Conjugated Dienoic Acid Peroxides as Substrates in Chaetopterus Bioluminescence System
Abstract
1. Introduction
2. Results and Discussion
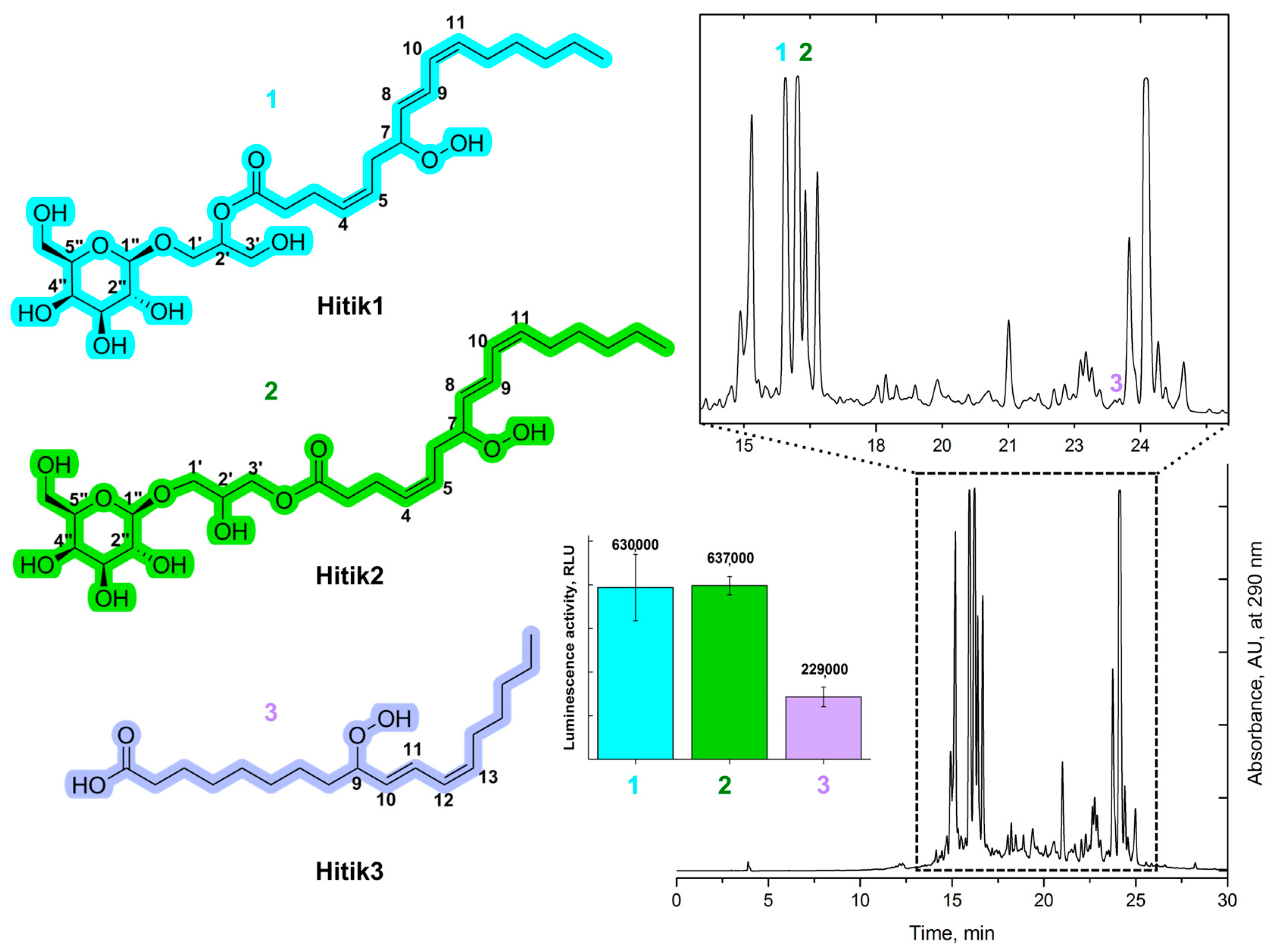
2.1. Extraction, Purification, and Structural Elucidation of Bioluminescent Compounds from Chaetomorpha Linum Biomass
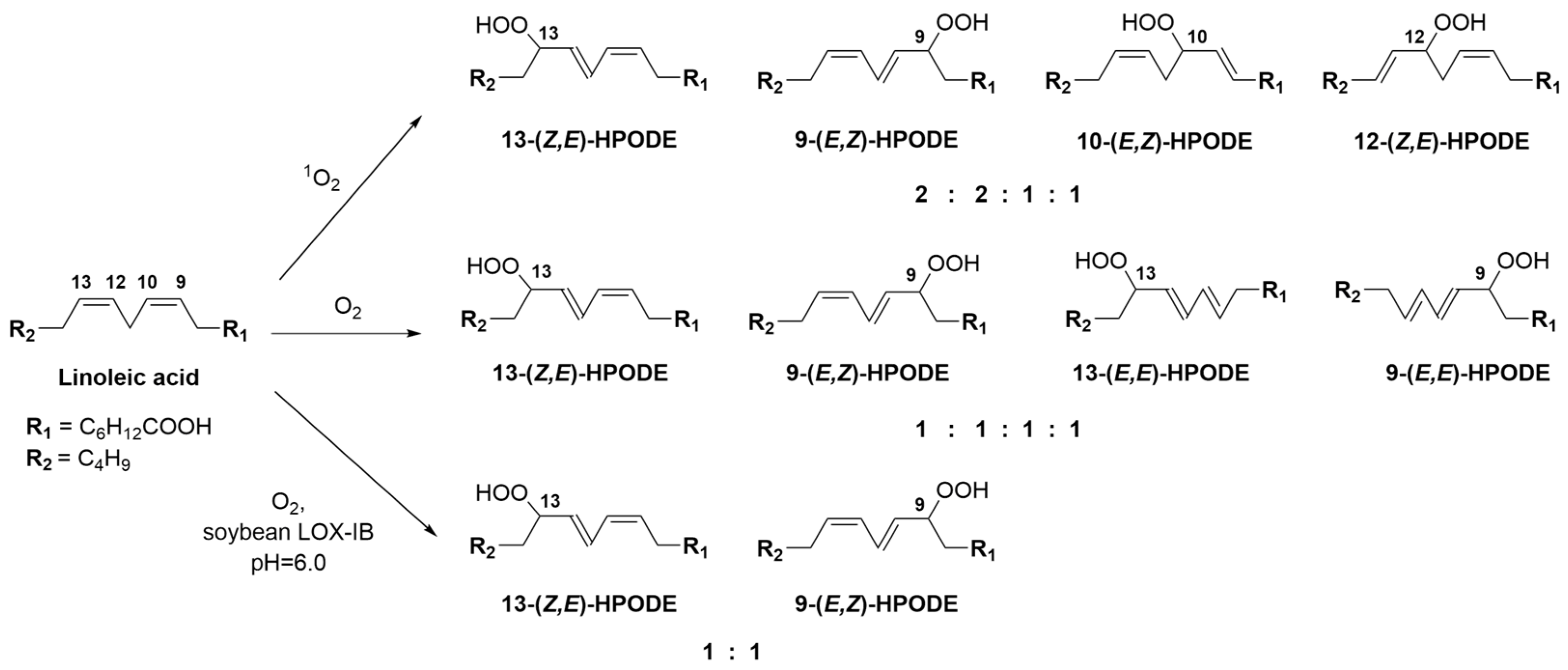
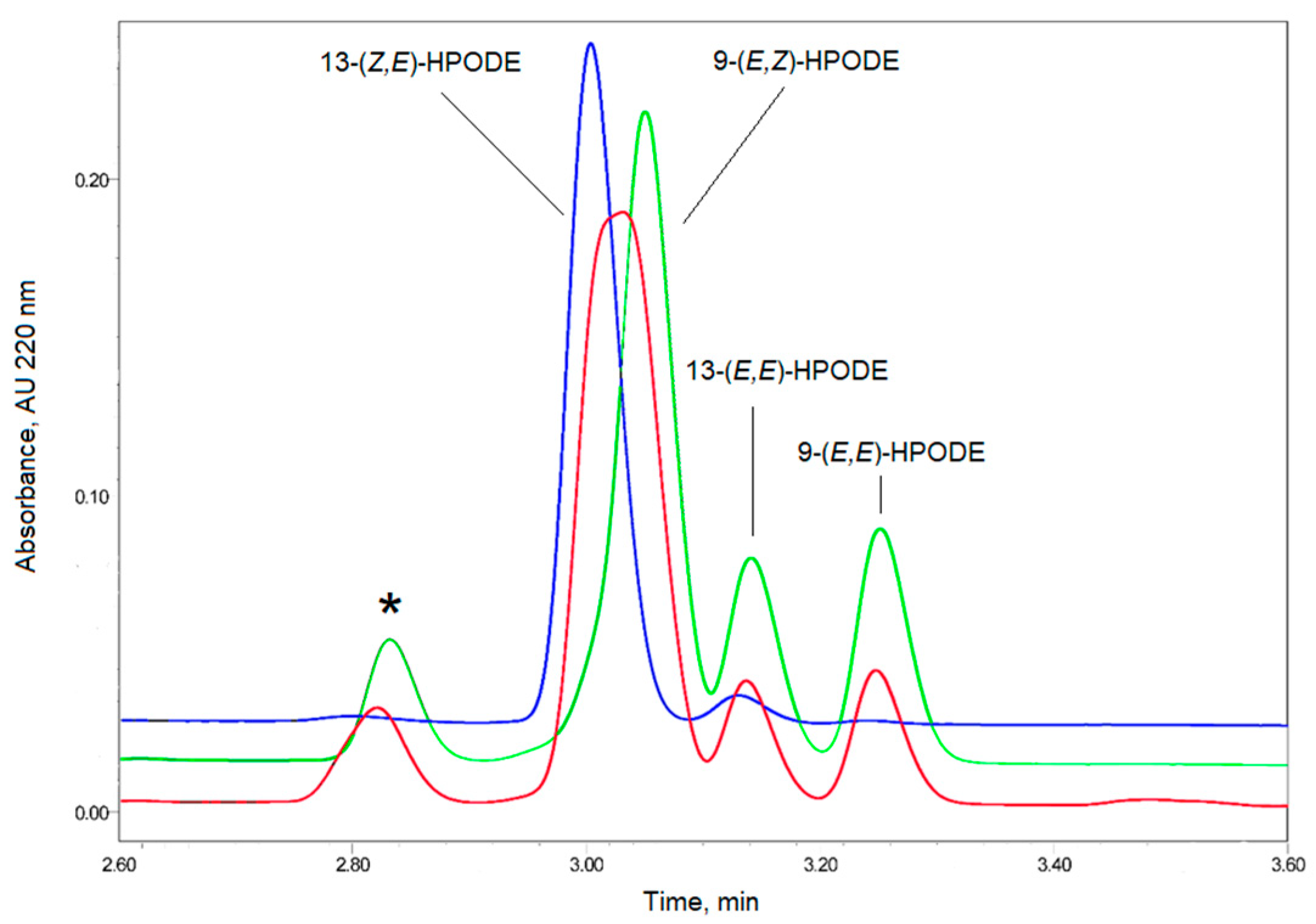
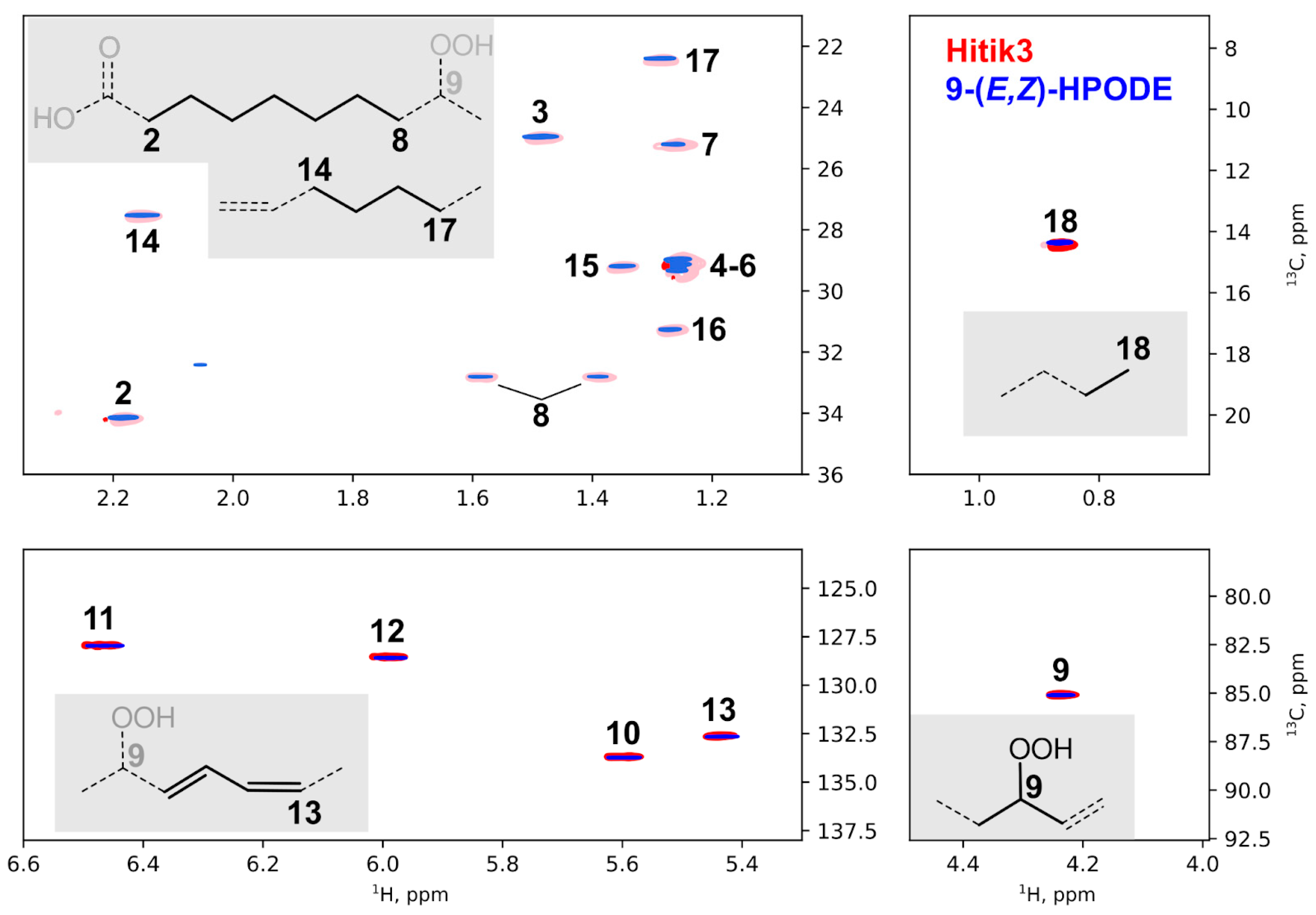
2.2. Synthesis of Bioluminescent Substrate Hitik3
2.3. Synthesis of Hitik Structural Analogues
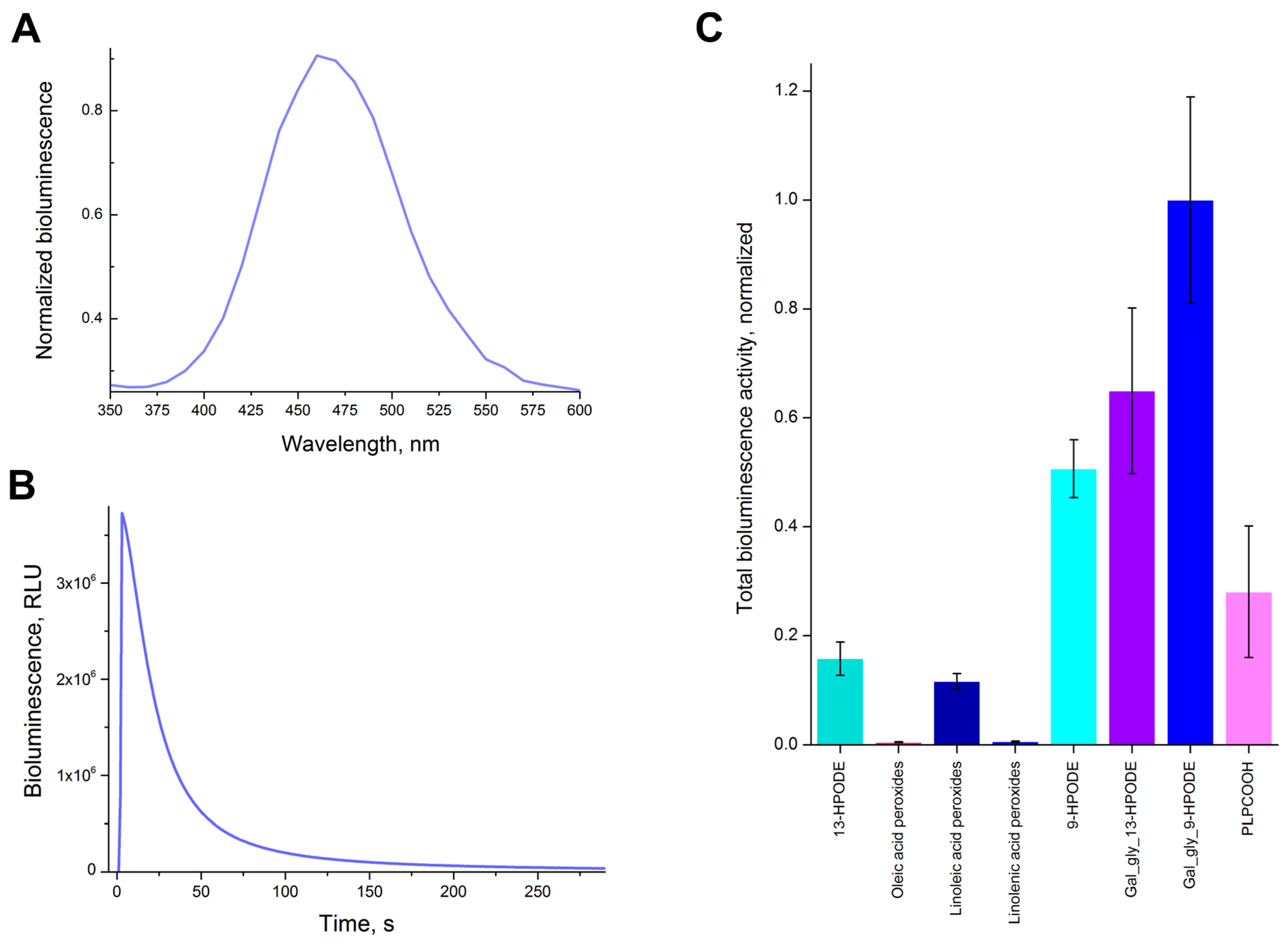
2.4. Bioluminescent Activity of the Synthesized Substrates
2.5. Mechanism of Chaetopterus Bioluminescence and Bioimaging of Ferroptosis
3. Materials and Methods
3.1. Chaetopterus Variopedatus Biomass and Luciferase
3.2. Hitik1, Hitik2, Hitik3 Purification Procedure
3.3. LOX-Catalyzed Peroxidation of PUFA
3.4. Methylene Blue Catalyzed Photo-Oxidation of PUFA or Their Derivatives
3.5. Bioluminescence Spectrum of 13-HPODE
3.6. Bioluminescence Activity Assay
3.7. Other Methods, Synthetic Procedures, and NMR Spectra
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Osamu Shimomura, I.V.Y. Bioluminescence: Chemical Principles and Methods, 3rd, ed.; World Scientific: Singapore, 2019; ISBN 981-327-712-2. [Google Scholar]
- Tsarkova, A.S. Luciferins Under Construction: A Review of Known Biosynthetic Pathways. Front. Ecol. Evol. 2021, 9, 667829. [Google Scholar] [CrossRef]
- Syed, A.J.; Anderson, J.C. Applications of Bioluminescence in Biotechnology and Beyond. Chem. Soc. Rev. 2021, 50, 5668–5705. [Google Scholar] [CrossRef]
- Oba, Y.; Stevani, C.V.; Oliveira, A.G.; Tsarkova, A.S.; Chepurnykh, T.V.; Yampolsky, I.V. Selected Least Studied but Not Forgotten Bioluminescent Systems. Photochem. Photobiol. 2017, 93, 405–415. [Google Scholar] [CrossRef]
- Panceri, P. Atti della R. Academia delle Scienze Fisiche e Matematiche. In Osservazioni Intorno A Nuove Forme di Vermi Nematodi Marini; Pera Studio Bibliografico: Lucca, Italy, 1878. [Google Scholar]
- Mirza, J.D.; Migotto, Á.E.; Yampolsky, I.V.; de Moraes, G.V.; Tsarkova, A.S.; Oliveira, A.G. Chaetopterus Variopedatus Bioluminescence: A Review of Light Emission within a Species Complex. Photochem. Photobiol. 2020, 96, 768–778. [Google Scholar] [CrossRef]
- Shimomura, O.; Johnson, F.H. Chaetopterus Photoprotein: Crystallization and Cofactor Requirements for Bioluminescence. Science 1968, 159, 1239–1240. [Google Scholar] [CrossRef]
- Deheyn, D.D.; Enzor, L.A.; Dubowitz, A.; Urbach, J.S.; Blair, D. Optical and Physicochemical Characterization of the Luminous Mucous Secreted by the Marine Worm Chaetopterus sp. Physiol. Biochem. Zool. 2013, 86, 702–705. [Google Scholar] [CrossRef]
- Purtov, K.V.; Petushkov, V.N.; Rodionova, N.S.; Pakhomova, V.G.; Myasnyanko, I.N.; Myshkina, N.M.; Tsarkova, A.S.; Gitelson, J.I. Luciferin–Luciferase System of Marine Polychaete Chaetopterus variopedatus. Dokl. Biochem. Biophys. 2019, 486, 209–212. [Google Scholar] [CrossRef]
- Purtov, K.V.; Petushkov, V.N.; Baranov, M.S.; Mineev, K.S.; Rodionova, N.S.; Kaskova, Z.M.; Tsarkova, A.S.; Petunin, A.I.; Bondar, V.S.; Rodicheva, E.K.; et al. The Chemical Basis of Fungal Bioluminescence. Angew. Chem. Int. Ed. 2015, 54, 8124–8128. [Google Scholar] [CrossRef]
- Lundborg, M.; Widmalm, G. NMR Chemical Shift Prediction of Glycans: Application of the Computer Program CASPER in Structural Analysis. In Glycoinformatics; Lütteke, T., Frank, M., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2015; Volume 1273, pp. 29–40. ISBN 978-1-4939-2342-7. [Google Scholar]
- Iacazio, G.; Langrand, G.; Baratti, J.; Buono, G.; Triantaphylides, C. Preparative, Enzymic Synthesis of Linoleic Acid (13S)-Hydroperoxide Using Soybean Lipoxygenase-1. J. Org. Chem. 1990, 55, 1690–1691. [Google Scholar] [CrossRef]
- Porter, N.A. Mechanisms for the Autoxidation of Polyunsaturated Lipids. Acc. Chem. Res. 1986, 19, 262–268. [Google Scholar] [CrossRef]
- Romero, N.A.; Nicewicz, D.A. Organic Photoredox Catalysis. Chem. Rev. 2016, 116, 10075–10166. [Google Scholar] [CrossRef]
- Di Mascio, P.; Martinez, G.R.; Miyamoto, S.; Ronsein, G.E.; Medeiros, M.H.G.; Cadet, J. Singlet Molecular Oxygen Reactions with Nucleic Acids, Lipids, and Proteins. Chem. Rev. 2019, 119, 2043–2086. [Google Scholar] [CrossRef]
- Gardner, H.W. Soybean Lipoxygenase-1 Enzymically Forms Both (9S)- and (13S)-Hydroperoxides from Linoleic Acid by a PH-Dependent Mechanism. Biochim. Biophys. Acta BBA-Lipids Lipid Metab. 1989, 1001, 274–281. [Google Scholar] [CrossRef]
- Ziegler, T. Preparation of Some Fully Chloroacetylated Glycopyranosyl Bromides: Useful Intermediates for the Synthesis of Base- and Hydrogenolysis-Sensitive Glycosides. Liebigs Ann. Chem. 1990, 1990, 1125–1131. [Google Scholar] [CrossRef]
- Manzo, E.; Ciavatta, M.L.; Pagano, D.; Fontana, A. An Efficient and Versatile Chemical Synthesis of Bioactive Glyco-Glycerolipids. Tetrahedron Lett. 2012, 53, 879–881. [Google Scholar] [CrossRef]
- Pagano, D.; Cutignano, A.; Manzo, E.; Tinto, F.; Fontana, A. Glycolipids Synthesis: Improved Hydrazinolysis Conditions for Preparation of 1,2-Polyunsaturated Fatty Acyl-β-Monogalactosyl-Glycerols. Carbohydr. Res. 2016, 424, 21–23. [Google Scholar] [CrossRef]
- von Minden, H.M.; Morr, M.; Milkereit, G.; Heinz, E.; Vill, V. Synthesis and Mesogenic Properties of Glycosyl Diacylglycerols. Chem. Phys. Lipids 2002, 114, 55–80. [Google Scholar] [CrossRef]
- Dussault, P.; Sahli, A. An Olefination-Based Route to Unsaturated Hydroperoxides. Tetrahedron Lett. 1990, 31, 5118–5120. [Google Scholar] [CrossRef]
- Ibusuki, D.; Nakagawa, K.; Asai, A.; Oikawa, S.; Masuda, Y.; Suzuki, T.; Miyazawa, T. Preparation of Pure Lipid Hydroperoxides. J. Lipid Res. 2008, 49, 2668–2677. [Google Scholar] [CrossRef]
- Branchini, B.R.; Behney, C.E.; Southworth, T.L.; Rawat, R.; Deheyn, D.D. Chemical Analysis of the Luminous Slime Secreted by the Marine Worm Chaetopterus (Annelida, Polychaeta). Photochem. Photobiol. 2014, 90, 247–251. [Google Scholar] [CrossRef]
- Brodl, E.; Winkler, A.; Macheroux, P. Molecular Mechanisms of Bacterial Bioluminescence. Comput. Struct. Biotechnol. J. 2018, 16, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Petushkov, V.N.; Vavilov, M.V.; Ivanov, I.A.; Ziganshin, R.H.; Rodionova, N.S.; Yampolsky, I.V.; Tsarkova, A.S.; Dubinnyi, M.A. Deazaflavin Cofactor Boosts Earthworms Henlea Bioluminescence. Org. Biomol. Chem. 2023, 21, 415–427. [Google Scholar] [CrossRef]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Friedmann Angeli, J.P.; Schneider, M.; Proneth, B.; Tyurina, Y.Y.; Tyurin, V.A.; Hammond, V.J.; Herbach, N.; Aichler, M.; Walch, A.; Eggenhofer, E.; et al. Inactivation of the Ferroptosis Regulator Gpx4 Triggers Acute Renal Failure in Mice. Nat. Cell Biol. 2014, 16, 1180–1191. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cao, F.; Yin, H.; Huang, Z.; Lin, Z.; Mao, N.; Sun, B.; Wang, G. Ferroptosis: Past, Present and Future. Cell Death Dis. 2020, 11, 88. [Google Scholar] [CrossRef]
- Zeng, F.; Nijiati, S.; Tang, L.; Ye, J.; Zhou, Z.; Chen, X. Ferroptosis Detection: From Approaches to Applications. Angew. Chem. Int. Ed. Engl. 2003; e202300379, ahead of print. [Google Scholar] [CrossRef]
- Li, H.; An, Y.; Gao, J.; Yang, M.; Luo, J.; Li, X.; Lv, J.; Li, X.; Yuan, Z.; Ma, H. Recent Advances of Fluorescence Probes for Imaging of Ferroptosis Process. Chemosensors 2022, 10, 233. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zagitova, R.I.; Purtov, K.V.; Shcheglov, A.S.; Mineev, K.S.; Dubinnyi, M.A.; Myasnyanko, I.N.; Belozerova, O.A.; Pakhomova, V.G.; Petushkov, V.N.; Rodionova, N.S.; et al. Conjugated Dienoic Acid Peroxides as Substrates in Chaetopterus Bioluminescence System. Int. J. Mol. Sci. 2023, 24, 9466. https://doi.org/10.3390/ijms24119466
Zagitova RI, Purtov KV, Shcheglov AS, Mineev KS, Dubinnyi MA, Myasnyanko IN, Belozerova OA, Pakhomova VG, Petushkov VN, Rodionova NS, et al. Conjugated Dienoic Acid Peroxides as Substrates in Chaetopterus Bioluminescence System. International Journal of Molecular Sciences. 2023; 24(11):9466. https://doi.org/10.3390/ijms24119466
Chicago/Turabian StyleZagitova, Renata I., Konstantin V. Purtov, Aleksandr S. Shcheglov, Konstantin S. Mineev, Maxim A. Dubinnyi, Ivan N. Myasnyanko, Olga A. Belozerova, Vera G. Pakhomova, Valentin N. Petushkov, Natalia S. Rodionova, and et al. 2023. "Conjugated Dienoic Acid Peroxides as Substrates in Chaetopterus Bioluminescence System" International Journal of Molecular Sciences 24, no. 11: 9466. https://doi.org/10.3390/ijms24119466
APA StyleZagitova, R. I., Purtov, K. V., Shcheglov, A. S., Mineev, K. S., Dubinnyi, M. A., Myasnyanko, I. N., Belozerova, O. A., Pakhomova, V. G., Petushkov, V. N., Rodionova, N. S., Lushpa, V. A., Guglya, E. B., Kovalchuk, S., Kozhemyako, V. B., Mirza, J. D., Oliveira, A. G., Yampolsky, I. V., Kaskova, Z. M., & Tsarkova, A. S. (2023). Conjugated Dienoic Acid Peroxides as Substrates in Chaetopterus Bioluminescence System. International Journal of Molecular Sciences, 24(11), 9466. https://doi.org/10.3390/ijms24119466







