Bacterial Therapy of Cancer: A Way to the Dustbin of History or to the Medicine of the Future?
Abstract
1. Introduction
2. Bacterial Dualism: What Can Kill Can Also Heal
2.1. In Pursuit of the “Cancer Germ”
2.2. Scientists Make Bacterial “Anti-Cancer Vaccines” Almost Blindly, and It Sometimes Works!
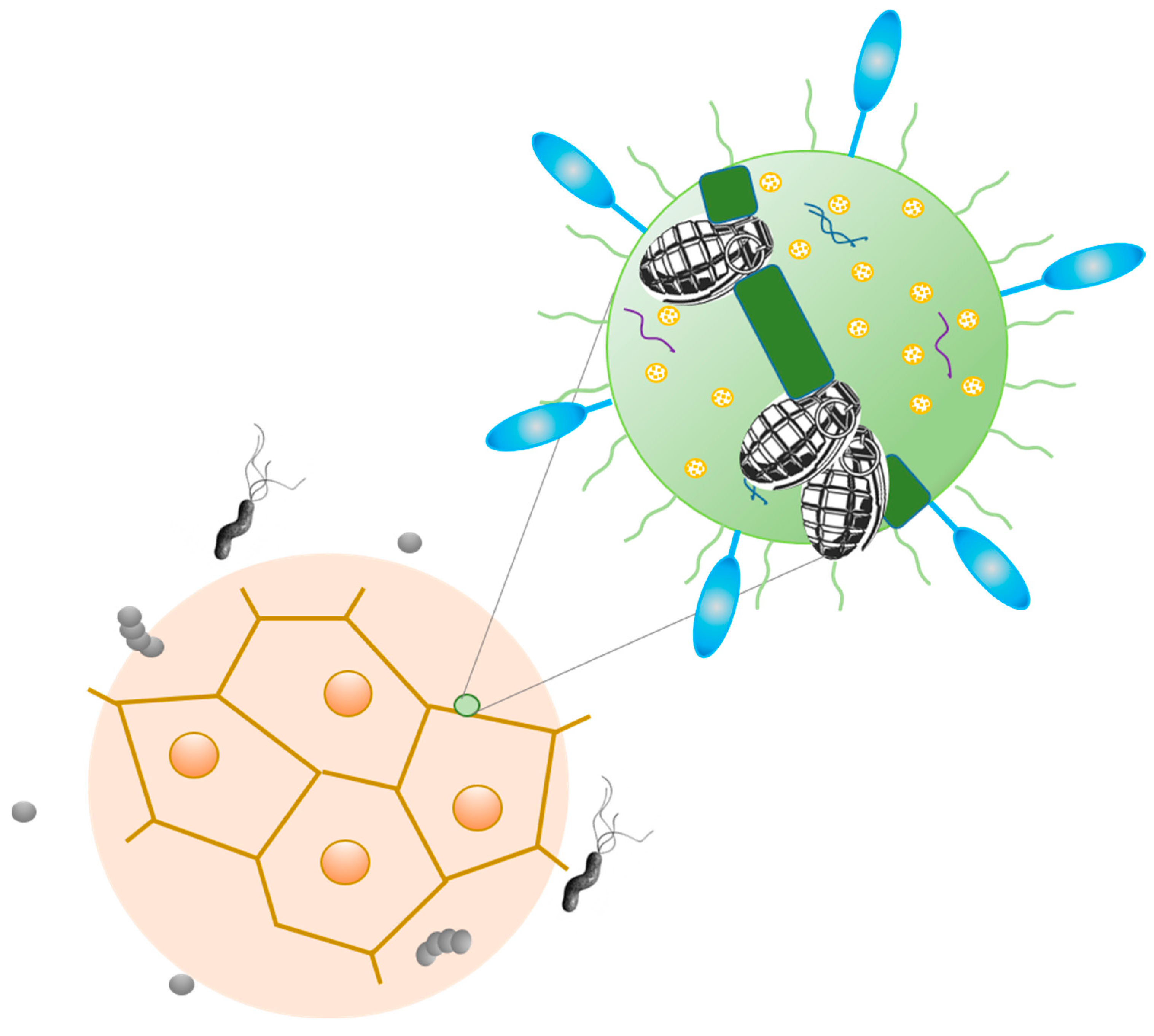
3. Tumor Microbiome: Peaceful Inhabitants or Malicious Instigators?
4. Tumorigenic Effect of Bacteria: Microbes, Which Are Typically Linked to Cancer, and Probable Mechanisms of Carcinogenic Action
5. Bacteria in Cancer Therapy
5.1. Scientists’ Favorite “Toys” in Looking for a Panacea
5.2. Mechanisms of Antitumor Action
5.2.1. Stimulation of the Immune Response
5.2.2. Production of the Cell-Death-Inducing Agents
6. Future Perspectives: Bacteriobots?
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Felgner, S.; Kocijancic, D.; Frahm, M.; Weiss, S. Bacteria in Cancer Therapy: Renaissance of an Old Concept. Int. J. Microbiol. 2016, 2016, 8451728. [Google Scholar] [CrossRef] [PubMed]
- Kucerova, P.; Cervinkova, M. Spontaneous regression of tumour and the role of microbial infection—Possibilities for cancer treatment. Anti-Cancer Drugs 2016, 27, 269. [Google Scholar] [CrossRef] [PubMed]
- Challis, G.B.; Stam, H.J. The spontaneous regression of cancer. A review of cases from 1900 to 1987. Acta Oncol. 1990, 29, 545–550. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Wang, Y.; Xu, H.; Guo, Y.; Xia, W.; Zhao, C.; Zhao, X.; Wu, J. Recent advances in bacterial therapeutics based on sense and response. Acta Pharm. Sin. B 2023, 13, 1014–1027. [Google Scholar] [CrossRef] [PubMed]
- Felgner, S.; Pawar, V.; Kocijancic, D.; Erhardt, M.; Weiss, S. Tumour-targeting bacteria-based cancer therapies for increased specificity and improved outcome. Microbial. Biotechnol. 2017, 10, 1074–1078. [Google Scholar] [CrossRef] [PubMed]
- Nallar, S.C.; Xu, D.-Q.; Kalvakolanu, D.V. Bacteria and genetically modified bacteria as cancer therapeutics: Current advances and challenges. Cytokine 2017, 89, 160–172. [Google Scholar] [CrossRef]
- Cummins, J.; Tangney, M. Bacteria and tumours: Causative agents or opportunistic inhabitants? Infect. Agents Cancer 2013, 8, 11. [Google Scholar] [CrossRef]
- Zu, C.; Wang, J. Tumor-colonizing bacteria: A potential tumor targeting therapy. Crit. Rev. Microbiol. 2013, 51, 1. [Google Scholar] [CrossRef]
- Patyar, S.; Joshi, R.; Prasad Byrav, D.S.; Prakash, A.; Medhi, B.; Das, B.K. Bacteria in cancer therapy: A novel experimental strategy. J. Biomed. Sci. 2010, 17, 21. Available online: http://www.jbiomedsci.com/content/17/1/21 (accessed on 1 April 2023). [CrossRef]
- Forbes, N.S. Engineering the perfect (bacterial) cancer therapy. Nat. Rev. Cancer 2010, 10, 785–794. [Google Scholar] [CrossRef]
- Ryan, R.M.; Green, J.; Lewis, C.E. Use of bacteria in anti-cancer therapies. BioEssays 2006, 28, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, A.M. Microorganisms and cancer: Quest for a therapy. J. Bacteriol. 2003, 185, 2683–2686. [Google Scholar] [CrossRef] [PubMed]
- De Martel, C.; Georges, D.; Bray, F.; Ferlay, J.; Clifford, G.M. Global burden of cancer attributable to infections in 2018: A worldwide incidence analysis. Lancet Glob. Health 2020, 8, e180–e190. [Google Scholar] [CrossRef] [PubMed]
- Rubtsov, B.V. Bacteriological examination of malignant tumors (retrospective overview). I.P. Pavlov Rus. Med.-Biol. Vestn. 2015, 23, 155–167. (In Russian) [Google Scholar] [CrossRef]
- Burke, J. Cancer and infection. Aust. J. Med. Med. Herbal. 2008, 20, 47–55. [Google Scholar]
- Robertson, W.F. The relation of carcinoma to infection. Br. Med. J. 1921, 2, 929. [Google Scholar] [CrossRef]
- Dudgeon, L.S.; Dunkley, E.V. The Micrococcus neoformans: Its cultural characters and pathogenicity and the results of the estimation of the opsonic and agglutinative properties of the serum of patients suffering from malignant disease on this organism and on the Staphylococcus Albus. J. Hyg. 1907, 7, 13–21. [Google Scholar] [CrossRef]
- Wainwright, M.; Al Talih, A. Is this the historical ‘cancer germ’? Med. Hypotheses 2003, 60, 290–292. [Google Scholar] [CrossRef]
- Nuzum, J. The experimental production of metastasing carcinoma in the breast of the dog and primary epithelioma in man by repeated inoculation of a Micrococcus isolated from human breast cancer. Surg. Gynecol. Obstet. 1925, 11, 343–352. [Google Scholar]
- Blumenthal, F. The specific parasites of cancer. Zbl. f. Bact. 1927, 104, 11–17. [Google Scholar]
- Livingston, V.W.; Livingston, A.M. Demonstration of progenitor cryptocides in the blood of patients with collagen and neoplastic diseases. Trans. N. Y. Acad. Sci. 1972, 34, 433–453. [Google Scholar] [CrossRef] [PubMed]
- Livingston, V.W.C.; Livingston, A.M. Some cultural, immunological, and biochemical properties of Progenitor cryptocides. Tr. N. Y. Acad. Sci. 1974, 36, 569–582. [Google Scholar] [CrossRef]
- Dróżdż, M.; Makuch, S.; Cieniuch, G.; Woźniak, M.; Ziółkowski, P. Obligate and facultative anaerobic bacteria in targeted cancer therapy: Current strategies and clinical applications. Life Sci. 2020, 261, 118296. [Google Scholar] [CrossRef] [PubMed]
- Sawant, S.S.; Patil, S.M.; Gupta, V.; Kunda, N.K. Microbes as medicines: Harnessing the power of bacteria in advancing cancer treatment. Int. J. Mol. Sci. 2020, 21, 7575. [Google Scholar] [CrossRef] [PubMed]
- Nauts, H.C.; McLaren, J.R. Coley toxins—The first century. In Consensus on Hyperthermia for the 1990s: Clinical Practice in Cancer Treatment; Springer: Boston, MA, USA, 1990; pp. 483–500. [Google Scholar] [CrossRef]
- Wiemann, B.; Starnes, C.O. Coley’s toxins, tumor necrosis factor and cancer research: A historical perspective. Pharmacol. Ther. 1994, 64, 529–564. [Google Scholar] [CrossRef]
- Bickels, J.; Kollender, Y.; Merinsky, O.; Meller, I. Coley’s toxin: Historical Perspective. Isr. Med. Assoc. J. 2002, 4, 471–472. [Google Scholar]
- Nybo, K. Part I: Fighting cancer with deadly bacteria. Biotechnology 2018, 64, 6–8. [Google Scholar] [CrossRef]
- Troitskaya, A.S. Microscopy data of a thick drop of blood in patients with malignant tumors (preliminary report). In Book of Scientific Papers of Clinicians of the Kaluga Region; Troitskaya, A.S., Ed.; Kaluga Publishing House: Kaluga, Russia, 1959; pp. 98–103. [Google Scholar]
- Troitskaya, A.S. Malignant tumors in mice infected with hemocultures isolated from patients with cancer and sarcoma. In Book of Scientific Papers of Clinicians of the Kaluga Region; Troitskaya, A.S., Nikolskaya, A.P., Eds.; Kaluga Publishing House: Kaluga, Russia, 1963; pp. 113–131. [Google Scholar]
- Krestovnikova, V.A. Microbiological Study of Tumors; Kalin, G.P., Ed.; Medgiz: Moscow, USSR, 1960; p. 188. [Google Scholar]
- Andreeva, Z.M.; Khramova, N.I.; Ershova, E.B.; Barkhudaryan, V.A. The Bacterial Strain Corynebacterium Krestovnikova-Troitskaja FSBI “SCEEMP” № 226 for the Preparation of an Immunostimulatory. Russian Federation Patent № 2027755, 27 January 1995. Kaluga Municipal Scientific and Technical Center “Impulse”: Kaluga, Russia. № 93026850/13, 27.01.1995. [Google Scholar]
- Andreeva, Z.M.; Khramova, N.I.; Ershova, E.B.; Barkhudaryan, V.A. The Bacterial Strain Corynebacterium Krestovnikova-Troitskaja FSBI “SCEEMP” № 227 for the Preparation of an Immunostimulatory. Russian Federation Patent № 2027756, 27 January 1995. The Patent № 2027756 of the Russian Federation. Kaluga Municipal Scientific and Technical Center “Impulse”: Kaluga, Russia. № 93026851/13, 27.01.1995. [Google Scholar]
- Bhatt, A.P.; Redinbo, M.R.; Bultman, S.J. The role of the microbiome in cancer development and therapy. CA Cancer. J. Clin. 2017, 67, 326–344. [Google Scholar] [CrossRef]
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020, 368, 973–980. [Google Scholar] [CrossRef]
- Livyatan, I.; Nejman, D.; Shental, N.; Straussman, R. Characterization of the human tumor microbiome reveals tumor-type specific intra-cellular bacteria. Oncoimmunology 2020, 9, 1800957. [Google Scholar] [CrossRef]
- Atreya, C.E.; Turnbaugh, P.J. Probing the tumor micro(b)environment. Science 2020, 368, 938. [Google Scholar] [CrossRef] [PubMed]
- Greathouse, K.L.; Stone, J.K.; Harris, C.C. Cancer-type-Specific Bacteria: Freeloaders or Partners? Cancer Cell 2020, 38, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Monographs on the Identification of Carcinogenic Hazards to Humans; IARC: Lyon, France, 2019; Available online: https://monographs.iarc.fr/wp-content/uploads/2019/07/Preamble-2019.pdf (accessed on 1 January 2019).
- Smith, M.T.; Guyton, K.Z.; Gibbons, C.F.; Fritz, J.M.; Portier, C.J.; Rusyn, I.; DeMarini, D.M.; Caldwell, J.C.; Kavlock, R.J.; Lambert, P.; et al. Key characteristics of carcinogens as a basis for organizing data on mechanisms of carcinogenesis. Environ. Health Perspect. 2016, 124, 713–721. [Google Scholar] [CrossRef] [PubMed]
- IARC Helicobacter pylori Working Group. Helicobacter Pylori Eradication as a Strategy for Preventing Gastric Cancer; IARC Working Group Reports, No. 8; International Agency for Research on Cancer: Lyon, France, 2014; Available online: http://www.iarc.fr/en/publications/pdfsonline/wrk/wrk8/index.php (accessed on 4 December 2013).
- Muzaheed. Helicobacter pylori oncogenicity: Mechanism, prevention, and risk factors. Sci. World J. 2020, 2020, 3018326. [Google Scholar] [CrossRef]
- Alipour, M. Molecular mechanism of Helicobacter pylori-induced gastric cancer. J. Gastrointest. Cancer. 2021, 52, 23–30. [Google Scholar] [CrossRef]
- Hartl, K.; Sigal, M. Microbe-Driven Genotoxicity in Gastrointestinal Carcinogenesis. Int. J. Mol. Sci. 2020, 21, 7439. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.H.; Parsonnet, J. Role of bacteria in oncogenesis. Clin. Microbiol. Rev. 2010, 23, 837–857. [Google Scholar] [CrossRef]
- Al-Hilu, S.A.; Al-Shujairi, W.H. Dual role of bacteria in carcinoma: Stimulation and inhibition. Int. J. Microbiol. 2020, 76, 1. [Google Scholar] [CrossRef]
- Song, S.; Vuai, M.S.; Zhong, M. The role of bacteria in cancer therapy–enemies in the past, but allies at present. Infect. Agents Cancer 2018, 13, 9. [Google Scholar] [CrossRef]
- Koshiol, J.; Wozniak, A.; Cook, P.; Adaniel, C.; Acevedo, J.; Azócar, L.; Hsing, A.W.; Roa, J.C.; Pasetti, M.F.; Miquel, J.F.; et al. Gallbladder Cancer Chile Working Group. Salmonella enterica serovar Typhi and gallbladder cancer: A case-control study and meta-analysis. Cancer Med. 2016, 5, 3235–3310. [Google Scholar] [CrossRef]
- Khan, A.A.; Bano, Y. Salmonella enterica subsp. enterica host-pathogen interactions and their implications in gallbladder cancer. Microb. Pathog. 2021, 157, 105011. [Google Scholar] [CrossRef]
- Di Domenico, E.G.; Cavallo, I.; Pontone, M.; Toma, L.; Ensoli, F. Biofilm producing Salmonella Typhi: Chronic colonization and development of gallbladder cancer. Int. J. Mol. Sci. 2017, 18, 1887. [Google Scholar] [CrossRef]
- He, Z.; Gharaibeh, R.Z.; Newsome, R.C.; Pope, J.L.; Dougherty, M.W.; Tomkovich, S.; Pons, B.; Mirey, G.; Vignard, J.; Hendrixson, D.R.; et al. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut 2019, 68, 289–300. [Google Scholar] [CrossRef]
- Ponzoni, M.; Ferreri, A.J. Bacteria associated with marginal zone lymphomas. Best Pract. Res. Clin. Haematol. 2017, 30, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Travaglino, A.; Pace, M.; Varricchio, S.; Della Pepa, R.; Iuliano, A.; Picardi, M.; Pane, F.; Staibano, S.; Mascolo, M. Prevalence of Chlamydia psittaci, Chlamydia pneumoniae, and Chlamydia trachomatis determined by molecular testing in ocular adnexa lymphoma specimens. Am. J. Clin. Pathol. 2020, 153, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.A.; Sheikh, J.A.; Ehtesham, N.Z.; Hira, S.; Hasnain, S.E. Can Mycobacterium tuberculosis infection lead to cancer? Call for a paradigm shift in understanding TB and cancer. Int. J. Med. Microbiol. 2022, 312, 151558. [Google Scholar] [CrossRef] [PubMed]
- Shang, F.M.; Liu, H.L. Fusobacterium nucleatum and colorectal cancer: A review. World J. Gastrointest. Oncol. 2018, 10, 71–81. [Google Scholar] [CrossRef]
- Hashemi Goradel, N.; Heidarzadeh, S.; Jahangiri, S.; Farhood, B.; Mortezaee, K.; Khanlarkhani, N.; Negahdari, B. Fusobacterium nucleatum and colorectal cancer: A mechanistic overview. J. Cell Physiol. 2019, 234, 2337–2344. [Google Scholar] [CrossRef]
- Li, S.; Liu, J.; Zheng, X.; Ren, L.; Yang, Y.; Li, W.; Fu, W.; Wang, J.; Du, G. Tumorigenic bacteria in colorectal cancer: Mechanisms and treatments. Cancer Biol. Med. 2021, 19, 147–162. [Google Scholar] [CrossRef]
- Mármol, I.; Sánchez-de-Diego, C.; Pradilla Dieste, A.; Cerrada, E.; Rodriguez Yoldi, M.J. Colorectal carcinoma: A general overview and future perspectives in colorectal cancer. Int. J. Mol. Sci. 2017, 18, 197. [Google Scholar] [CrossRef]
- Cheng, Y.; Ling, Z.; Li, L. The intestinal microbiota and colorectal cancer. Front. Immunol. 2020, 11, 615056. [Google Scholar] [CrossRef] [PubMed]
- Galdy, S.; Nastasi, G. Streptococcus bovis endocarditis and colon cancer: Myth or reality? A case report and literature review. BMJ Case Rep. 2012, 2012, bcr2012006961. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; Cerqueira, F.; Medeiros, R. Chlamydia trachomatis infection: Implications for HPV status and cervical cancer. Arch. Gynecol. Obstet. 2014, 289, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Irfan, M.; Delgado, R.Z.R.; Frias-Lopez, J. The oral microbiome and cancer. Front. Immunol. 2020, 11, 591088. [Google Scholar] [CrossRef] [PubMed]
- Lamont, R.J.; Fitzsimonds, Z.R.; Wang, H.; Gao, S. Role of Porphyromonas gingivalis in oral and orodigestive squamous cell carcinoma. Periodontology 2022, 89, 154–165. [Google Scholar] [CrossRef]
- Duong, M.T.-Q.; Qin, Y.; You, S.-H.; Min, J.-J. Bacteria-cancer interactions: Bacteria-based cancer therapy. Exp. Mol. Med. 2019, 51, 152. [Google Scholar] [CrossRef]
- Kramer, M.G.; Masner, M.; Ferreira, F.A.; Hoffman, R.M. Bacterial therapy of cancer: Promises, limitations, and insights for future directions. Front. Microbiol. 2018, 9, 16. [Google Scholar] [CrossRef]
- Kouhsari, E.; Ghadimi-Daresajini, A.; Abdollahi, H.; Amirmozafari, N.; Mahdavi, S.R.; Abbasian, S.; Mousavi, S.H.; Yaseri, H.F.; Moghaderi, M. The potential roles of bacteria to improve radiation treatment outcome. Clin. Transl. Oncol. 2018, 20, 127–139. [Google Scholar] [CrossRef]
- Laliani, G.; Sorboni, S.G.; Lari, R.; Yaghoubi, A.; Soleimanpour, S.; Khazaei, M.; Hasanian, S.M.; Avan, A. Bacteria and cancer: Different sides of the same coin. Life Sci. 2020, 246, 117398. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, W.; Wu, X.; Zhang, Y.; Mannion, C.; Brouchkov, A.; Man, Y.; Chen, T. Oncolytic bacteria and their potential role in bacterium-mediated tumour therapy: A conceptual analysis. J. Cancer. 2019, 10, 4442. [Google Scholar] [CrossRef]
- Chien, T.; Doshi, A.; Danino, T. Advances in bacterial cancer therapies using synthetic biology. Curr. Opin. Syst. Biol. 2017, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Divyashree, M.; Prakash, S.K.; Aditya, V.; Aljabali, A.A.; Alzahrani, K.J.; Azevedo, V.; Góes-Neto, A.; Tambuwala, M.M.; Barh, D. Bugs as drugs: Neglected but a promising future therapeutic strategy in cancer. Future Oncol. 2022, 18, 1609–1626. [Google Scholar] [CrossRef] [PubMed]
- Sieow, B.F.L.; Wun, K.S.; Yong, W.P.; Hwang, I.Y.; Chang, M.W. Tweak to treat: Reprograming bacteria for cancer treatment. Trends Cancer. 2021, 7, 447–464. [Google Scholar] [CrossRef] [PubMed]
- Łukasiewicz, K.; Fol, M. Microorganisms in the treatment of cancer: Advantages and limitations. J. Immunol. Res. 2018, 2018, 2397808. [Google Scholar] [CrossRef] [PubMed]
- Fol, M.; Kozinski, P.; Kulesza, J.; Białecki, P.; Druszczynska, M. Dual nature of relationship between mycobacteria and cancer. Int. J. Mol. Sci. 2021, 22, 8332. [Google Scholar] [CrossRef]
- Pettenati, C.; Ingersoll, M.A. Mechanisms of BCG immunotherapy and its outlook for bladder cancer. Nat. Rev. Urol. 2018, 15, 615–625. [Google Scholar] [CrossRef]
- Baindara, P.; Mandal, S.M. Bacteria and bacterial anticancer agents as a promising alternative for cancer therapeutics. Biochimie 2020, 177, 164–189. [Google Scholar] [CrossRef]
- Staedtke, V.; Roberts, N.J.; Bai, R.-Y.; Zhou, S. Clostridium novyi-NT in cancer therapy. Genes Dis. 2016, 3, 144–152. [Google Scholar] [CrossRef]
- Zhou, S.; Gravekamp, C.; Bermudes, D.; Liu, K. Tumor-targeting bacteria engineered to fight cancer. Nat. Rev. Cancer 2018, 18, 727–743. [Google Scholar] [CrossRef]
- Roberts, N.J.; Zhang, L.; Janku, F.; Collins, A.; Bai, R.-Y.; Staedtke, V.; Rusk, A.W.; Tung, D.; Miller, M.; Roix, J.; et al. Intratumoral injection of Clostridium novyi-NT spores induces antitumor responses. Sci. Transl. Med. 2014, 6, 249ra111. [Google Scholar] [CrossRef]
- Feng, X.; He, P.; Zeng, C.; Li, Y.; Das, S.K.; Li, B.; Yang, H.-F.; Du, Y. Novel insights into the role of Clostridium novyi-NT related combination bacteriolytic therapy in solid tumors. Oncol. Lett. 2021, 21, 110. [Google Scholar] [CrossRef] [PubMed]
- Staedtke, V.; Baim, R.Y.; Sun, W.; Huang, J.; Kibler, K.K.; Tyler, B.M.; Gallia, G.L.; Kinzler, K.; Vogelstein, B.; Zhou, S.; et al. Clostridium novyi-NT can cause regression of orthotopically implanted glioblastomas in rats. Oncotargets 2015, 6, 5536–5546. [Google Scholar] [CrossRef] [PubMed]
- Liang, K.; Liu, Q.; Kong, Q. New technologies in developing recombinant—Attenuated bacteria for cancer therapy. Biotechnol. Bioeng. 2021, 118, 513–530. [Google Scholar] [CrossRef] [PubMed]
- Schmitz-Winnenthal, F.H.; Hohmann, N.; Schmidt, T.; Podola, L.; Friedrich, T.; Lubenau, H.; Springer, M.; Wieckowski, S.; Breiner, K.M.; Mikus, G.; et al. A phase 1 trial extension to assess immunologic efficacy and safety of prime-boost vaccination with VXM01, an oral T cell vaccine against VEGFR2, in patients with advanced pancreatic cancer. Oncoimmunology 2018, 7, e1303584. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.H.; Nowicki, C.; Giurini, E.F.; Marzo, A.L.; Zloza, A. Bacterial-based cancer therapy (BBCT): Recent advances, current challenges, and future prospects for cancer immunotherapy. Vaccines 2021, 9, 1497. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, B.; Vadalà, M.; Roncati, L.; Garelli, A.; Scandone, F.; Bondi, M.; Cermelli, C. The long—Standing history of Corynebacterium parvum, immunity, and viruses. J. Med. Virol. 2020, 92, 2429. [Google Scholar] [CrossRef]
- Morrow, Z.T.; Powers, Z.M.; Sauer, J.D. Listeria monocytogenes cancer vaccines: Bridging innate and adaptive immunity. Curr. Clin. Micro. Rpt. 2019, 6, 213–224. [Google Scholar] [CrossRef]
- Gentschev, I.; Petrov, I.; Ye, M.; Kafuri Cifuentes, L.; Toews, R.; Cecil, A.; Oelschaeger, T.A.; Szalay, A.A. Tumor colonization and therapy by Escherichia coli Nissle 1917 strain in syngeneic tumor-bearing mice is strongly affected by the gut microbiome. Cancers 2022, 4, 6033. [Google Scholar] [CrossRef]
- Chen, J.; Li, X.; Liu, Y.; Su, T.; Lin, C.; Shao, L.; Li, L.; Li, W.; Niu, G.; Yu, J.; et al. Engineering a probiotic strain of Escherichia coli to induce the regression of colorectal cancer through production of 5-aminolevulinic acid. Microb. Biotechnol. 2021, 14, 2130–2139. [Google Scholar] [CrossRef]
- Pang, Z.; Gu, M.D.; Tang, T. Pseudomonas aeruginosa in cancer therapy: Current knowledge, challenges and future perspectives. Front. Oncol. 2022, 12, 891187. [Google Scholar] [CrossRef]
- Wei, X.; Du, M.; Chen, Z.; Yuan, Z. Recent Advances in Bacteria-Based Cancer Treatment. Cancers 2022, 14, 4945. [Google Scholar] [CrossRef] [PubMed]
- Dunn, G.P.; Old, L.J.; Schreiber, R.D. The three Es of cancer immunoediting. Annu. Rev. Immunol. 2004, 22, 329–360. [Google Scholar] [CrossRef] [PubMed]
- Torres, W.; Lameda, V.; Olivar, L.C.; Navarro, C.; Fuenmayor, J.; Pérez, A.; Mindiola, A.; Rojas, M.; Martínez, M.S.; Velasco, M.; et al. Bacteria in cancer therapy: Beyond immunostimulation. J. Cancer Metastasis Treat. 2018, 4, 4. [Google Scholar] [CrossRef]
- Huang, X.; Pan, J.; Xu, F.; Shao, B.; Wang, Y.; Guo, X.; Zhou, S. Bacteria-Based Cancer Immunotherapy. Adv. Sci. 2021, 7, 200532. [Google Scholar] [CrossRef]
- Carroll, C.S.E.; Andrew, E.R.; Malik, L.; Elliott, K.F.; Brennan, M.; Meyer, J.; Hintze, A.; Almonte, A.A.; Lappin, C.; MacPherson, P.; et al. Simple and effective bacterial-based intratumoral cancer immunotherapy. J. Immunother. Cancer 2021, 9, e002688. [Google Scholar] [CrossRef]
- Sfakianos, J.P.; Salome, B.; Daza, J.; Farkas, A.; Bhardwaj, N.; Horowitz, A. Bacillus Calmette-Guerin (BCG): Its fight against pathogens and cancer. Urol. Oncol. Semin. Orig. Investig. 2021, 39, 121–129. [Google Scholar] [CrossRef]
- Bisiaux, A.; Thiounn, N.; Timsit, M.O.; Eladaoui, A.; Chang, H.H.; Mapes, J.; Mogenet, A.; Bresson, J.L.; Prié, D.; Béchet, S.; et al. Molecular analyte profiling of the early events and tissue conditioning following intravesical Bacillus Calmette-Guerin therapy in patients with superficial bladder cancer. J. Urol. 2009, 181, 1571–1580. [Google Scholar] [CrossRef]
- Forbes, N.S.; Coffin, R.S.; Deng, L.; Evgin, L.; Fiering, S.; Giacalone, M.; Gravekamp, C.; Gulley, J.L.; Gunn, H.; Hoffman, R.M.; et al. White paper on microbial anti-cancer therapy and prevention. J. Immunother. Cancer 2018, 6, 78. [Google Scholar] [CrossRef]
- Jiang, S.N.; Park, S.H.; Lee, H.J.; Zheng, J.H.; Kim, H.S.; Bom, H.S.; Hong, Y.; Szardenings, M.; Shin, M.G.; Kim, S.C.; et al. Engineering of bacteria for the visualization of targeted delivery of a cytolytic anticancer agent. Mol. Ther. 2013, 21, 1985. [Google Scholar] [CrossRef]
- Liu, X.; Jiang, S.; Piao, L.; Yuan, F. Radiotherapy combined with an engineered Salmonella typhimurium inhibits tumor growth in a mouse model of colon cancer. Exp. Anim. 2016, 65, 413–418. [Google Scholar] [CrossRef]
- Jiang, S.N.; Phan, T.X.; Nam, T.K.; Nguyen, V.H.; Kim, H.S.; Bom, H.S.; Choy, H.E.; Hong, Y.; Min, J.J. Inhibition of tumor growth and metastasis by a combination of escherichia coli-mediated cytolytic therapy and radiotherapy. Mol. Ther. 2010, 18, 635. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Schulte, W.; Pink, D.; Phipps, K.; Zijlstra, A.; Lewis, J.D.; Waisman, D.M. Sensitivity of cancer cells to truncated diphtheria toxin. PLoS ONE 2010, 5, 10498. [Google Scholar] [CrossRef]
- Shafiee, F.; Aucoin, M.G.; Jahanian-Najafabadi, A. Targeted diphtheria toxin-based therapy: A review article. Front Microbiol. 2019, 10, 2340. [Google Scholar] [CrossRef] [PubMed]
- Proietti, S.; Nardicchi, V.; Porena, M.; Giannantoni, A. Botulinum toxin type-A toxinactivity in prostate cancer cell lines. Urologia J. 2012, 79, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Karpiński, T.M.; Adamczak, A. Anticancer activity of bacterial proteins and peptides. Pharmacy 2018, 10, 54. [Google Scholar] [CrossRef]
- Bandala, C.; Perez-Santos, J.L.; Lara-Padilla, E.; Delgado Lopez, G.; Anaya-Ruiz, M. Effect of botulinum toxin A on proliferation and apoptosis in the T47D breast cancer cell line. Asian Pac. J. Cancer Prev. 2013, 14, 891. [Google Scholar] [CrossRef]
- Michl, P.; Buchholz, M.; Rolke, M.; Kunsch, S.; Löhr, M.; McClane, B.; Tsukita, S.; Leder, G.; Adler, G.; Gress, T.M. Claudin-4: A new target for pancreatic cancer treatment using Clostridium perfringens enterotoxin. Gastroenterology 2001, 121, 678–684. [Google Scholar] [CrossRef]
- Bhattacharjee, R.N.; Park, K.S.; Uematsu, S.; Okada, K.; Hoshino, K.; Takeda, K.; Takeuchi, O.; Akira, S.; Iida, T.; Honda, T. Escherichia coli verotoxin 1 mediates apoptosis in human HCT116 colon cancer cells by inducing overexpression of the GADD family of genes and S phase arrest. FEBS Lett. 2005, 579, 6604. [Google Scholar] [CrossRef]
- Rommasi, F. Bacterial-based methods for cancer treatment: What we know and where we are. Oncol. Ther. 2022, 10, 23–54. [Google Scholar] [CrossRef]
- Michalska, M.; Wolf, P. Pseudomonas Exotoxin A: Optimized by evolution for effective killing. Front. Microbiol. 2015, 6, 963. [Google Scholar] [CrossRef]
- Nguyen, H.A.; Su, Y.; Zhang, J.Y.; Antanasijevic, A.; Caffrey, M.; Schalk, A.M.; Liu, L.; Rondelli, D.; Oh, A.; Mahmud, D.L.; et al. A novel l-Asparaginase with low l-Glutaminase coactivity is highly efficacious against both T-and B-cell acute lymphoblastic leukemias in vivo ASNase efficacy with low l-Glutaminase coactivity. Cancer Res. 2018, 78, 1549–1560. [Google Scholar] [CrossRef]
- Ghasemian, A.; Al-marzoqi, A.H.; Al-abodi, H.R.; Alghanimi, Y.K.; Kadhum, S.A.; Shokouhi Mostafavi, S.K.; Fattahi, A. Bacterial l-asparaginases for cancer therapy: Current knowledge and future perspectives. J. Cell. Physiol. 2019, 234, 19271–19279. [Google Scholar] [CrossRef] [PubMed]
- Mayakrishnan, V.; Kannappan, P.; Tharmalingam, N.; Bose, R.J.C.; Madheswaran, T.; Ramasamy, M. Bacterial cancer therapy: A turning point for new paradigms. Drug Discovery Today 2022, 27, 2043–2050. [Google Scholar] [CrossRef] [PubMed]
- Zam, W. Arginine enzymatic deprivation and diet restriction for cancer treatment. Braz. J. Pharm. Sci. 2017, 3, 53. [Google Scholar] [CrossRef]
- Fiedler, T.; Strauss, M.; Hering, S.; Redanz, U.; William, D.; Rosche, Y.; Classen, C.F.; Kreikemeyer, B.; Linnebacher, M.; Maletzki, C. Arginine deprivation by arginine deiminase of Streptococcus pyogenes controls primary glioblastoma growth in vitro and in vivo. Cancer Biol. Ther. 2015, 16, 1047. [Google Scholar] [CrossRef]
- Kunda, N.K. Antimicrobial peptides as novel therapeutics for non-small cell lung cancer. Drug Discovery Today 2020, 25, 238–247. [Google Scholar] [CrossRef]
- Kaur, S.; Kaur, S. Bacteriocins as potential anticancer agents. Front. Pharmacol. 2015, 6, 272. [Google Scholar] [CrossRef]
- Gudina, E.J.; Rangarajan, V.; Sen, R.; Rodrigues, L.R. Potential therapeutic applications of biosurfactants. Trends Pharmacol. Sci. 2013, 34, 667. [Google Scholar] [CrossRef]
- Duarte, C.; Gudiña, E.J.; Lima, C.F.; Rodrigues, L.R. Effects of biosurfactants on the viability and proliferation of human breast cancer cells. AMB Express 2014, 4, 1. [Google Scholar] [CrossRef]
- Marchant, R.; Banat, I.M. Microbial biosurfactants: Challenges and opportunities for future exploitation. Trends Biotechnol. 2012, 30, 558–565. [Google Scholar] [CrossRef]
- Abdelli, F.; Jardak, M.; Elloumi, J.; Stien, D.; Cherif, S.; Mnif, S.; Aifa, S. Antibacterial, anti-adherent and cytotoxic activities of surfactin(s) from a lipolytic strain Bacillus safensis F. Biodegradation 2019, 30, 287. [Google Scholar] [CrossRef] [PubMed]
- Saini, H.S.; Barragán-Huerta, B.E.; Lebrón-Paler, A.; Pemberton, J.E.; Vázquez, R.R.; Burns, A.M.; Marron, M.T.; Seliga, C.J.; Gunatilaka, A.A.L.; Maier, R.M. Efficient purification of the biosurfactant viscosin from Pseudomonas libanensis strain M9-3 and its physicochemical and biological properties. J. Nat. Prod. 2008, 71, 1011–1015. [Google Scholar] [CrossRef] [PubMed]
- De Vleeschouwer, M.; Van Kersavond, T.; Verleysen, Y.; Sinnaeve, D.; Coenye, T.; Martins, J.C.; Madder, A. Identification of the molecular determinants involved in antimicrobial activity of pseudodesmin a, a cyclic lipopeptide from the viscosin group. Front. Microbiol. 2020, 11, 646. [Google Scholar] [CrossRef] [PubMed]
- Diaz, L.A., Jr.; Cheong, I.; Foss, C.A.; Zhang, X.; Peters, B.A.; Agrawal, N.; Bettegowda, C.; Karim, B.; Liu, G.; Khan, K.; et al. Pharmacologic and Toxicologic Evaluation of C. novyi-NT Spores. Toxicol. Sci. 2005, 88, 562. [Google Scholar] [CrossRef] [PubMed]
- Umer, B.; Good, D.; Anné, J.; Duan, W.; Wei, M.Q. Clostridial spores for cancer therapy: Targeting solid tumour microenvironment. J. Toxicol. 2012, 2012, 862764. [Google Scholar] [CrossRef]
- Rice, K.C.; Bayles, K.W. Molecular control of bacterial death and lysis. Microbiol. Mol. Biol. Rev. 2008, 72, 85–109. [Google Scholar] [CrossRef]
- van Pijkeren, J.P.; Morrissey, D.; Monk, I.R.; Cronin, M.; Rajendran, S.; O’Sullivan, G.C.; Gahan, C.G.; Tangney, M. A novel Listeria monocytogenes-based DNA delivery system for cancer gene therapy. Hum. Gene Ther. 2010, 21, 405–416. [Google Scholar] [CrossRef]
- Riglar, D.T.; Silver, P.A. Engineering bacteria for diagnostic and therapeutic applications. Nat. Rev. Microbiol. 2018, 16, 215. [Google Scholar] [CrossRef]
- Lim, D.; Song, M. Development of bacteria as diagnostics and therapeutics by genetic engineering. J. Microbiol. 2019, 57, 637. [Google Scholar] [CrossRef]
- Schmidt, C.K.; Medina-Sánchez, M.; Edmondson, R.J.; Schmidt, O.G. Engineering microrobots for targeted cancer therapies from a medical perspective. Nat. Commun. 2020, 11, 5618. [Google Scholar] [CrossRef]
- Alizadeh, S.; Esmaeili, A.; Barzegari, A.; Rafi, M.A.; Omidi, Y. Bioengineered smart bacterial carriers for combinational targeted therapy of solid tumors. J. Drug Target. 2020, 28, 700–713. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, Z.-H.; Zeng, X.; Zhang, X.-Z. Bacteria-based bioactive materials for cancer imaging and therapy. Adv. Drug Delivery Rev. 2023, 193, 114696. [Google Scholar] [CrossRef] [PubMed]
- Hosseinidoust, Z.; Mostaghaci, B.; Yasa, O.; Park, B.-W.; Singh, A.V.; Sitti, M. Bioengineered and biohybrid bacteria-based systems for drug delivery. Adv. Drug Delivery Rev. 2016, 106, 27–44. [Google Scholar] [CrossRef]
- Cao, Z.; Liu, J. Bacteria and bacterial derivatives as drug carriers for cancer therapy. J. Control Release 2020, 326, 396–407. [Google Scholar] [CrossRef]
- Bazylinski, D.A.; Williams, T.J.; Lefèvre, C.T.; Berg, R.J.; Zhang, C.L.; Bowser, S.S.; Dean, A.J.; Beveridge, T.J. Magnetococcus marinus gen. nov., sp. nov., a marine, magnetotactic bacterium that represents a novel lineage (Magnetococcaceae fam. nov., Magnetococcales ord. nov.) at the base of the Alphaproteobacteria. Int. J. Syst. Evol. Microbiol. 2013, 63, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Felfoul, O.; Mohammadi, M.; Taherkhani, S.; de Lanauze, D.; Zhong, X.Y.; Loghin, D.; Essa, S.; Jancik, S.; Houle, D.; Lafleur, M.; et al. Magneto-aerotactic bacteria deliver drug-containing nanoliposomes to tumour hypoxic regions. Nat. Nanotechnol. 2016, 11, 941–947. [Google Scholar] [CrossRef]
- Aubry, M.; Wang, W.A.; Guyodo, Y.; Delacou, E.; Guigner, J.M.; Espeli, O.; Lebreton, A.; Guyot, F.; Gueroui, Z.; Engineering, E. coli for magnetic control and the spatial localization of functions. ACS Synth. Biol. 2020, 9, 3030–3041. [Google Scholar] [CrossRef]
- Park, S.H.; Zheng, J.H.; Nguyen, V.H.; Jiang, S.N.; Kim, D.Y.; Szardenings, M.; Min, J.H.; Hong, Y.; Choy, H.E.; Min, J.J. RGD peptide cell-surface display enhances the targeting and therapeutic efficacy of attenuated Salmonella-mediated cancer therapy. Theranostics. 2016, 6, 1672. [Google Scholar] [CrossRef]
- Massa, P.E.; Paniccia, A.; Monegal, A.; De Marco, A.; Rescigno, M. Salmonella engineered to express CD20-targeting antibodies and a drug-converting enzyme can eradicate human lymphomas. J. Am. Soc. Hematol. 2013, 122, 705–714. [Google Scholar] [CrossRef]
- Yoon, W.S.; Chae, Y.S.; Hong, J.; Park, Y.K. Antitumor therapeutic effects of a genetically engineered Salmonella typhimurium harboring TNF-α in mice. Appl. Microbiol. Biotech. 2011, 89, 1807–1819. [Google Scholar] [CrossRef]
- Ganai, S.; Arenas, R.B.; Forbes, N.S. Tumour-targeted delivery of TRAIL using Salmonella typhimurium enhances breast cancer survival in mice. Br. J. Cancer 2009, 101, 1683–1691. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Park, S.H.; Cho, S.; Kim, D.M.; Lee, Y.; Ko, S.Y.; Hong, Y.; Choy, H.E.; Park, J.; Park, S. New paradigm for tumor theranostic methodology using bacteria-based microrobot. Sci. Rep. 2013, 3, 3394. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.K.; Liu, Q.; Liang, K.; Li, P.; Kong, Q. Bacteria-derived minicells for cancer therapy. Cancer Lett. 2020, 491, 11–21. [Google Scholar] [CrossRef] [PubMed]


| Microorganism | Cancer Type Associated | References |
|---|---|---|
| Helicobacter pylori | gastric carcinoma | [42,43] |
| Salmonella enterica serovar Typhi | gallbladder cancer | [48,49,50] |
| Campylobacter jejuni | small intestinal lymphomas, colorectal cancer | [51,52] |
| Chlamydia psittaci | ocular lymphomas | [52,53] |
| Mycobacterium tuberculosis | lung cancer | [54] |
| Chlamydia pneumonia | lung cancer, lymphomas | [53] |
| Fusobacterium nucleatum | colorectal cancer | [55,56] |
| Bacteroides fragilis | colorectal cancer | [57,58] |
| Escherichia coli | colorectal cancer | [57,58] |
| Streptococcus bovis | colorectal cancer | [59,60] |
| Chlamydia trachomatis | cervical cancer, lymphomas | [53,54,55,56,57,58,59,60,61] |
| Porphyromonas gingivalis | pancreatic and oral cancer | [62,63] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ikryannikova, L.N.; Gorokhovets, N.V.; Belykh, D.A.; Kurbatov, L.K.; Zamyatnin, A.A., Jr. Bacterial Therapy of Cancer: A Way to the Dustbin of History or to the Medicine of the Future? Int. J. Mol. Sci. 2023, 24, 9726. https://doi.org/10.3390/ijms24119726
Ikryannikova LN, Gorokhovets NV, Belykh DA, Kurbatov LK, Zamyatnin AA Jr. Bacterial Therapy of Cancer: A Way to the Dustbin of History or to the Medicine of the Future? International Journal of Molecular Sciences. 2023; 24(11):9726. https://doi.org/10.3390/ijms24119726
Chicago/Turabian StyleIkryannikova, Larisa N., Neonila V. Gorokhovets, Darya A. Belykh, Leonid K. Kurbatov, and Andrey A. Zamyatnin, Jr. 2023. "Bacterial Therapy of Cancer: A Way to the Dustbin of History or to the Medicine of the Future?" International Journal of Molecular Sciences 24, no. 11: 9726. https://doi.org/10.3390/ijms24119726
APA StyleIkryannikova, L. N., Gorokhovets, N. V., Belykh, D. A., Kurbatov, L. K., & Zamyatnin, A. A., Jr. (2023). Bacterial Therapy of Cancer: A Way to the Dustbin of History or to the Medicine of the Future? International Journal of Molecular Sciences, 24(11), 9726. https://doi.org/10.3390/ijms24119726






