Extracellular Vesicles as Markers of Liver Function: Optimized Workflow for Biomarker Identification in Liver Disease
Abstract
1. Introduction
2. Results
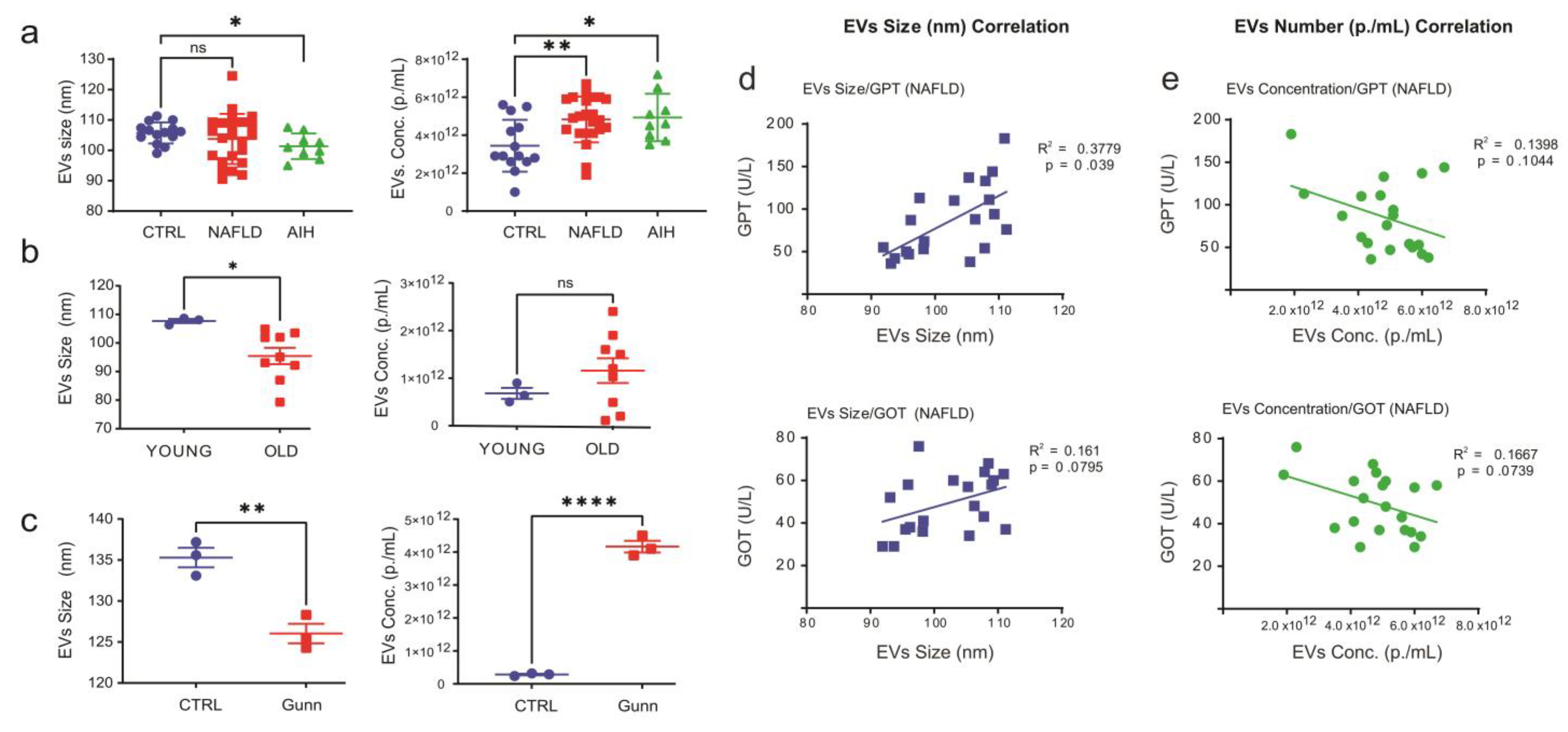
2.1. Characterization of Extracellular Vesicles in Crude Sera
2.2. Assessing the Effect of Bilirubin, Hemoglobin and Proteins Aggregates on NTA Measurements
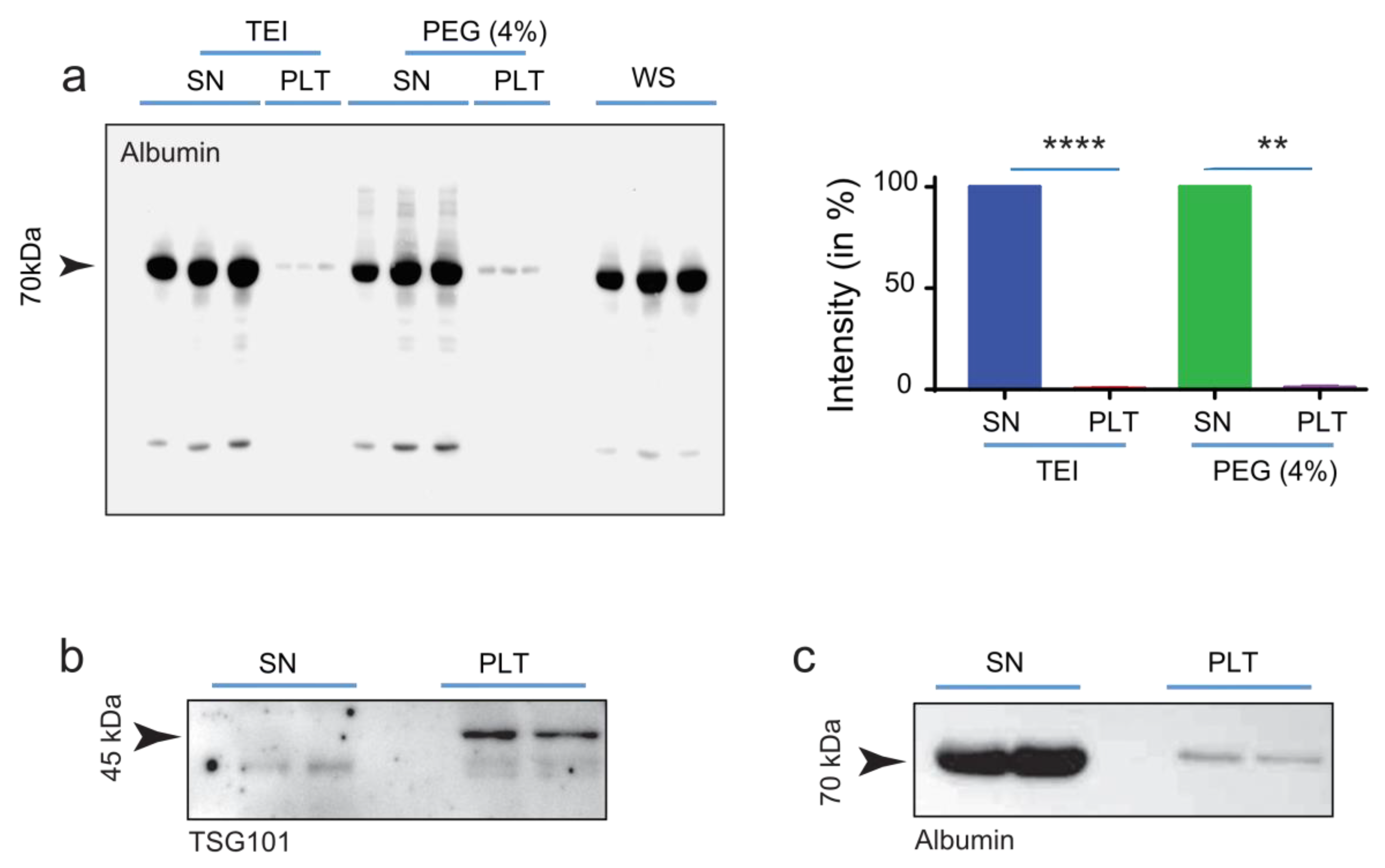
2.3. Separation of EVs from Serum-Protein by Using an Optimized PEG Approach
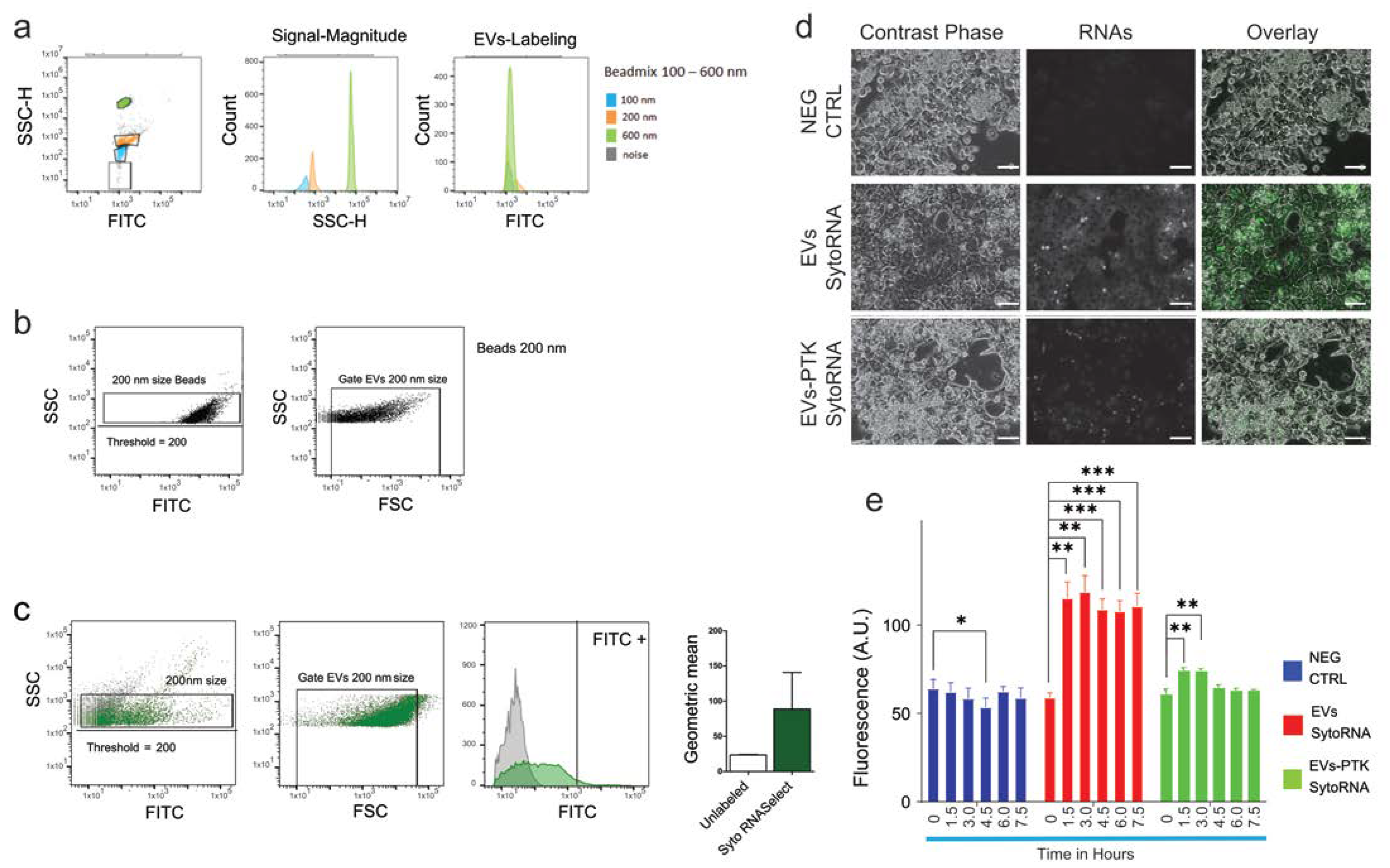
2.4. Evaluation of Extracellular Vesicles Integrity and Functionality
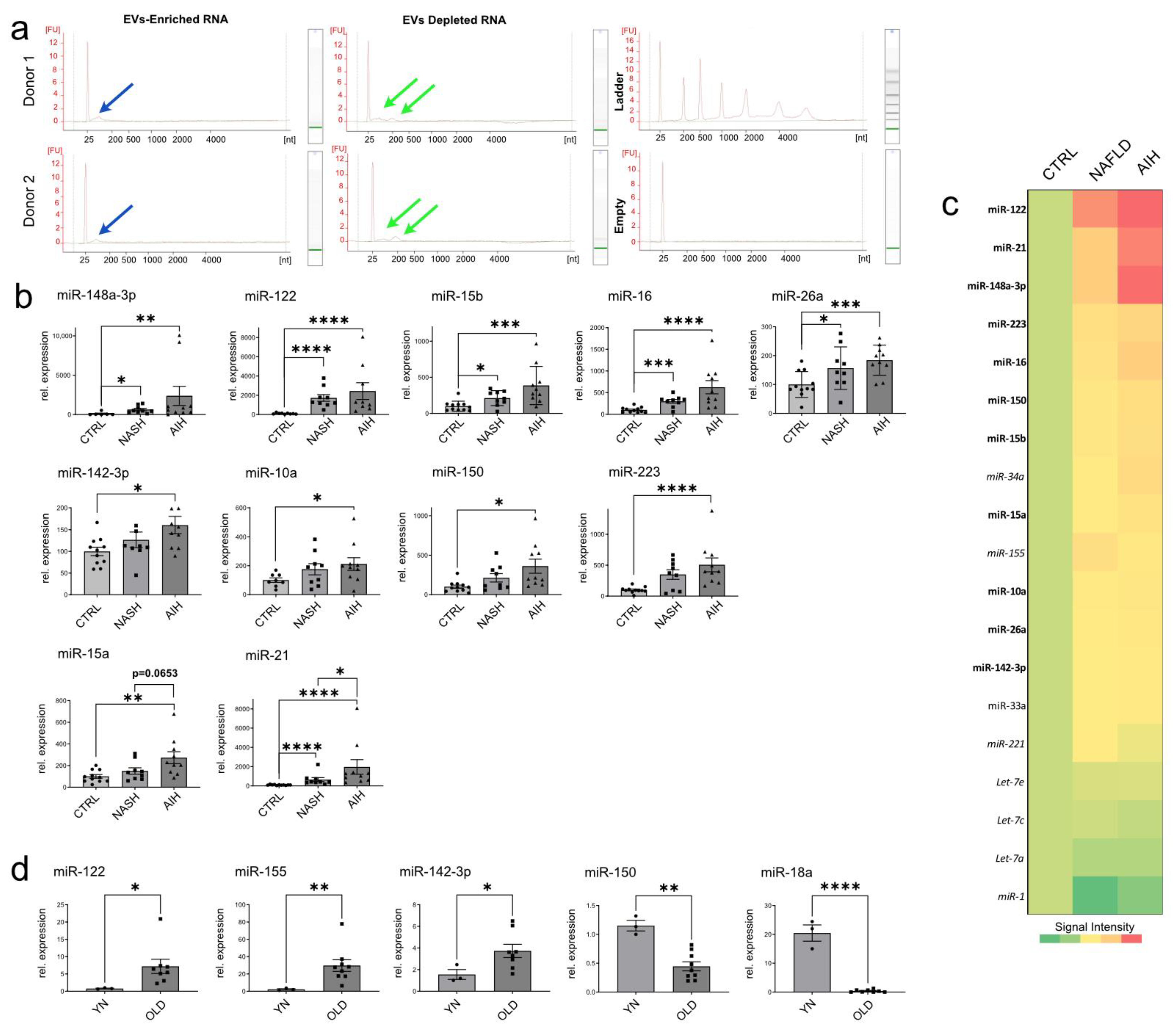
2.5. Analysis of miRNA Expression in EVs Isolated from Patients with Liver Diseases and Aged Animals
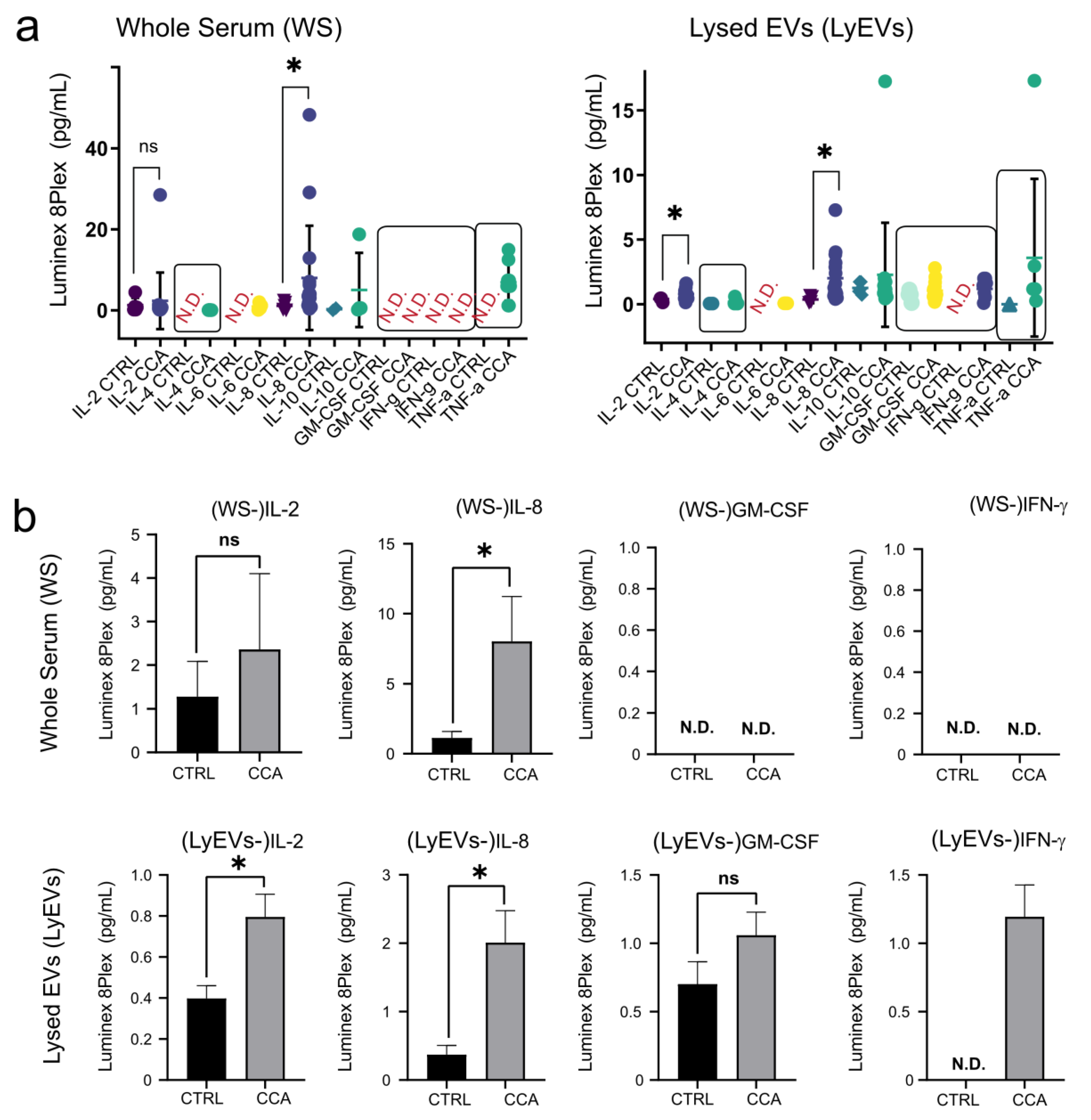
2.6. Luminex-Based Measurement of Cytokines in EV-Enriched Pellets and EVs-Depleted Supernatants
3. Discussion
4. Materials & Methods
4.1. Quantification of EVs by Using Nanoparticle Tracking Analyzer and Flow Cytometry
4.2. Isolation of extracellular vesicles with PEG and TEI, and labeling
4.3. Western Blot Analyses and Antibodies
4.4. RNA Isolation, Visualization and miRNA Analysis by miQPCR
4.5. Preparation of Primary Cells from Rat Liver and Liver Samples from Gunn and Aging Rats
4.6. Visualization and Quantification of EVs Uptake
4.7. Patient’s Material and Cytokine Quantification by Luminex
4.8. Statistical Analysis and Imaging Software
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Pitt, J.M.; Kroemer, G.; Zitvogel, L. Extracellular vesicles: Masters of intercellular communication and potential clinical interventions. J. Clin. Investig. 2016, 126, 1139–1143. [Google Scholar] [CrossRef]
- Murphy, D.E.; de Jong, O.G.; Brouwer, M.; Wood, M.J.; Lavieu, G.; Schiffelers, R.M.; Vader, P. Extracellular vesicle-based therapeutics: Natural versus engineered targeting and trafficking. Exp. Mol. Med. 2019, 51, 1–12. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- Yanez-Mo, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed]
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience 2015, 65, 783–797. [Google Scholar] [CrossRef]
- Tucher, C.; Bode, K.; Schiller, P.; Classen, L.; Birr, C.; Souto-Carneiro, M.M.; Blank, N.; Lorenz, H.M.; Schiller, M. Extracellular Vesicle Subtypes Released From Activated or Apoptotic T-Lymphocytes Carry a Specific and Stimulus-Dependent Protein Cargo. Front. Immunol. 2018, 9, 534. [Google Scholar] [CrossRef] [PubMed]
- Liangsupree, T.; Multia, E.; Riekkola, M.L. Modern isolation and separation techniques for extracellular vesicles. J. Chromatogr. A 2021, 1636, 461773. [Google Scholar] [CrossRef]
- Sidhom, K.; Obi, P.O.; Saleem, A. A Review of Exosomal Isolation Methods: Is Size Exclusion Chromatography the Best Option? Int. J. Mol. Sci. 2020, 21, 6466. [Google Scholar] [CrossRef]
- Chowdhury, J.R.; Kondapalli, R.; Chowdhury, N.R. Gunn rat: A model for inherited deficiency of bilirubin glucuronidation. Adv. Vet. Sci. Comp. Med. 1993, 37, 149–173. [Google Scholar]
- Yamamoto, S.; Niida, S.; Azuma, E.; Yanagibashi, T.; Muramatsu, M.; Huang, T.T.; Sagara, H.; Higaki, S.; Ikutani, M.; Nagai, Y.; et al. Inflammation-induced endothelial cell-derived extracellular vesicles modulate the cellular status of pericytes. Sci. Rep. 2015, 5, 8505. [Google Scholar] [CrossRef]
- Baran, J.; Baj-Krzyworzeka, M.; Weglarczyk, K.; Szatanek, R.; Zembala, M.; Barbasz, J.; Czupryna, A.; Szczepanik, A.; Zembala, M. Circulating tumour-derived microvesicles in plasma of gastric cancer patients. Cancer Immunol. Immunother. CII 2010, 59, 841–850. [Google Scholar] [CrossRef]
- Bagci, C.; Sever-Bahcekapili, M.; Belder, N.; Bennett, A.P.S.; Erdener, S.E.; Dalkara, T. Overview of extracellular vesicle characterization techniques and introduction to combined reflectance and fluorescence confocal microscopy to distinguish extracellular vesicle subpopulations. Neurophotonics 2022, 9, 021903. [Google Scholar] [CrossRef]
- Mehdiani, A.; Maier, A.; Pinto, A.; Barth, M.; Akhyari, P.; Lichtenberg, A. An innovative method for exosome quantification and size measurement. J. Vis. Exp. 2015, 95, 50974. [Google Scholar] [CrossRef]
- Auger, C.; Brunel, A.; Darbas, T.; Akil, H.; Perraud, A.; Begaud, G.; Bessette, B.; Christou, N.; Verdier, M. Extracellular Vesicle Measurements with Nanoparticle Tracking Analysis: A Different Appreciation of Up and Down Secretion. Int. J. Mol. Sci. 2022, 23, 2310. [Google Scholar] [CrossRef]
- Bachurski, D.; Schuldner, M.; Nguyen, P.H.; Malz, A.; Reiners, K.S.; Grenzi, P.C.; Babatz, F.; Schauss, A.C.; Hansen, H.P.; Hallek, M.; et al. Extracellular vesicle measurements with nanoparticle tracking analysis—An accuracy and repeatability comparison between NanoSight NS300 and ZetaView. J. Extracell. Vesicles 2019, 8, 1596016. [Google Scholar] [CrossRef] [PubMed]
- Osteikoetxea, X.; Sodar, B.; Nemeth, A.; Szabo-Taylor, K.; Paloczi, K.; Vukman, K.V.; Tamasi, V.; Balogh, A.; Kittel, A.; Pallinger, E.; et al. Differential detergent sensitivity of extracellular vesicle subpopulations. Org. Biomol. Chem. 2015, 13, 9775–9782. [Google Scholar] [CrossRef]
- Tcaciuc, E.; Podurean, M.; Tcaciuc, A. Management of Crigler-Najjar syndrome. Med. Pharm. Rep. 2021, 94, S64–S67. [Google Scholar] [PubMed]
- Philipson, L.; Albertsson, P.A.; Frick, G. The purification and concentration of viruses by aqueous polymerphase systems. Virology 1960, 11, 553–571. [Google Scholar] [CrossRef]
- Rider, M.A.; Hurwitz, S.N.; Meckes, D.G., Jr. ExtraPEG: A Polyethylene Glycol-Based Method for Enrichment of Extracellular Vesicles. Sci. Rep. 2016, 6, 23978. [Google Scholar] [CrossRef]
- Nielsen, M.H.; Beck-Nielsen, H.; Andersen, M.N.; Handberg, A. A flow cytometric method for characterization of circulating cell-derived microparticles in plasma. J. Extracell. Vesicles 2014, 3, 20795. [Google Scholar] [CrossRef]
- Turchinovich, A.; Baranova, A.; Drapkina, O.; Tonevitsky, A. Cell-Free Circulating Nucleic Acids as Early Biomarkers for NAFLD and NAFLD-Associated Disorders. Front. Physiol. 2018, 9, 1256. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Kondo, S.; Matsuzaki, J.; Esaki, M.; Okusaka, T.; Shimada, K.; Murakami, Y.; Enomoto, M.; Tamori, A.; Kato, K.; et al. Highly Sensitive Circulating MicroRNA Panel for Accurate Detection of Hepatocellular Carcinoma in Patients With Liver Disease. Hepatol. Commun. 2020, 4, 284–297. [Google Scholar] [CrossRef] [PubMed]
- Rohn, F.; Kordes, C.; Buschmann, T.; Reichert, D.; Wammers, M.; Poschmann, G.; Stuhler, K.; Benk, A.S.; Geiger, F.; Spatz, J.P.; et al. Impaired integrin alpha5 /beta1 -mediated hepatocyte growth factor release by stellate cells of the aged liver. Aging Cell 2020, 19, e13131. [Google Scholar] [CrossRef] [PubMed]
- Giloteaux, L.; O’Neal, A.; Castro-Marrero, J.; Levine, S.M.; Hanson, M.R. Cytokine profiling of extracellular vesicles isolated from plasma in myalgic encephalomyelitis/chronic fatigue syndrome: A pilot study. J. Transl. Med. 2020, 18, 387. [Google Scholar] [CrossRef]
- Fitzgerald, W.; Freeman, M.L.; Lederman, M.M.; Vasilieva, E.; Romero, R.; Margolis, L. A System of Cytokines Encapsulated in ExtraCellular Vesicles. Sci. Rep. 2018, 8, 8973. [Google Scholar] [CrossRef]
- Banales, J.M.; Marin, J.J.G.; Lamarca, A.; Rodrigues, P.M.; Khan, S.A.; Roberts, L.R.; Cardinale, V.; Carpino, G.; Andersen, J.B.; Braconi, C.; et al. Cholangiocarcinoma 2020: The next horizon in mechanisms and management. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 557–588. [Google Scholar] [CrossRef]
- Rodrigues, P.M.; Vogel, A.; Arrese, M.; Balderramo, D.C.; Valle, J.W.; Banales, J.M. Next-Generation Biomarkers for Cholangiocarcinoma. Cancers 2021, 13, 3222. [Google Scholar] [CrossRef]
- Dana, P.; Kariya, R.; Lert-Itthiporn, W.; Seubwai, W.; Saisomboon, S.; Wongkham, C.; Okada, S.; Wongkham, S.; Vaeteewoottacharn, K. Homophilic Interaction of CD147 Promotes IL-6-Mediated Cholangiocarcinoma Invasion via the NF-kappaB-Dependent Pathway. Int. J. Mol. Sci. 2021, 22, 13496. [Google Scholar] [CrossRef]
- Webber, J.; Clayton, A. How pure are your vesicles? J. Extracell. Vesicles 2013, 2, 19861. [Google Scholar] [CrossRef]
- Takov, K.; Yellon, D.M.; Davidson, S.M. Comparison of small extracellular vesicles isolated from plasma by ultracentrifugation or size-exclusion chromatography: Yield, purity and functional potential. J. Extracell. Vesicles 2019, 8, 1560809. [Google Scholar] [CrossRef] [PubMed]
- Hirsova, P.; Ibrahim, S.H.; Krishnan, A.; Verma, V.K.; Bronk, S.F.; Werneburg, N.W.; Charlton, M.R.; Shah, V.H.; Malhi, H.; Gores, G.J. Lipid-Induced Signaling Causes Release of Inflammatory Extracellular Vesicles From Hepatocytes. Gastroenterology 2016, 150, 956–967. [Google Scholar] [CrossRef] [PubMed]
- Kakazu, E.; Mauer, A.S.; Yin, M.; Malhi, H. Hepatocytes release ceramide-enriched pro-inflammatory extracellular vesicles in an IRE1alpha-dependent manner. J. Lipid Res. 2016, 57, 233–245. [Google Scholar] [CrossRef]
- Castoldi, M.; Kordes, C.; Sawitza, I.; Haussinger, D. Isolation and characterization of vesicular and non-vesicular microRNAs circulating in sera of partially hepatectomized rats. Sci. Rep. 2016, 6, 31869. [Google Scholar] [CrossRef]
- Scholer, D.; Castoldi, M.; Jordens, M.S.; Schulze-Hagen, M.; Kuhl, C.; Keitel, V.; Luedde, T.; Roderburg, C.; Loosen, S.H. Enlarged extracellular vesicles are a negative prognostic factor in patients undergoing TACE for primary or secondary liver cancer-a case series. PLoS ONE 2021, 16, e0255983. [Google Scholar] [CrossRef]
- Chahar, H.S.; Corsello, T.; Kudlicki, A.S.; Komaravelli, N.; Casola, A. Respiratory Syncytial Virus Infection Changes Cargo Composition of Exosome Released from Airway Epithelial Cells. Sci. Rep. 2018, 8, 387. [Google Scholar] [CrossRef]
- Eldh, M.; Lotvall, J.; Malmhall, C.; Ekstrom, K. Importance of RNA isolation methods for analysis of exosomal RNA: Evaluation of different methods. Mol. Immunol. 2012, 50, 278–286. [Google Scholar] [CrossRef]
- Jung, H.H.; Kim, J.Y.; Lim, J.E.; Im, Y.H. Cytokine profiling in serum-derived exosomes isolated by different methods. Sci. Rep. 2020, 10, 14069. [Google Scholar] [CrossRef] [PubMed]
- El-Hefnawy, T.; Raja, S.; Kelly, L.; Bigbee, W.L.; Kirkwood, J.M.; Luketich, J.D.; Godfrey, T.E. Characterization of amplifiable, circulating RNA in plasma and its potential as a tool for cancer diagnostics. Clin. Chem. 2004, 50, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Rykova, E.Y.; Wunsche, W.; Brizgunova, O.E.; Skvortsova, T.E.; Tamkovich, S.N.; Senin, I.S.; Laktionov, P.P.; Sczakiel, G.; Vlassov, V.V. Concentrations of circulating RNA from healthy donors and cancer patients estimated by different methods. Ann. N. Y. Acad. Sci. 2006, 1075, 328–333. [Google Scholar] [CrossRef]
- Benes, V.; Collier, P.; Kordes, C.; Stolte, J.; Rausch, T.; Muckentaler, M.U.; Haussinger, D.; Castoldi, M. Identification of cytokine-induced modulation of microRNA expression and secretion as measured by a novel microRNA specific qPCR assay. Sci. Rep. 2015, 5, 11590. [Google Scholar] [CrossRef]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Gunn, C.K. Hereditary Acholuric Jaundice in the Rat. Can. Med. Assoc. J. 1944, 50, 230–237. [Google Scholar] [PubMed]

| NAFLD | (n = 24) | AIH | (n = 9) | Statistical | |
|---|---|---|---|---|---|
| Range | Mean ± SD | Range | Mean ± SD | Analysis | |
| GOT (U/L) | 23–76 | 44.3 ± 16.3 | 27–337 | 98.2 ± 98.9 | n.s. |
| GPT (U/L) | 12–144 | 65.7 ± 41.9 | 29–451 | 120.1 ± 130.6 | n.s. |
| GGT (U/L) | 8–657 | 132.5 ± 156.3 | 14–589 | 201.6 ± 173.4 | n.s. |
| AP (U/L) | 37–195 | 75.5 ± 34.6 | 60–158 | 108.1 ± 32.0 | p = 0.0037 |
| Albumin | 4.0–5.1 | 4.56 ± 0.31 | 4.0–4.6 | 4.29 ± 0.26 | p = 0.0153 |
| Bilirubin (mg/dL) | 0.17–2.51 | 0.55 ± 0.48 | 0.27–0.58 | 0.44 ± 0.10 | n.s. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paluschinski, M.; Loosen, S.; Kordes, C.; Keitel, V.; Kuebart, A.; Brandenburger, T.; Schöler, D.; Wammers, M.; Neumann, U.P.; Luedde, T.; et al. Extracellular Vesicles as Markers of Liver Function: Optimized Workflow for Biomarker Identification in Liver Disease. Int. J. Mol. Sci. 2023, 24, 9631. https://doi.org/10.3390/ijms24119631
Paluschinski M, Loosen S, Kordes C, Keitel V, Kuebart A, Brandenburger T, Schöler D, Wammers M, Neumann UP, Luedde T, et al. Extracellular Vesicles as Markers of Liver Function: Optimized Workflow for Biomarker Identification in Liver Disease. International Journal of Molecular Sciences. 2023; 24(11):9631. https://doi.org/10.3390/ijms24119631
Chicago/Turabian StylePaluschinski, Martha, Sven Loosen, Claus Kordes, Verena Keitel, Anne Kuebart, Timo Brandenburger, David Schöler, Marianne Wammers, Ulf P. Neumann, Tom Luedde, and et al. 2023. "Extracellular Vesicles as Markers of Liver Function: Optimized Workflow for Biomarker Identification in Liver Disease" International Journal of Molecular Sciences 24, no. 11: 9631. https://doi.org/10.3390/ijms24119631
APA StylePaluschinski, M., Loosen, S., Kordes, C., Keitel, V., Kuebart, A., Brandenburger, T., Schöler, D., Wammers, M., Neumann, U. P., Luedde, T., & Castoldi, M. (2023). Extracellular Vesicles as Markers of Liver Function: Optimized Workflow for Biomarker Identification in Liver Disease. International Journal of Molecular Sciences, 24(11), 9631. https://doi.org/10.3390/ijms24119631








