfkbiz-a1.4 | TTCTGAGCTGACAAACAGTGTTC, ACTAAACCCTCTAACATGAGTTCT | XM_036982244, XM_036982245 | 86 |
| bcl3-a | GCCAGTCGTACAGTGGGAACA, CAAGAACAAGAWGGTAACAGATGT | XM_021624309, XM_021591521 | 160 |
| cxcl8a | CCATTACTGAGGGGATGAGTCTG, GAGACACTGAGATCATTGCCACTC | XM_021625342 a, XM_024415648 | 153 |
| cxcl8b | CTACATGATACAAGGGAGAGG, GGAAGAAGTCATTGTCACAC | XM_036989276, XM_024434566 | 146 |
| il1b | GCTGTGGAAGAACATATAGTGTTGG, GCTACCACAAAGTGCATTTG | XM_036979104, XM_024418276 | 198 |
| il10 | ATGAACAACAGAACACAGAACAACA, CCAATGTAGGAACTACTTCTCCTG | NM_001245099 a, NM_001246350 c, XM_042324963 | 113 |
| nfkbia | GCACAGGAACAATGTAGCG, GATGAACAGATGTACGATGACATTAC | XM_021600117, XM_024377738 | 281 |
| nfkbie | AGGAGCGGTTGGATTCTGCTTAT, CCTTCTCACCACCATCACTGAA | XM_021600115 b, XM_021611602, XM_024419748, XM_024377733 a | 158 |
| tgfb | CATTCCAAGGTGCTAGGTCTGT, ACATCGGCAAGACCCCCAAGA | XM_024386204, XM_024402049 a, XM_021563342, XM_021596503 a | 121 |
| tnf | TTTACCTGGCACTCCAAGGATC, GCATACCCTGAGACAACTCTCT | XM_024407165, XM_021572765 c, XM_036971683 c | 93 |
| Gene/Construct Name | Primer Sequence 5′→3′ (Sense, Antisense) | NCBI-Nucleotide Accession Code | Fragment Length [bp] |
|---|---|---|---|
| nfkbia | CCCAAGCTTGATATGGATGTTTATAGAGTTTCAAACG a, GATGACATTACATTTGGTCAGAATGAATTCTCAAC a | NM_001124368 (position 50–991) | 960 |
| nfkbia-AR12 | CCCAAGCTTATGGATGTTTATAGAGTTTCAAACG a, GTGACCCGCGGATAGCAGACGAATTCGGG a | NM_001124368 (position 50–493) | 465 |
| nfkbia-AR456 | CCCAAGCTTATGAGCGGACACAACTGCCTC a, GATGACATTACATTTGGTCAGAATGAATTCGGG a | NM_001124368 (position 611–991) | 402 |
| nfkbie | CCCAAGCTTCTGATATGCAAAGCGCCGAAGATGCG a, CCCGAATTCCTGATCAGAATGGCCCTCCAACCAC a | XM_021611602 (position 450–1511) | 1080 |
| nfkbie-AR12 | CCCAAGCTTATGCAAAGCGCCGAAGATGCG a, GGGGCCAGCCTGGAGCTGAGATCTGGG a | XM_021611602 (position 450–977) | 546 |
| nfkbie-AR456 | CCCAAGCTTATGAGAGGTCTCACCTGTCTC a, TCAGTGGTTGGAGGGCCATTCAGATCTGGG a | XM_021611602 (position 1104–1511) | 429 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van Muilekom, D.R.; Collet, B.; Rebl, H.; Zlatina, K.; Sarais, F.; Goldammer, T.; Rebl, A. Lost and Found: The Family of NF-κB Inhibitors Is Larger than Assumed in Salmonid Fish. Int. J. Mol. Sci. 2023, 24, 10229. https://doi.org/10.3390/ijms241210229
van Muilekom DR, Collet B, Rebl H, Zlatina K, Sarais F, Goldammer T, Rebl A. Lost and Found: The Family of NF-κB Inhibitors Is Larger than Assumed in Salmonid Fish. International Journal of Molecular Sciences. 2023; 24(12):10229. https://doi.org/10.3390/ijms241210229
Chicago/Turabian Stylevan Muilekom, Doret R., Bertrand Collet, Henrike Rebl, Kristina Zlatina, Fabio Sarais, Tom Goldammer, and Alexander Rebl. 2023. "Lost and Found: The Family of NF-κB Inhibitors Is Larger than Assumed in Salmonid Fish" International Journal of Molecular Sciences 24, no. 12: 10229. https://doi.org/10.3390/ijms241210229
APA Stylevan Muilekom, D. R., Collet, B., Rebl, H., Zlatina, K., Sarais, F., Goldammer, T., & Rebl, A. (2023). Lost and Found: The Family of NF-κB Inhibitors Is Larger than Assumed in Salmonid Fish. International Journal of Molecular Sciences, 24(12), 10229. https://doi.org/10.3390/ijms241210229









