The Curcumin Analog PAC Is a Potential Solution for the Treatment of Triple-Negative Breast Cancer by Modulating the Gene Expression of DNA Repair Pathways
Abstract
1. Introduction
2. Results
2.1. PAC Induces Morphological Changes in MCF-7 and MDA-MB-231 Cells
2.2. PAC Inhibits Cell Proliferation and Potent Cytotoxicity against MDA-MB-231 and MCF-7 Cells
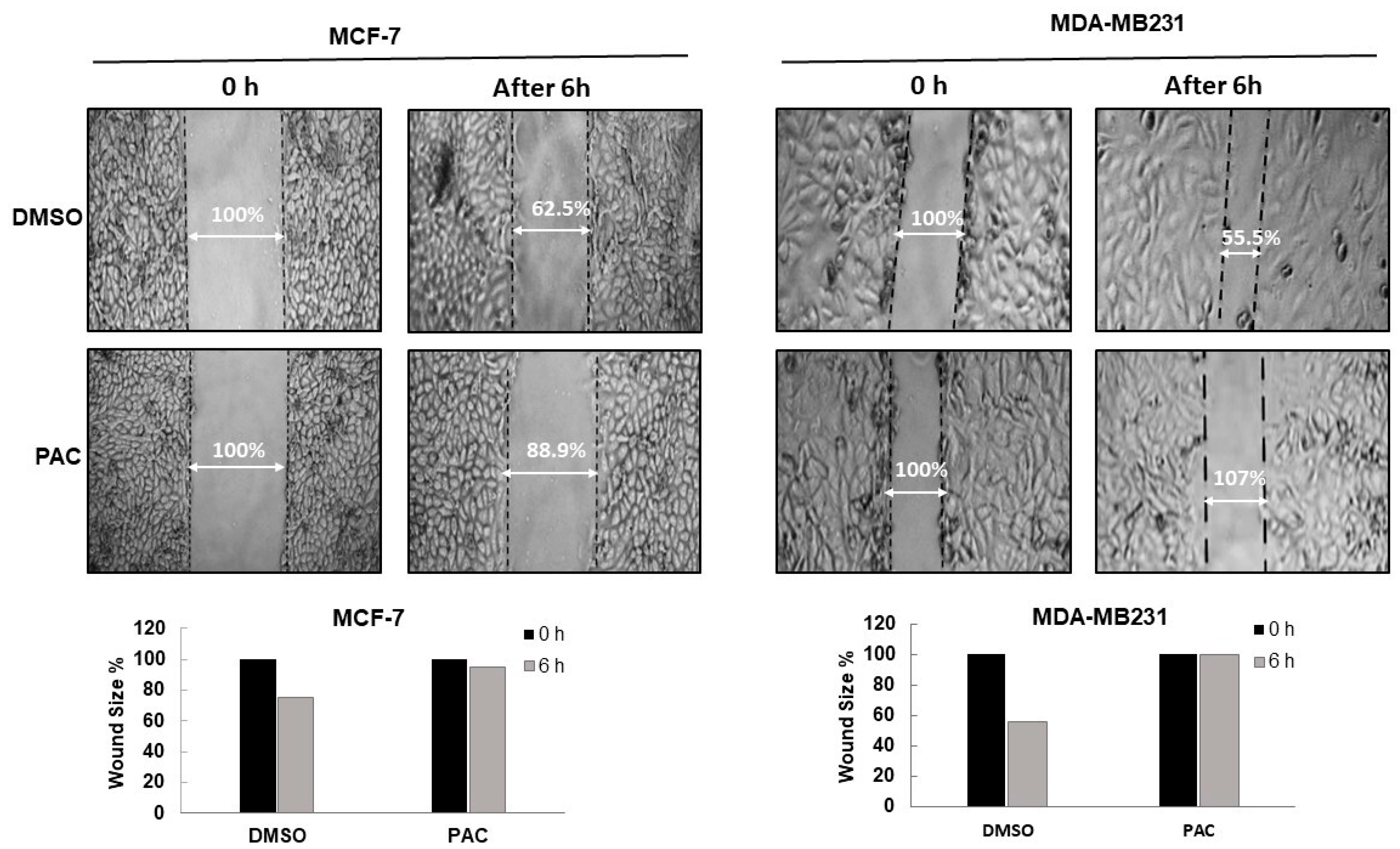
2.3. PAC Suppresses the Migration in Breast-Cancer Cell Lines
2.4. PAC Induces Greater Cell Apoptosis in Triple-Negative Breast Cancer
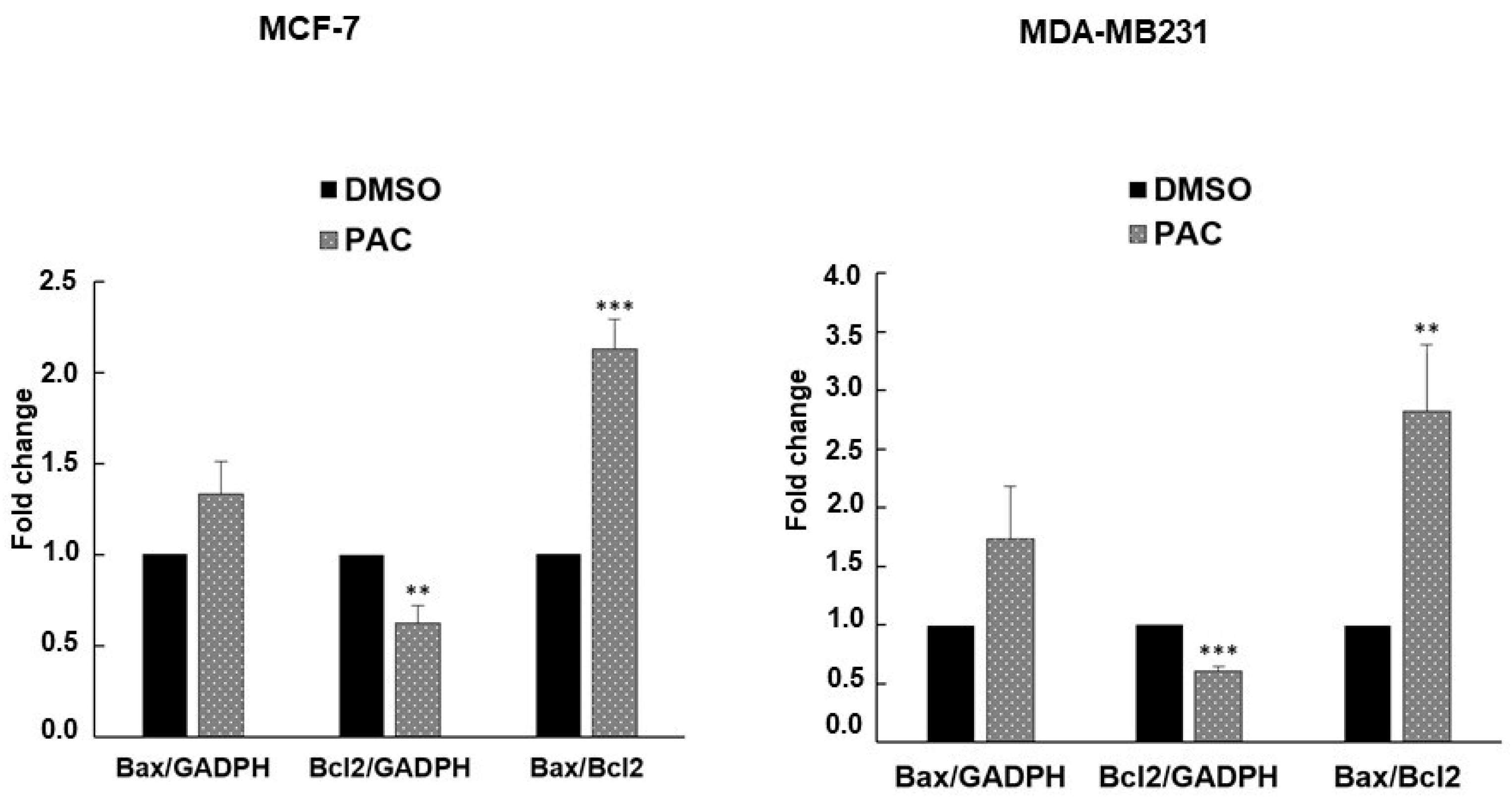
2.5. PAC Induces Apoptosis in MCF-7 and MDA-MB-231 through an Increase in the Bax/Bcl-2 Ratio
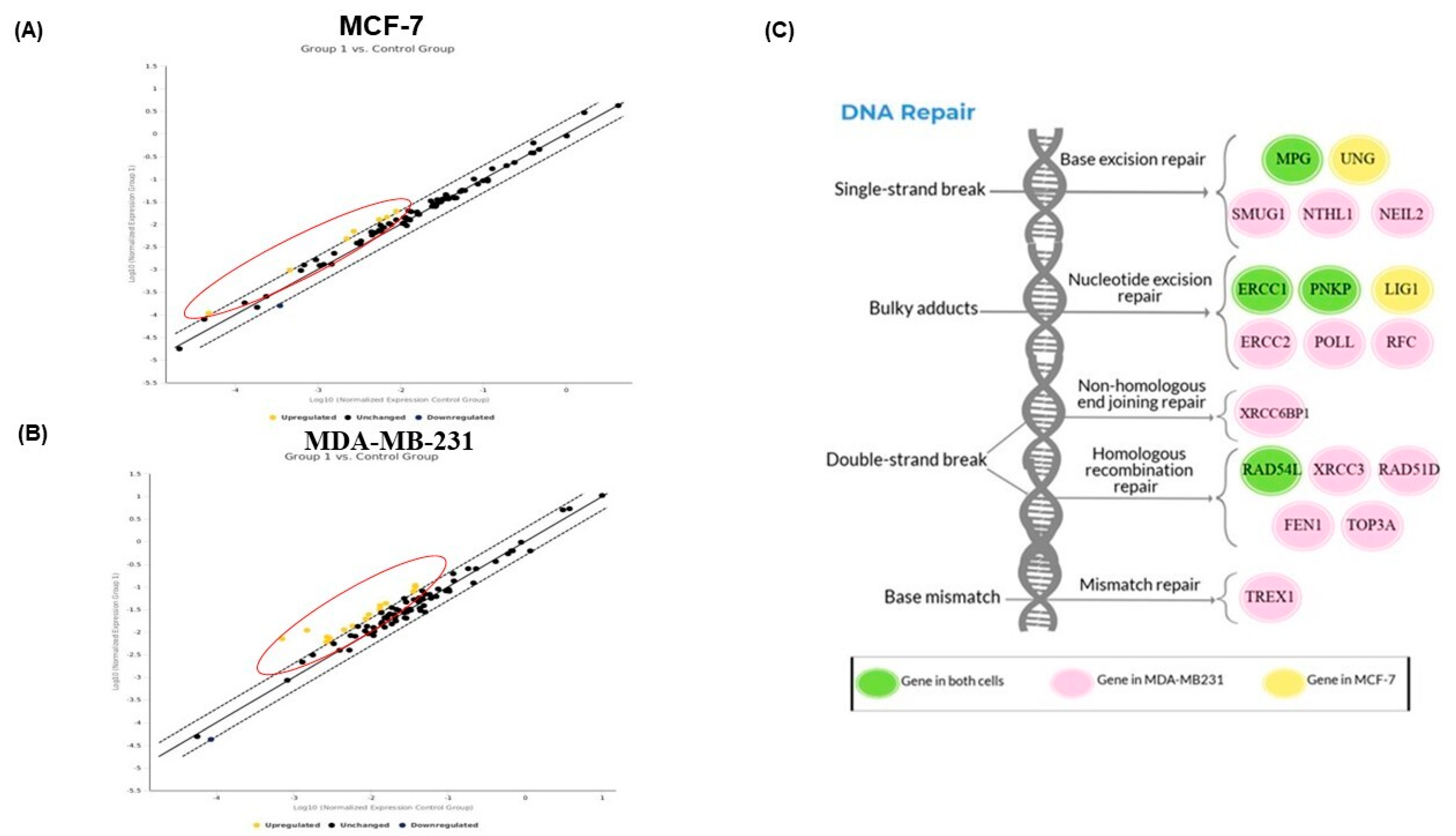
2.6. PAC Modulates the DNA Repair Gene Expression in MCF-7 and MDA-MB-231 Cells
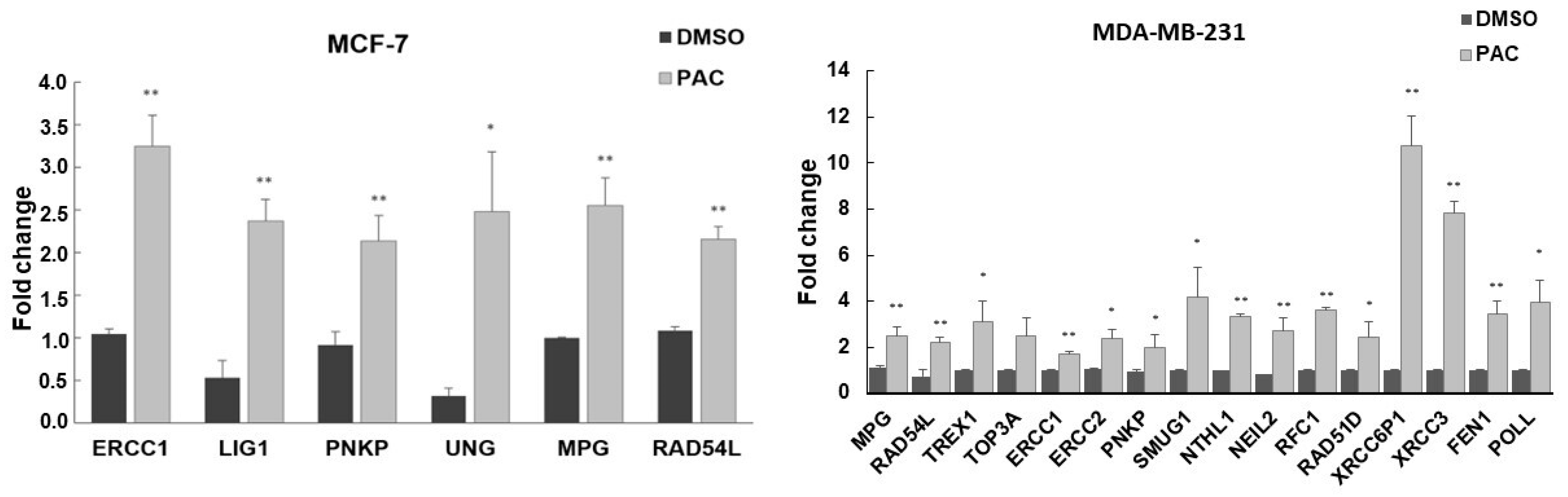
2.7. Validation of DNA Repair in Gene Expression by RT-PCR Analysis
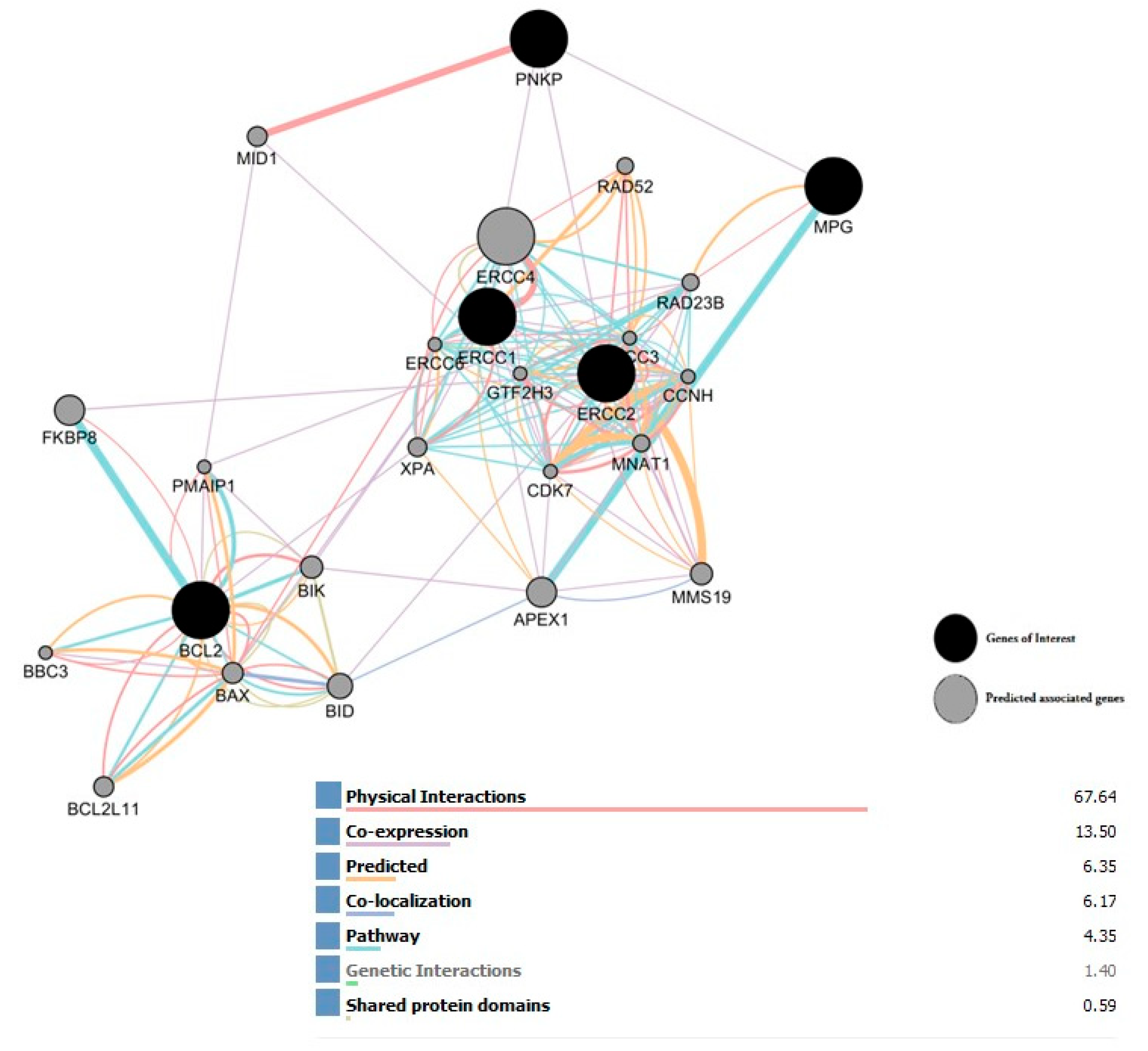
2.8. Interaction Network of All Associated Genes in Human DNA Repair
3. Discussion
4. Materials and Methods
4.1. Reagents and Chemicals
4.2. Cell Line and Culture
4.3. Morphological Observation by Light and Fluorescence Microscopy
4.4. Cell Viability by MTT Assay
4.5. Cytotoxicity Assay
4.6. Cellular Migration Analysis
4.7. Annexin V-FITC Propidium Iodide Assay
4.8. RNA Extraction and cDNA Preparation
4.9. Quantitative RT-PCR
4.10. Screening with RT2 Profiler PCR Array
4.11. In Silico Analysis
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fitzmaurice, C.; Dicker, D.; Pain, A.; Hamavid, H.; Moradi-Lakeh, M.; MacIntyre, M.F.; Allen, C.; Hansen, G.; Woodbrook, R.; Wolfe, C.; et al. Global Burden of Disease Cancer 2013. JAMA Oncol. 2015, 1, 505–527. [Google Scholar] [PubMed]
- Doyle, L.A.; Yang, W.; Abruzzo, L.V.; Krogmann, T.; Gao, Y.; Rishi, A.K.; Ross, D.D. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc. Natl. Acad. Sci. USA 1998, 95, 15665–15670. [Google Scholar] [CrossRef] [PubMed]
- Ullah, M.F. Cancer multidrug resistance (MDR): A major impediment to effective chemotherapy. Asian Pac. J. Cancer Prev. 2008, 9, 1–6. [Google Scholar] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef]
- Anand, P.; Sundaram, C.; Jhurani, S.; Kunnumakkara, A.B.; Aggarwal, B.B. Curcumin and cancer: An “old-age” disease with an “age-old” solution. Cancer Lett. 2008, 267, 133–164. [Google Scholar] [CrossRef]
- Friedman, L.; Lin, L.; Ball, S.; Bekaii-Saab, T.; Fuchs, J.; Li, P.K.; Li, C.; Lin, J. Curcumin analogs exhibit enhanced growth suppressive activity in human pancreatic cancer cells. Anticancer Drugs 2009, 20, 444–449. [Google Scholar] [CrossRef]
- Cridge, B.J.; Larsen, L.; Rosengren, R.J. Curcumin and its derivatives in breast cancer: Current developments and potential for the treatment of drug-resistant cancers. Oncol. Discov. 2013, 1, 1–9. [Google Scholar] [CrossRef]
- Shishodia, S.; Sethi, G.; Aggarwal, B.B. Curcumin: Getting back to the roots. Ann. N. Y. Acad. Sci. 2005, 1056, 206–217. [Google Scholar] [CrossRef]
- Youssef, K.M.; El-Sherbeny, M.A.; El-Shafie, F.S.; Farag, H.A.; Al-Deeb, O.A.; Awadalla, S.A.A. Synthesis of curcumin analogs as potential antioxidant, cancer chemopreventive agents. Arch. Der Pharm. Int. J. Pharm. Med. Chem. 2004, 337, 42–54. [Google Scholar] [CrossRef]
- Al-Howail, H.A.; Hakami, H.A.; Al-Otaibi, B.; Al-Mazrou, A.; Daghestani, M.H.; Al-Jammaz, I.; Al-Khalaf, H.H.; Aboussekhra, A. PAC down-regulates estrogen receptor alpha and suppresses epithelial-to-mesenchymal transition in breast cancer cells. BMC Cancer 2016, 16, 540. [Google Scholar] [CrossRef]
- Al-Hujaily, E.M.; Mohamed, A.G.; Al-Sharif, I.; Youssef, K.M.; Manogaran, P.S.; Al-Otaibi, B.; Al-Haza’a, A.; Al-Jammaz, I.; Al-Hussein, K.; Aboussekhra, A. PAC, a novel curcumin analog, has anti-breast cancer properties with higher efficiency on ER-negative cells. Breast Cancer Res. Treat. 2011, 128, 97–107. [Google Scholar] [CrossRef]
- Al-Qasem, A.; Al-Howail, H.A.; Al-Swailem, M.; Al-Mazrou, A.; Al-Otaibi, B.; Al-Jammaz, I.; Al-Khalaf, H.H.; Aboussekhra, A. PAC exhibits potent anti-colon cancer properties through targeting cyclin D1 and suppressing epithelial-to-mesenchymal transition. Mol. Carcinog. 2016, 55, 233–244. [Google Scholar] [CrossRef]
- Helleday, T.; Petermann, E.; Lundin, C.; Hodgson, B.; Sharma, R.A. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer 2008, 8, 193–204. [Google Scholar] [CrossRef]
- Iyama, T.; Wilson, D.M., 3rd. DNA repair mechanisms in dividing and non-dividing cells. DNA Repair 2013, 12, 620–636. [Google Scholar] [CrossRef]
- Lauren, P. Molecular Biology of Cancer: Mechanisms, Targets, and Therapeutics, 3rd ed.; Oxford University Press: Oxford, UK, 2012; pp. 1–342. [Google Scholar]
- Torgovnick, A.; Schumacher, B. DNA repair mechanisms in cancer development and therapy. Front. Genet. 2015, 6, 157. [Google Scholar] [CrossRef]
- Adams, B.K.; Cai, J.; Armstrong, J.; Herold, M.; Lu, Y.J.; Sun, A.; Snyder, J.P.; Liotta, D.C.; Jones, D.P.; Shoji, M. EF24, a novel synthetic curcumin analog, induces apoptosis in cancer cells via a redox-dependent mechanism. Anticancer Drugs 2005, 16, 263–275. [Google Scholar] [CrossRef]
- Smigal, C.; Jemal, A.; Ward, E.; Cokkinides, V.; Smith, R.; Howe, H.L.; Thun, M. Trends in breast cancer by race and ethnicity: Update 2006. CA Cancer J. Clin. 2006, 56, 168–183. [Google Scholar] [CrossRef]
- He, Y.; Li, W.; Hu, G.; Sun, H.; Kong, Q. Bioactivities of EF24, a Novel Curcumin Analog: A Review. Front. Oncol. 2018, 8, 614. [Google Scholar] [CrossRef]
- Selvendiran, K.; Tong, L.; Vishwanath, S.; Bratasz, A.; Trigg, N.J.; Kutala, V.K.; Hideg, K.; Kuppusamy, P. EF24 induces G(2)/M arrest and apoptosis in cisplatin-resistant human ovarian cancer cells by increasing PTEN expression. J. Biol. Chem. 2007, 282, 28609–28618. [Google Scholar] [CrossRef]
- Clementi, E.; Inglin, L.; Beebe, E.; Gsell, C.; Garajova, Z.; Markkanen, E. Persistent DNA damage triggers activation of the integrated stress response to promote cell survival under nutrient restriction. BMC Biol. 2020, 18, 36. [Google Scholar] [CrossRef]
- Pearl, L.H.; Schierz, A.C.; Ward, S.E.; Al-Lazikani, B.; Pearl, F.M. Therapeutic opportunities within the DNA damage response. Nat. Rev. Cancer 2015, 15, 166–180. [Google Scholar] [CrossRef] [PubMed]
- Li, L.Y.; Guan, Y.D.; Chen, X.S.; Yang, J.M.; Cheng, Y. DNA Repair Pathways in Cancer Therapy and Resistance. Front. Pharmacol. 2020, 11, 629266. [Google Scholar] [CrossRef] [PubMed]
- Hosoya, N.; Miyagawa, K. Targeting DNA damage response in cancer therapy. Cancer Sci. 2014, 105, 370–388. [Google Scholar] [CrossRef]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, R.N.; Wang, X.H.; Jelezcova, E.; Goellner, E.M.; Tang, J.B.; Sobol, R.W. Human methyl purine DNA glycosylase and DNA polymerase beta expression collectively predict sensitivity to temozolomide. Mol. Pharmacol. 2008, 74, 505–516. [Google Scholar] [CrossRef]
- Fishel, M.L.; He, Y.; Smith, M.L.; Kelley, M.R. Manipulation of base excision repair to sensitize ovarian cancer cells to alkylating agent temozolomide. Clin. Cancer Res. 2007, 13, 260–267. [Google Scholar] [CrossRef]
- Kemmerich, K.; Dingler, F.A.; Rada, C.; Neuberger, M.S. Germline ablation of SMUG1 DNA glycosylase causes loss of 5-hydroxymethyl-uracil- and UNG-backup uracil-excision activities and increases cancer predisposition of Ung-/-Msh2-/- mice. Nucleic Acids Res. 2012, 40, 6016–6025. [Google Scholar] [CrossRef]
- Wallace, S.S.; Murphy, D.L.; Sweasy, J.B. Base excision repair and cancer. Cancer Lett. 2012, 327, 73–89. [Google Scholar] [CrossRef]
- Broderick, P.; Bagratuni, T.; Vijayakrishnan, J.; Lubbe, S.; Chandler, I.; Houlston, R.S. Evaluation of NTHL1, NEIL1, NEIL2, MPG, TDG, UNG and SMUG1 genes in familial colorectal cancer predisposition. BMC Cancer 2006, 6, 243. [Google Scholar] [CrossRef]
- Das, L.; Quintana, V.G.; Sweasy, J.B. NTHL1 in genomic integrity, aging and cancer. DNA Repair 2020, 93, 102920. [Google Scholar] [CrossRef]
- Magrin, L.; Fanale, D.; Brando, C.; Fiorino, A.; Corsini, L.R.; Sciacchitano, R.; Filorizzo, C.; Dimino, A.; Russo, A.; Bazan, V. POLE, POLD1, and NTHL1: The last but not the least hereditary cancer-predisposing genes. Oncogene 2021, 40, 5893–5901. [Google Scholar] [CrossRef]
- Grolleman, J.E.; De Voer, R.M.; Elsayed, F.A.; Nielsen, M.; Weren, R.D.; Palles, C.; Ligtenberg, M.J.; Vos, J.R.; Ten Broeke, S.W.; De Miranda, N.F.; et al. Mutational Signature Analysis Reveals NTHL1 Deficiency to Cause a Multi-tumor Phenotype. Cancer Cell 2019, 35, 256–266.e5. [Google Scholar] [CrossRef]
- Zhai, X.; Zhao, H.; Liu, Z.; Wang, L.E.; El-Naggar, A.K.; Sturgis, E.M.; Wei, Q. Functional variants of the NEIL1 and NEIL2 genes and risk and progression of squamous cell carcinoma of the oral cavity and oropharynx. Clin. Cancer Res. 2008, 14, 4345–4352. [Google Scholar] [CrossRef] [PubMed]
- Shinmura, K.; Kato, H.; Kawanishi, Y.; Igarashi, H.; Goto, M.; Tao, H.; Inoue, Y.; Nakamura, S.; Misawa, K.; Mineta, H.; et al. Abnormal Expressions of DNA Glycosylase Genes NEIL1, NEIL2, and NEIL3 Are Associated with Somatic Mutation Loads in Human Cancer. Oxidative Med. Cell. Longev. 2016, 2016, 1546392. [Google Scholar] [CrossRef]
- Buschta-Hedayat, N.; Buterin, T.; Hess, M.T.; Missura, M.; Naegeli, H. Recognition of non-hybridizing base pairs during nucleotide excision repair of DNA. Proc. Natl. Acad. Sci. USA 1999, 96, 6090–6095. [Google Scholar] [CrossRef] [PubMed]
- Ban, S.; Kuramoto, K.; Oda, K.; Tanaka, H.; Kimura, A.; Suzuki, G.; Imai, T. Radiosensitivity and expression of nucleotide excision repair genes in peripheral blood mononuclear cells of myelodysplastic syndrome patients. Int. Congr. Ser. 2002, 1236, 67–69. [Google Scholar] [CrossRef]
- Leibeling, D.; Laspe, P.; Emmert, S. Nucleotide excision repair and cancer. J. Mol. Histol. 2006, 37, 225–238. [Google Scholar] [CrossRef]
- Saldivar, J.S.; Wu, X.; Follen, M.; Gershenson, D. Nucleotide excision repair pathway review I: Implications in ovarian cancer and platinum sensitivity. Gynecol. Oncol. 2007, 107 (Suppl. 1), S56–S71. [Google Scholar] [CrossRef]
- Friboulet, L.; Olaussen, K.A.; Pignon, J.P.; Shepherd, F.A.; Tsao, M.S.; Graziano, S.; Kratzke, R.; Douillard, J.Y.; Seymour, L.; Pirker, R.; et al. ERCC1 isoform expression and DNA repair in non-small-cell lung cancer. N. Engl. J. Med. 2013, 368, 1101–1110. [Google Scholar] [CrossRef]
- Ryu, J.S.; Memon, A.; Lee, S.K. ERCC1 and personalized medicine in lung cancer. Ann. Transl. Med. 2014, 2, 32. [Google Scholar]
- Lord, R.V.; Brabender, J.; Gandara, D.; Alberola, V.; Camps, C.; Domine, M.; Cardenal, F.; Sánchez, J.M.; Gumerlock, P.H.; Tarón, M.; et al. Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clin. Cancer Res. 2002, 8, 2286–2291. [Google Scholar] [PubMed]
- Li, Q.; Damish, A.W.; Frazier, Z.; Liu, D.; Reznichenko, E.; Kamburov, A.; Bell, A.; Zhao, H.; Jordan, E.J.; Gao, S.P.; et al. ERCC2 Helicase Domain Mutations Confer Nucleotide Excision Repair Deficiency and Drive Cisplatin Sensitivity in Muscle-Invasive Bladder Cancer. Clin. Cancer Res. 2019, 25, 977–988. [Google Scholar] [CrossRef]
- Van Allen, E.M.; Mouw, K.W.; Kim, P.; Iyer, G.; Wagle, N.; Al-Ahmadie, H.; Zhu, C.; Ostrovnaya, I.; Kryukov, G.V.; O’Connor, K.W.; et al. Somatic ERCC2 Mutations Correlate with Cisplatin Sensitivity in Muscle-Invasive Urothelial Carcinoma. Cancer Discov. 2014, 4, 1140–1153. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Qiu, J.; Wang, D.; Tao, Y.; Song, Y.; Wang, H.; Tang, J.; Wang, X.; Sun, Y.U.; Yang, Z.; et al. Traditional Chinese Medicine Curcumin Sensitizes Human Colon Cancer to Radiation by Altering the Expression of DNA Repair-related Genes. Anticancer Res. 2018, 38, 131–136. [Google Scholar] [PubMed]
- Tsukada, K.; Shimada, M.; Imamura, R.; Saikawa, K.; Ishiai, M.; Matsumoto, Y. The FHA domain of PNKP is essential for its recruitment to DNA damage sites and maintenance of genome stability. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2021, 822, 111727. [Google Scholar] [CrossRef]
- Li, D.; Li, R.; Zhang, J.; Li, K.; Wu, Y. Association Between the LIG1 Polymorphisms and Lung Cancer Risk: A Meta-analysis of Case-Control Studies. Cell Biochem. Biophys. 2015, 73, 381–387. [Google Scholar] [CrossRef]
- Cortese, A.; Simone, R.; Sullivan, R.; Vandrovcova, J.; Tariq, H.; Yau, W.Y.; Humphrey, J.; Jaunmuktane, Z.; Sivakumar, P.; Polke, J.; et al. Biallelic expansion of an intronic repeat in RFC1 is a common cause of late-onset ataxia. Nat. Genet. 2019, 51, 649–658. [Google Scholar] [CrossRef]
- Barr, M.; Farrell, R.; Singh, S.; Foley, E.; He, Y.; Brady, L.; Young, V.; Ryan, R.; Nicholson, S.; Leonard, N.; et al. XRCC6BP1: A DNA Repair Gene in Cisplatin Resistant Lung Cancer Stem Cells That May Predict Survival Outcomes in Patients. J. Thorac. Oncol. 2018, 13, S377. [Google Scholar] [CrossRef]
- Leone, P.E.; Mendiola, M.; Alonso, J.; Paz-y-Miño, C.; Pestaña, A. Implications of a RAD54L polymorphism (2290C/T) in human meningiomas as a risk factor and/or a genetic marker. BMC Cancer 2003, 3, 6. [Google Scholar] [CrossRef]
- Rasko, D.A.; Sperandio, V. Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 2010, 9, 117–128. [Google Scholar] [CrossRef]
- Matsuda, M.; Miyagawa, K.; Takahashi, M.; Fukuda, T.; Kataoka, T.; Asahara, T.; Inui, H.; Watatani, M.; Yasutomi, M.; Kamada, N.; et al. Mutations in the RAD54 recombination gene in primary cancers. Oncogene 1999, 18, 3427–3430. [Google Scholar] [CrossRef]
- AACR Project GENIE Consortium; André, F.; Arnedos, M.; Baras, A.S.; Baselga, J.; Bedard, P.L.; Berger, M.F.; Bierkens, M.; Calvo, F.; Cerami, E. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017, 7, 818–831. [Google Scholar] [CrossRef]
- Roos, W.P.; Frohnapfel, L.; Quiros, S.; Ringel, F.; Kaina, B. XRCC3 contributes to temozolomide resistance of glioblastoma cells by promoting DNA double-strand break repair. Cancer Lett. 2018, 424, 119–126. [Google Scholar] [CrossRef]
- Avadanei, E.R.; Giusca, S.E.; Negura, L.; Caruntu, I.D. Single Nucleotide Polymorphisms of Xrcc3 Gene in Hepatocellular Carcinoma—Relationship with Clinicopathological Features. Pol. J. Pathol. 2018, 69, 73–81. [Google Scholar] [CrossRef]
- Soukupová, J.; Lhotová, K.; Janatová, M.; Kleiblová, P.; Vočka, M.; Foretová, L.; Zikán, M.; Kleibl, Z. Germline mutations in RAD51C and RAD51D and hereditary predisposition to ovarian cancer. Klin. Onkol. 2021, 34, 26–32. [Google Scholar] [CrossRef]
- Yang, F.; Hu, Z.; Guo, Z. Small-Molecule Inhibitors Targeting FEN1 for Cancer Therapy. Biomolecules 2022, 12, 1007. [Google Scholar] [CrossRef]
- He, L.; Luo, L.; Zhu, H.; Yang, H.; Zhang, Y.; Wu, H.; Sun, H.; Jiang, F.; Kathera, C.S.; Liu, L.; et al. FEN1 promotes tumor progression and confers cisplatin resistance in non-small-cell lung cancer. Mol. Oncol. 2017, 11, 640–654. [Google Scholar] [CrossRef]
- Zhong, G.; Wang, Y.; Wei, H.; Chen, M.; Lin, H.; Huang, Z.; Huang, J.; Wang, S.; Lin, J. The Clinical Significance of the Expression of FEN1 in Primary Osteosarcoma. Int. J. Gen. Med. 2021, 14, 6477–6485. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Zhang, Y.; Sun, H.; Jiang, F.; Yang, H.; Wu, H.; Zhou, T.; Hu, S.; Kathera, C.S.; Wang, X.; et al. Targeting DNA Flap Endonuclease 1 to Impede Breast Cancer Progression. Biomedicine 2016, 14, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Liu, R.; Wang, M.; Kumar, A.K.; Pan, F.; He, L.; Hu, Z.; Guo, Z. MicroRNA-140 impedes DNA repair by targeting FEN1 and enhances chemotherapeutic response in breast cancer. Oncogene 2020, 39, 234–247. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yuan, P.; Du, F.; Cui, L.; Zheng, F.; Yue, J.; Zhang, B.; Wang, J.; Fan, Y.; Ma, F.; et al. PD-L1 expression and TOP3A mutation as prognostic factors for adjuvant chemotherapy in negative negative triple-negative breast cancer. J. Clin. Oncol. 2021, 39, 541. [Google Scholar] [CrossRef]
- Bai, Y.; Li, L.D.; Li, J.; Lu, X. Targeting of topoisomerases for prognosis and drug resistance in ovarian cancer. J. Ovarian Res. 2016, 9, 35. [Google Scholar] [CrossRef]
- Vanpouille-Box, C.; Alard, A.; Aryankalayil, M.J.; Sarfraz, Y.; Diamond, J.M.; Schneider, R.J.; Inghirami, G.; Coleman, C.N.; Formenti, S.C.; Demaria, S. DNA exonuclease Trex1 regulates radiotherapy-induced tumortumor immunogenicity. Nat. Commun. 2017, 8, 15618. [Google Scholar] [CrossRef]
- Semlali, A.; Beji, S.; Ajala, I.; Rouabhia, M. Effects of tetrahydrocannabinol on human oral cancer cell proliferation, apoptosis, autophagy, oxidative stress, and DNA damage. Arch. Oral Biol. 2021, 129, 105200. [Google Scholar] [CrossRef]
- Semlali, A.; Contant, C.; Al-Otaibi, B.; Al-Jammaz, I.; Chandad, F. The curcumin analog (PAC) suppressed cell survival and induced apoptosis and autophagy in oral cancer cells. Sci. Rep. 2021, 11, 11701. [Google Scholar] [CrossRef]
- Contant, C.; Rouabhia, M.; Loubaki, L.; Chandad, F.; Semlali, A. Anethole induces anti-oral cancer activity by triggering apoptosis, autophagy, and oxidative stress and by modulation of multiple signaling pathways. Sci. Rep. 2021, 11, 13087. [Google Scholar] [CrossRef]
- Semlali, A.; Al Mutairi, M.; Oqla Alanazi, I.; Awad Aljohi, H.; Reddy Parine, N.; Alhadheq, A.; Al-Jafari, A.A.; Mobeirek, A.F.; Al Amri, A.; Shaik, J.P.; et al. Toll-like receptor 4 polymorphisms in Saudi population with cardiovascular diseases. Mol. Genet. Genom. Med. 2019, 7, e852. [Google Scholar] [CrossRef]
- Semlali, A.; Almutairi, M.; Pathan, A.A.K.; Azzi, A.; Parine, N.R.; AlAmri, A.; Arafah, M.; Aljebreen, A.M.; Almadi, M.A.; Azzam, N.A.; et al. Toll-like receptor 6 expression, sequence variants, and their association with colorectal cancer risk. J. Cancer 2019, 10, 2969–2981. [Google Scholar] [CrossRef]
- Lampron, M.C.; Paré, I.; Al-Zharani, M.; Semlali, A.; Loubaki, L. Cannabinoid Mixture Affects the Fate and Functions of B Cells through the Modulation of the Caspase and MAP Kinase Pathways. Cells 2023, 12, 588. [Google Scholar] [CrossRef]
- Tazi, N.; Semlali, A.; Loubaki, L.; Alamri, A.; Rouabhia, M. Cannabis smoke condensate induces human gingival epithelial cell damage through apoptosis, autophagy, and oxidative stress. Arch. Oral Biol. 2022, 141, 105498. [Google Scholar] [CrossRef]



| Cell Line | Genes | Name | Fold Regulation by RT2 Profiler PCR Array | Fold Regulation by RT-PCR |
|---|---|---|---|---|
| MDA-MB 231 | ERCC1 | Excision repair 1,endonuclease non-catalytic subunit | 2.79 | 1.69 |
| ERCC2 | Excision repair 1,TFIIH core complex helicase subunit | 2.19 | 2.39 | |
| PNKP | Polynucleotide kinase 3′-phosphatase | 2.86 | 2.00 | |
| POLL | DNA polymerase lambda | 2.44 | 3.86 | |
| MPG | N-methylpurine DNA glycosylase | 2.54 | 2.46 | |
| NEIL2 | Nei like DNA glycosylase 2 | 2.28 | 2.74 | |
| NTHL1 | Nth like DNA glycosylase 1 | 2.81 | 3.34 | |
| SMUG1 | Single strand selective monofunctional uracil-DNA glycosylase 1 | 2.24 | 4.18 | |
| RAD51D | RAD51 paralog D | 2.23 | 2.44 | |
| RAD54L | RAD54 like | 2.43 | 2.21 | |
| RFC1 | Replication factor C subunit 1 | 2.23 | 3.63 | |
| TOP3A | DNA topoisomerase III alpha | 2.69 | 4.04 | |
| XRCC3 | X-ray repair cross complementing 3 | 7.32 | 7.82 | |
| XRCC6BP1 | XRCC6 binding protein 1 | 9.98 | 10.78 | |
| FEN1 | lap structure-specific endonuclease 1 | 2.57 | 3.45 | |
| TREX1 | Three prime repair exonucleases 1 | 2.23 | 2.91 | |
| MCF-7 | ERCC1 | excision repair 1, endonuclease non-catalytic subunit | 2.53 | 2.71 |
| LIG1 | DNA ligase 1 | 2.08 | 2.43 | |
| PNKP | polynucleotide kinase 3′-phosphatase | 2.30 | 2.13 | |
| UNG | uracil DNA glycosylase | 2.12 | 2.48 | |
| MPG | N-methylpurine DNA glycosylase | 2.15 | 2.22 | |
| RAD54L | RAD54 like | 2.18 | 2.15 |
| Gene | Strand | Primers Sequences | Amplicon Size (bp) | Tm |
|---|---|---|---|---|
| GAPDH | Forward | GGTATCGTGGAAGGACTCATGAC | 188 | 66 |
| Reverse | ATGCCAGTGAGCTTCCCGTTCAGC | |||
| Bax | Forward | AGTGTCTCAGGCGAATTGGC | 102 | 60 |
| Reverse | CACGGAAGAAGACCTCTCGG | |||
| BcL2 | Forward | ACTGAGTACCTGAACCGGCATC | 108 | 61 |
| Reverse | GGAGAAATCAAACAGAGGTCGC |
| Gene | Strand | Primers Sequences | Amplicon Size (bp) | Tm |
|---|---|---|---|---|
| ERCC1 | Forward | GGGTGACTGAATGTCTGACCA | 191 | 54 |
| Reverse | GGGTACTTTCAAGAAGGGCTC | |||
| ERCC2 | Forward | GACCTGGTGTCCAAGGAACTG | 165 | 57 |
| Reverse | GATCCTGAGCACCGTCTTCTG | |||
| FEN1 | Forward | AAGTCTATGCTGCGGCTACC | 83 | 60 |
| Reverse | TTCACTGGCAGTCAGGTGTC | |||
| MPG | Forward | GAAGCAGCG ACCAGCTAGAGC | 83 | 60 |
| Reverse | CGAGCTGTGTGGCTGCTCT | |||
| NTHL1 | Forward | CAAGATGGCACACCTGGCTA | 174 | 60 |
| Reverse | TCGTGCCACAGCTCCCTA | |||
| PNKP | Forward | GTTACGGACCGGAAGTGCTCCA | 120 | 60 |
| Reverse | CTTCAACTCCTGGGTCCCGGTA | |||
| POLL | Forward | CATTTGCCCTCCTTCCCCC | 144 | 60 |
| Reverse | GCCCCAAGAGTCTCTCCAAC | |||
| RAD51D | Forward | AGTGGTGGACCTGGTTTCTG | 139 | 60 |
| Reverse | CTCGTAGAGATCAGCGCCATT | |||
| RAD54L | Forward | TTCGATTGACCCGGTCTTGG | 197 | 60 |
| Reverse | TCAGACTCAGGGAGGTCGAG | |||
| SMUG1 | Forward | AAGGAACCGGAAACGGGATG | 109 | 59 |
| Reverse | GCAGCAGTCTCTTGATGAGG | |||
| NEIL2 | Forward | CCCTTCCGCGTCTCATCTTT | 336 | 60.5 |
| Reverse | TTCCATGGACCTGGGTGTCC | |||
| TREX1 | Forward | GAGGGAGACCCTCTCAGACA | 146 | 60 |
| Reverse | TGGGCAGAAGCACATCTCAG | |||
| RFC1 | Forward | ACAAACTCCTACGCTGGCTC | 76 | 59 |
| Reverse | AATTTACCAAACTTTGCGTGTTTTT | |||
| TOP3A | Forward | ATGAGGCGGAGAGAAGGACT | 132 | 60 |
| Reverse | CTGCATCTGGAAATCATGAGCC | |||
| XRCC6P1 | Forward | CTCCAGCTTCTTCACCAGCA | 113 | 59.5 |
| Reverse | CAGCACAACCTGAGTGTTTCA | |||
| XRCC3 | Forward | CGGGAGGATGTGCACAGAG | 171 | 60 |
| Reverse | CTGTCACTCTCTGGGGCTTG | |||
| UNG | Forward | CTCTGTGCAGGGTTCCCAGTC | 345 | 63 |
| Reverse | GCTCACAACCAGACAGCTCC | |||
| LIG1 | Forward | AGGAGGCATCCAATAGCAGCA | 87 | 61 |
| Reverse | GGACTTCCTGAATGGTCCGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almalki, E.; Al-Amri, A.; Alrashed, R.; AL-Zharani, M.; Semlali, A. The Curcumin Analog PAC Is a Potential Solution for the Treatment of Triple-Negative Breast Cancer by Modulating the Gene Expression of DNA Repair Pathways. Int. J. Mol. Sci. 2023, 24, 9649. https://doi.org/10.3390/ijms24119649
Almalki E, Al-Amri A, Alrashed R, AL-Zharani M, Semlali A. The Curcumin Analog PAC Is a Potential Solution for the Treatment of Triple-Negative Breast Cancer by Modulating the Gene Expression of DNA Repair Pathways. International Journal of Molecular Sciences. 2023; 24(11):9649. https://doi.org/10.3390/ijms24119649
Chicago/Turabian StyleAlmalki, Esraa, Abdullah Al-Amri, Reem Alrashed, Mohamed AL-Zharani, and Abdelhabib Semlali. 2023. "The Curcumin Analog PAC Is a Potential Solution for the Treatment of Triple-Negative Breast Cancer by Modulating the Gene Expression of DNA Repair Pathways" International Journal of Molecular Sciences 24, no. 11: 9649. https://doi.org/10.3390/ijms24119649
APA StyleAlmalki, E., Al-Amri, A., Alrashed, R., AL-Zharani, M., & Semlali, A. (2023). The Curcumin Analog PAC Is a Potential Solution for the Treatment of Triple-Negative Breast Cancer by Modulating the Gene Expression of DNA Repair Pathways. International Journal of Molecular Sciences, 24(11), 9649. https://doi.org/10.3390/ijms24119649







