An In Vivo Whole-Transcriptomic Approach to Assess Developmental and Reproductive Impairments Caused by Flumequine in Daphnia magna
Abstract
1. Introduction
2. Results
2.1. Phenotypical Responses
2.2. Transcriptomic Effects of FLU in D. magna
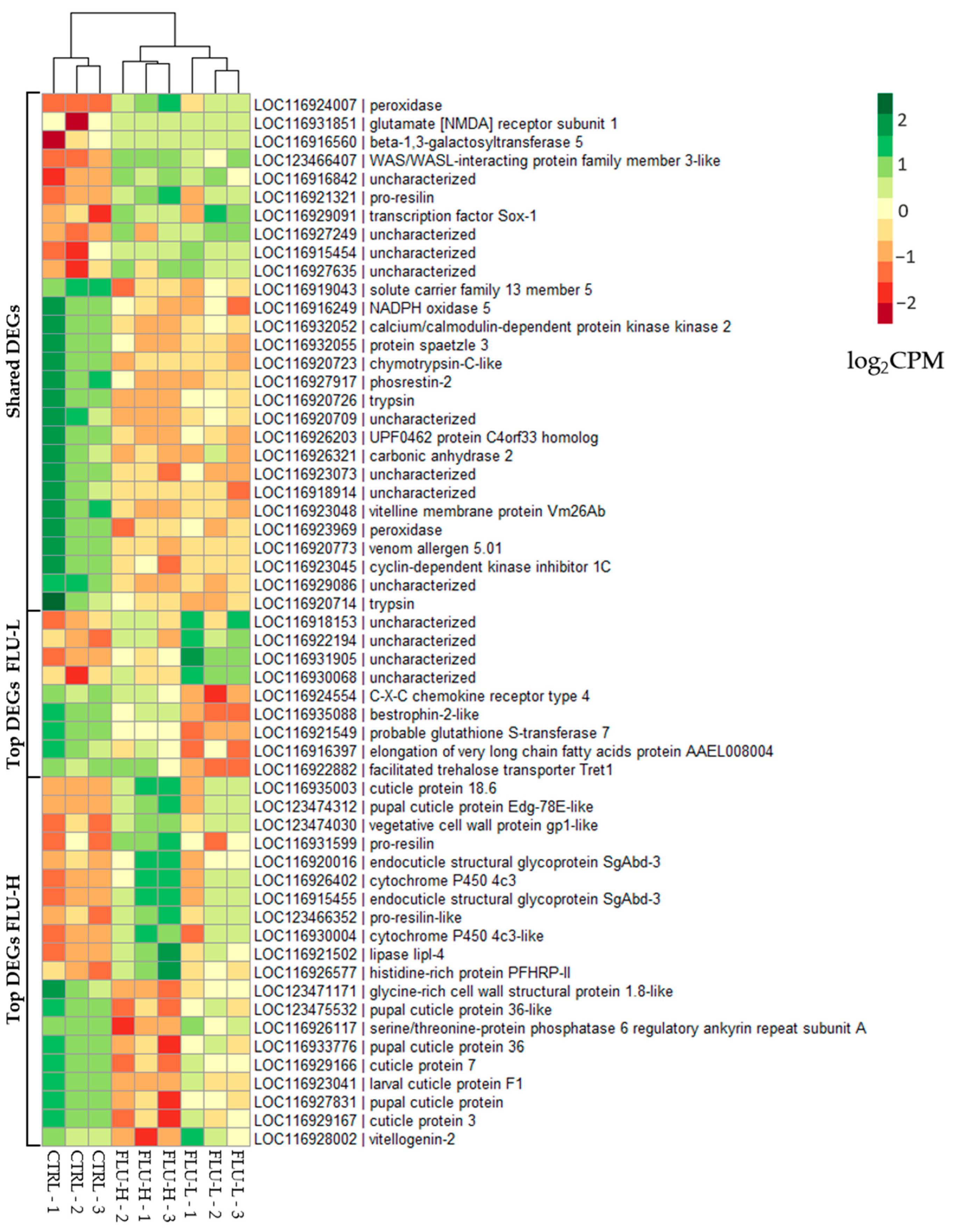
2.2.1. Differentially Expressed Genes (DEGs)
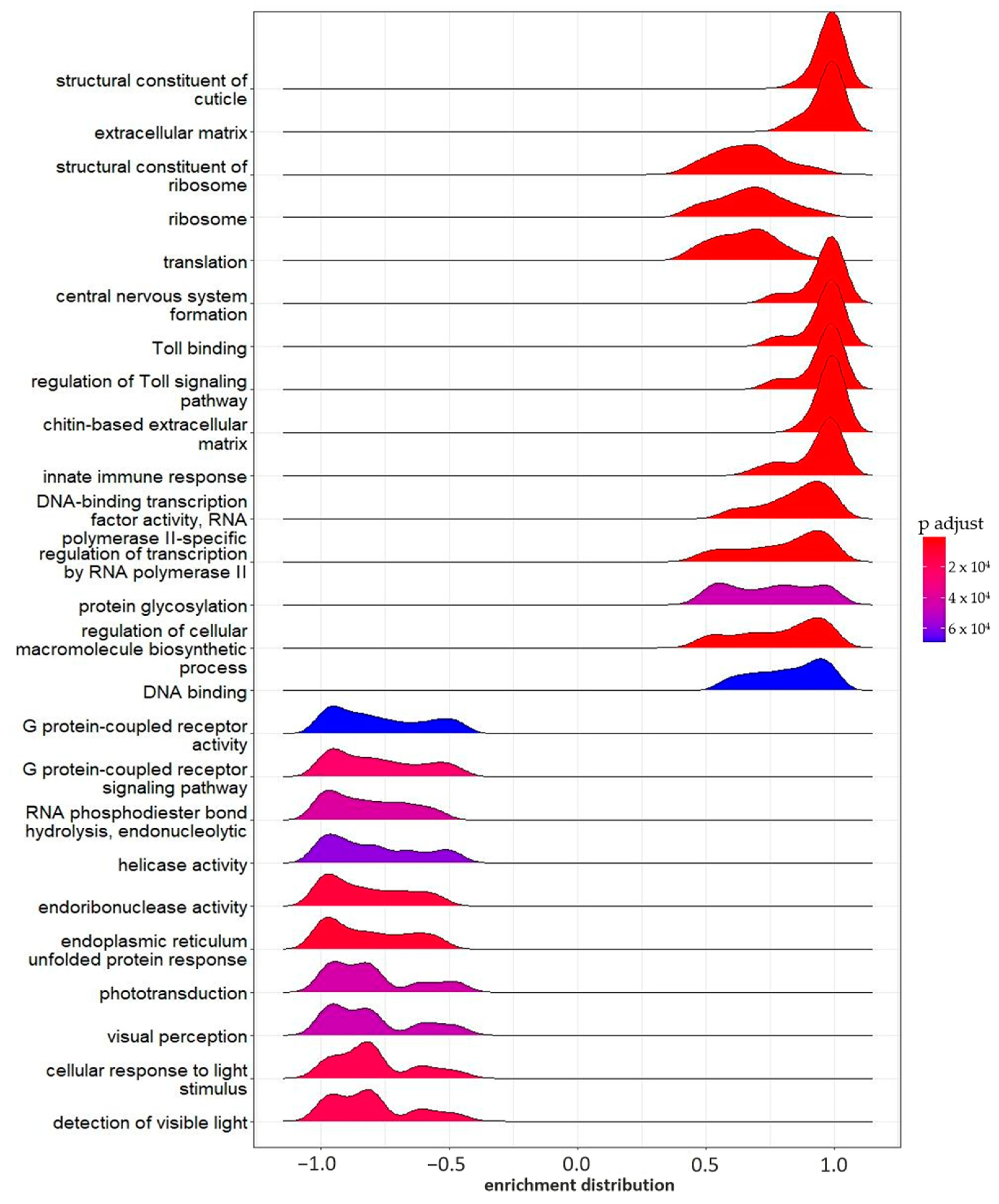
2.2.2. Functional Enrichment Analysis
2.2.3. Gene Set Enrichment Analysis (GSEA)
2.2.4. Targeted Gene Expression
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Test Organism and Culture Conditions
4.3. Daphnia magna Reproduction Test
4.4. RNA Isolation and RNA Sequencing
4.5. Transcriptome Mapping and Differential Expression Analysis
4.6. Enrichment Analysis
4.7. Quantitative Real-Time PCR (qPCR)
4.8. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Emmerson, A.M. The Quinolones: Decades of Development and Use. J. Antimicrob. Chemother. 2003, 51, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Marchant, J. When Antibiotics Turn Toxic. Nature 2018, 555, 431–433. [Google Scholar] [CrossRef] [PubMed]
- Idowu, T.; Schweizer, F. Ubiquitous Nature of Fluoroquinolones: The Oscillation between Antibacterial and Anticancer Activities. Antibiotics 2017, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Blondeau, J.M. Fluoroquinolones: Mechanism of Action, Classification, and Development of Resistance. Surv. Ophthalmol. 2004, 49, S73–S78. [Google Scholar] [CrossRef]
- Dalhoff, A. Selective Toxicity of Antibacterial Agents—Still a Valid Concept or Do We Miss Chances and Ignore Risks? Infection 2021, 49, 29–56. [Google Scholar] [CrossRef]
- Isidori, M.; Lavorgna, M.; Nardelli, A.; Pascarella, L.; Parrella, A. Toxic and Genotoxic Evaluation of Six Antibiotics on Non-Target Organisms. Sci. Total Environ. 2005, 346, 87–98. [Google Scholar] [CrossRef]
- Itoh, T.; Mitsumori, K.; Kawaguchi, S.; Sasaki, Y.F. Genotoxic Potential of Quinolone Antimicrobials in the in Vitro Comet Assay and Micronucleus Test. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2006, 603, 135–144. [Google Scholar] [CrossRef]
- Goto, K.; Yabe, K.; Suzuki, T.; Takasuna, K.; Jindo, T.; Manabe, S. Gene Expression Profiles in the Articular Cartilage of Juvenile Rats Receiving the Quinolone Antibacterial Agent Ofloxacin. Toxicology 2008, 249, 204–213. [Google Scholar] [CrossRef]
- Thomé, S.; Bizarro, C.R.; Lehmann, M.; de Abreu, B.R.R.; de Andrade, H.H.R.; Cunha, K.S.; Dihl, R.R. Recombinagenic and Mutagenic Activities of Fluoroquinolones in Drosophila melanogaster. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2012, 742, 43–47. [Google Scholar] [CrossRef]
- Bhattacharya, P.; Mukherjee, S.; Mandal, S.M. Fluoroquinolone Antibiotics Show Genotoxic Effect through DNA-Binding and Oxidative Damage. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 227, 117634. [Google Scholar] [CrossRef]
- Kashida, Y. Mechanistic Study on Flumequine Hepatocarcinogenicity Focusing on DNA Damage in Mice. Toxicol. Sci. 2002, 69, 317–321. [Google Scholar] [CrossRef] [PubMed]
- Zounková, R.; Klimešová, Z.; Nepejchalová, L.; Hilscherová, K.; Bláha, L. Complex Evaluation of Ecotoxicity and Genotoxicity of Antimicrobials Oxytetracycline and Flumequine Used in Aquaculture. Environ. Toxicol. Chem. 2011, 30, 1184–1189. [Google Scholar] [CrossRef]
- Ma, F.; Yuan, G.; Meng, L.; Oda, Y.; Hu, J. Contributions of Flumequine and Nitroarenes to the Genotoxicity of River and Ground Waters. Chemosphere 2012, 88, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Tennyson, L.E.; Averch, T.D. An Update on Fluoroquinolones: The Emergence of a Multisystem Toxicity Syndrome. Urol. Pract. 2017, 4, 383–387. [Google Scholar] [CrossRef]
- Badal, S.; Her, Y.F.; Maher, L.J. Nonantibiotic Effects of Fluoroquinolones in Mammalian Cells. J. Biol. Chem. 2015, 290, 22287–22297. [Google Scholar] [CrossRef] [PubMed]
- Hektoen, H.; Berge, J.A.; Hormazabal, V.; Yndestad, M. Persistence of Antibacterial Agents in Marine Sediments. Aquaculture 1995, 133, 175–184. [Google Scholar] [CrossRef]
- Van Doorslaer, X.; Dewulf, J.; Van Langenhove, H.; Demeestere, K. Fluoroquinolone Antibiotics: An Emerging Class of Environmental Micropollutants. Sci. Total Environ. 2014, 500–501, 250–269. [Google Scholar] [CrossRef]
- Gao, L.; Shi, Y.; Li, W.; Liu, J.; Cai, Y. Occurrence, Distribution and Bioaccumulation of Antibiotics in the Haihe River in China. J. Environ. Monit. 2012, 14, 1248. [Google Scholar] [CrossRef]
- Yang, J.-F.; Ying, G.-G.; Zhao, J.-L.; Tao, R.; Su, H.-C.; Chen, F. Simultaneous Determination of Four Classes of Antibiotics in Sediments of the Pearl Rivers Using RRLC–MS/MS. Sci. Total Environ. 2010, 408, 3424–3432. [Google Scholar] [CrossRef]
- Golet, E.M.; Alder, A.C.; Giger, W. Environmental Exposure and Risk Assessment of Fluoroquinolone Antibacterial Agents in Wastewater and River Water of the Glatt Valley Watershed, Switzerland. Environ. Sci. Technol. 2002, 36, 3645–3651. [Google Scholar] [CrossRef]
- Huang, L.; Mo, Y.; Wu, Z.; Rad, S.; Song, X.; Zeng, H.; Bashir, S.; Kang, B.; Chen, Z. Occurrence, Distribution, and Health Risk Assessment of Quinolone Antibiotics in Water, Sediment, and Fish Species of Qingshitan Reservoir, South China. Sci. Rep. 2020, 10, 15777. [Google Scholar] [CrossRef] [PubMed]
- Fick, J.; Söderström, H.; Lindberg, R.H.; Phan, C.; Tysklind, M.; Larsson, D.G.J. Contamination of Surface, Ground, and Drinking Water from Pharmaceutical Production. Environ. Toxicol. Chem. 2009, 28, 2522. [Google Scholar] [CrossRef]
- Larsson, D.G.J.; de Pedro, C.; Paxeus, N. Effluent from Drug Manufactures Contains Extremely High Levels of Pharmaceuticals. J. Hazard. Mater. 2007, 148, 751–755. [Google Scholar] [CrossRef] [PubMed]
- Gothwal, R.; Shashidhar. Occurrence of High Levels of Fluoroquinolones in Aquatic Environment Due to Effluent Discharges from Bulk Drug Manufacturers. J. Hazard. Toxic Radioact. Waste 2017, 21, 05016003. [Google Scholar] [CrossRef]
- Le, T.X.; Munekage, Y. Residues of Selected Antibiotics in Water and Mud from Shrimp Ponds in Mangrove Areas in Viet Nam. Mar. Pollut. Bull. 2004, 49, 922–929. [Google Scholar] [CrossRef]
- Tuc Dinh, Q.; Alliot, F.; Moreau-Guigon, E.; Eurin, J.; Chevreuil, M.; Labadie, P. Measurement of Trace Levels of Antibiotics in River Water Using On-Line Enrichment and Triple-Quadrupole LC–MS/MS. Talanta 2011, 85, 1238–1245. [Google Scholar] [CrossRef] [PubMed]
- Mompelat, S.; Thomas, O.; Le Bot, B. Contamination Levels of Human Pharmaceutical Compounds in French Surface and Drinking Water. J. Environ. Monit. 2011, 13, 2929. [Google Scholar] [CrossRef]
- Sargenti, M.; Bartolacci, S.; Luciani, A.; Di Biagio, K.; Baldini, M.; Galarini, R.; Giusepponi, D.; Capuccella, M. Investigation of the Correlation between the Use of Antibiotics in Aquaculture Systems and Their Detection in Aquatic Environments: A Case Study of the Nera River Aquafarms in Italy. Sustainability 2020, 12, 5176. [Google Scholar] [CrossRef]
- González-Gaya, B.; García-Bueno, N.; Buelow, E.; Marin, A.; Rico, A. Effects of Aquaculture Waste Feeds and Antibiotics on Marine Benthic Ecosystems in the Mediterranean Sea. Sci. Total Environ. 2022, 806, 151190. [Google Scholar] [CrossRef]
- Dalla Bona, M.; Lizzi, F.; Borgato, A.; De Liguoro, M. Increasing Toxicity of Enrofloxacin over Four Generations of Daphnia magna. Ecotoxicol. Environ. Saf. 2016, 132, 397–402. [Google Scholar] [CrossRef]
- De Liguoro, M.; Maraj, S.; Merlanti, R. Transgenerational Toxicity of Flumequine over Four Generations of Daphnia magna. Ecotoxicol. Environ. Saf. 2019, 169, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Ellis, L.J.A.; Kissane, S.; Lynch, I. Maternal Responses and Adaptive Changes to Environmental Stress via Chronic Nanomaterial Exposure: Differences in Inter and Transgenerational Interclonal Broods of Daphnia magna. Int. J. Mol. Sci. 2020, 22, 15. [Google Scholar] [CrossRef] [PubMed]
- Jeremias, G.; Barbosa, J.; Marques, S.M.; De Schamphelaere, K.A.C.; Van Nieuwerburgh, F.; Deforce, D.; Gonçalves, F.J.M.; Pereira, J.L.; Asselman, J. Transgenerational Inheritance of DNA Hypomethylation in Daphnia magna in Response to Salinity Stress. Environ. Sci. Technol. 2018, 52, 10114–10123. [Google Scholar] [CrossRef] [PubMed]
- Hearn, J.; Pearson, M.; Blaxter, M.; Wilson, P.J.; Little, T.J. Genome-Wide Methylation Is Modified by Caloric Restriction in Daphnia magna. BMC Genom. 2019, 20, 197. [Google Scholar] [CrossRef] [PubMed]
- Ebert, D. A Genome for the Environment. Science 2011, 331, 539–540. [Google Scholar] [CrossRef]
- Lee, B.-Y.; Choi, B.-S.; Kim, M.-S.; Park, J.C.; Jeong, C.-B.; Han, J.; Lee, J.-S. The Genome of the Freshwater Water Flea Daphnia magna: A Potential Use for Freshwater Molecular Ecotoxicology. Aquat. Toxicol. 2019, 210, 69–84. [Google Scholar] [CrossRef]
- Liu, Z.; Li, Y.; Pérez, E.; Jiang, Q.; Chen, Q.; Jiao, Y.; Huang, Y.; Yang, Y.; Zhao, Y. Polystyrene Nanoplastic Induces Oxidative Stress, Immune Defense, and Glycometabolism Change in Daphnia pulex: Application of Transcriptome Profiling in Risk Assessment of Nanoplastics. J. Hazard. Mater. 2021, 402, 123778. [Google Scholar] [CrossRef]
- Li, J.; Li, H.; Lin, D.; Li, M.; Wang, Q.; Xie, S.; Zhang, Y.; Liu, F. Effects of Butyl Benzyl Phthalate Exposure on Daphnia magna Growth, Reproduction, Embryonic Development and Transcriptomic Responses. J. Hazard. Mater. 2021, 404, 124030. [Google Scholar] [CrossRef]
- Mielecki, D.; Grzesiuk, E.; Bednarska, A.; Garbicz, D.; Świderska, B.; Grzesiuk, M. Contamination of Aquatic Environment with Anticancer Reagents Influences Daphnia magna—Ecotoxicogenomics Approach. Ecotoxicol. Environ. Saf. 2023, 249, 114372. [Google Scholar] [CrossRef]
- Ren, J.; Yang, F.; Ding, N.; Mo, J.; Guo, J. Transcriptomic Responses to Cytotoxic Drug Cisplatin in Water Flea Daphnia magna. Environ. Toxicol. Pharmacol. 2022, 95, 103964. [Google Scholar] [CrossRef]
- Di Delupis, G.D. Saggi di Ecotossicita con Daphnia magna Basati sul Comportamento Fototattico. Master’s Thesis, Istituto Superiore di Sanita, Rome, Italy, 1997. [Google Scholar]
- Kato, K.; Okamura, K.; Hiki, K.; Kintsu, H.; Nohara, K.; Yamagishi, T.; Nakajima, N.; Watanabe, H.; Yamamoto, H. Potential Differences in Chitin Synthesis Ability Cause Different Sensitivities to Diflubenzuron among Three Strains of Daphnia magna. Aquat. Toxicol. 2022, 243, 106071. [Google Scholar] [CrossRef] [PubMed]
- Vanvelk, H.; Govaert, L.; van den Berg, E.M.; Brans, K.I.; De Meester, L. Interspecific differences, plastic, and evolutionary responses to a heat wave in three co-occurring Daphnia species. Limnol. Oceano 2021, 66, 1201–1220. [Google Scholar] [CrossRef]
- Reinikainen, M.; Hietala, J.; Walls, M. Adaptations and resistance of zooplankton to stress: Effects of genetic, environmental, and physiological factors. Ecotoxicol. Environ. Saf. 1998, 40, 77–80. [Google Scholar] [CrossRef]
- Kim, J.; Coutellec, M.A.; Lee, S.; Choi, J. Insights into the mechanisms of within-species variation in sensitivity to chemicals: A case study using daphnids exposed to CMIT/MIT biocide. Ecotoxicol. Environ. Saf. 2023, 258, 114967. [Google Scholar] [CrossRef]
- Pfaff, J.; Reinwald, H.; Ayobahan, S.U.; Alvincz, J.; Göckener, B.; Shomroni, O.; Salinas, G.; Düring, R.A.; Schäfers, C.; Eilebrecht, S. Toxicogenomic differentiation of functional responses to fipronil and imidacloprid in Daphnia magna. Aquat. Toxicol. 2021, 238, 105927. [Google Scholar] [CrossRef] [PubMed]
- Fuertes, I.; Campos, B.; Rivetti, C.; Piña, B.; Barata, C. Effects of Single and Combined Low Concentrations of Neuroactive Drugs on Daphnia magna Reproduction and Transcriptomic Responses. Environ. Sci. Technol. 2019, 53, 11979–11987. [Google Scholar] [CrossRef] [PubMed]
- Russo, C.; Isidori, M.; Deaver, J.A.; Poynton, H.C. Toxicogenomic responses of low level anticancer drug exposures in Daphnia magna. Aquat. Toxicol. 2018, 203, 40–50. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, Z.; Tang, S.; Li, D.; Jiang, Q.; Zhang, T. Transcriptional Response Provides Insights into the Effect of Chronic Polystyrene Nanoplastic Exposure on Daphnia pulex. Chemosphere 2020, 238, 124563. [Google Scholar] [CrossRef]
- Andersen, S.O. Cuticular Sclerotization and Tanning. In Insect Molecular Biology and Biochemistry; Elsevier: Amsterdam, The Netherlands, 2012; pp. 167–192. [Google Scholar] [CrossRef]
- Qin, G.; Hu, X.; Cebe, P.; Kaplan, D.L. Mechanism of Resilin Elasticity. Nat. Commun. 2012, 3, 1003. [Google Scholar] [CrossRef]
- Johnstone, I.L. Cuticle Collagen Genes: Expression in Caenorhabditis elegans. Trends Genet. 2000, 16, 21–27. [Google Scholar] [CrossRef]
- Kawasaki, H.; Hirose, S.; Ueda, H. ΒFTZ-F1 Dependent and Independent Activation of Edg78E, a Pupal Cuticle Gene, during the Early Metamorphic Period in Drosophila melanogaster. Dev. Growth Differ. 2002, 44, 419–425. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, G.A. Crustacean Endocrine Toxicology: A Review. Ecotoxicology 2007, 16, 61–81. [Google Scholar] [CrossRef] [PubMed]
- Bindoli, A.; Rigobello, M.P. Peroxidase Biochemistry and Redox Signaling. In Encyclopedia of Biological Chemistry, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2013; pp. 407–412. [Google Scholar]
- Gomes, M.P.; Kitamura, R.S.A.; Marques, R.Z.; Barbato, M.L.; Zámocký, M. The Role of H2O2-Scavenging Enzymes (Ascorbate Peroxidase and Catalase) in the Tolerance of Lemna Minor to Antibiotics: Implications for Phytoremediation. Antioxidants 2022, 11, 151. [Google Scholar] [CrossRef] [PubMed]
- Finet, C.; Slavik, K.; Pu, J.; Carroll, S.B.; Chung, H. Birth-and-Death Evolution of the Fatty Acyl-CoA Reductase (FAR) Gene Family and Diversification of Cuticular Hydrocarbon Synthesis in Drosophila. Genome Biol. Evol. 2019, 11, 1541–1551. [Google Scholar] [CrossRef]
- Tong, H.; Wang, Y.; Wang, S.; Omar, M.A.A.; Li, Z.; Li, Z.; Ding, S.; Ao, Y.; Wang, Y.; Li, F.; et al. Fatty acyl-CoA reductase influences wax biosynthesis in the cotton mealybug, Phenacoccus solenopsis Tinsley. Commun. Biol. 2022, 5, 1108. [Google Scholar] [CrossRef]
- Davidsson, J.; Jahnke, K.; Forsgren, M.; Collin, A.; Soller, M. Dup(19)(Q12q13.2): Array-Based Genotype-Phenotype Correlation of a New Possibly Obesity-Related Syndrome. Obesity 2010, 18, 580–587. [Google Scholar] [CrossRef]
- Nauen, R.; Bass, C.; Feyereisen, R.; Vontas, J. The Role of Cytochrome P450s in Insect Toxicology and Resistance. Annu. Rev. Entomol. 2022, 67, 105–124. [Google Scholar] [CrossRef]
- Xing, X.; Yan, M.; Pang, H.; Wu, F.; Wang, J.; Sheng, S. Cytochrome P450s Are Essential for Insecticide Tolerance in the Endoparasitoid Wasp Meteorus pulchricornis (Hymenoptera: Braconidae). Insects 2021, 12, 651. [Google Scholar] [CrossRef]
- Lane, C.S.; Wang, Y.; Betts, R.; Griffiths, W.J.; Patterson, L.H. Comparative Cytochrome P450 Proteomics in the Livers of Immune-Deficient Mice Using 18 O Stable Isotope Labeling. Mol. Cell. Proteom. 2007, 6, 953–962. [Google Scholar] [CrossRef]
- Zhu, P.; Wang, H.; Zeng, Q. Comparative Transcriptome Reveals the Response of Oriental River Prawn (Macrobrachium nipponense) to Sulfide Toxicity at Molecular Level. Aquat. Toxicol. 2021, 230, 105700. [Google Scholar] [CrossRef]
- Coady, K.K.; Burgoon, L.; Doskey, C.; Davis, J.W. Assessment of Transcriptomic and Apical Responses of Daphnia magna Exposed to a Polyethylene Microplastic in a 21-d Chronic Study. Environ. Toxicol Chem 2020, 39, 1578–1589. [Google Scholar] [CrossRef] [PubMed]
- Zancanella, V.; Giantin, M.; Lopparelli, R.M.; Patarnello, T.; Dacasto, M.; Negrisolo, E. Proposed New Nomenclature for Bos Taurus Cytochromes P450 Involved in Xenobiotic Drug Metabolism: Bos Taurus P450 Nomenclature. J. Vet. Pharmacol. Ther. 2010, 33, 528–536. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, H.F.; McLean, V.; McLean, M. Molecular and Cellular Aspects of Thiol-Disulfide Exchange. Adv. Enzymol. Relat. Areas Mol. Biol. 1963, 63, 69. [Google Scholar]
- Kirsch, M.; Groot, H. NAD(P)H, a Directly Operating Antioxidant? FASEB J. 2001, 15, 1569–1574. [Google Scholar] [CrossRef]
- Ibi, M.; Yabe-Nishimura, C. The Role of Reactive Oxygen Species in the Pathogenic Pathways of Depression. In Oxidative Stress and Dietary Antioxidants in Neurological Diseases; Elsevier: Amsterdam, The Netherlands, 2020; pp. 3–16. [Google Scholar] [CrossRef]
- Baird, T.T.; Craik, C.S. Trypsin. In Encyclopedia of Genetics; Elsevier: Amsterdam, The Netherlands, 2001; pp. 2071–2075. [Google Scholar] [CrossRef]
- Locher, K.P. Structure and Mechanism of ATP-Binding Cassette Transporters. In Philosophical Transactions of the Royal Society B: Biological Sciences; Royal Society: London, UK, 2009; pp. 239–245. [Google Scholar] [CrossRef]
- Mayer, M.L. A Calcium-Activated Chloride Current Generates the After-Depolarization of Rat Sensory Neurones in Culture. J. Physiol. 1985, 364, 217–239. [Google Scholar] [CrossRef]
- Flores-Soto, M.E.; Chaparro-Huerta, V.; Escoto-Delgadillo, M.; Vazquez-Valls, E.; González-Castañeda, R.E.; Beas-Zarate, C. Neurología Structure and Function of NMDA-Type Glutamate Receptor Subunits PALABRAS CLAVE Excitotoxicidad; Isoformas; Receptor N-Metil-d-Aspartato; Subunidad NR1; Subunidad NR2. Neurologia 2012, 27, 301–310. [Google Scholar] [CrossRef]
- Lyu, S.; Terao, N.; Nakashima, H.; Itoh, M.; Tonoki, A. Neuropeptide Diuretic Hormone 31 Mediates Memory and Sleep via Distinct Neural Pathways in Drosophila. Neurosci. Res. 2023, in press. [Google Scholar] [CrossRef]
- Win-Shwe, T.T.; Tsukahara, S.; Yamamoto, S.; Fukushima, A.; Kunugita, N.; Arashidani, K.; Fujimaki, H. Up-Regulation of Neurotrophin-Related Gene Expression in Mouse Hippocampus Following Low-Level Toluene Exposure. Neurotoxicology 2010, 31, 85–93. [Google Scholar] [CrossRef]
- Janssen, R.; Andersson, E.; Betnér, E.; Bijl, S.; Fowler, W.; Höök, L.; Leyhr, J.; Mannelqvist, A.; Panara, V.; Smith, K.; et al. Embryonic Expression Patterns and Phylogenetic Analysis of Panarthropod Sox Genes: Insight into Nervous System Development, Segmentation and Gonadogenesis. BMC Evol. Biol. 2018, 18, 88. [Google Scholar] [CrossRef]
- Matsumoto, H.; Yamada, T. Phosrestins I and II Arrestin Homologs Which Undergo Differential Light-Induced Phos-Phorylation in the Drosophila Photoreceptor in Vivo. Biochem. Biophys. Res. Commun. 1991, 177, 1306–1312. [Google Scholar] [CrossRef]
- Tsukimura, B. Crustacean Vitellogenesis: Its Role in Oocyte Development. Am. Zool. 2001, 41, 465–476. [Google Scholar] [CrossRef]
- Avarre, J.-C.; Michelis, R.; Tietz, A.; Lubzens, E. Relationship Between Vitellogenin and Vitellin in a Marine Shrimp (Penaeus semisulcatus) and Molecular Characterization of Vitellogenin Complementary DNAs1. Biol. Reprod. 2003, 69, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Manogaran, A.L.; Beauchamp, J.M.; Waring, G.L. Drosophila Vitelline Membrane Assembly: A Critical Role for an Evolutionarily Conserved Cysteine in the “VM Domain” of SV23. Dev. Biol. 2010, 347, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Hannas, B.R.; Wang, Y.H.; Thomson, S.; Kwon, G.; Li, H.; LeBlanc, G.A. Regulation and Dysregulation of Vitellogenin MRNA Accumulation in Daphnids (Daphnia magna). Aquat. Toxicol. 2011, 101, 351–357. [Google Scholar] [CrossRef]
- Fagutao, F.F.; Yasuike, M.; Santos, M.D.; Ruangpan, L.; Sangrunggruang, K.; Tassanakajon, A.; Takahashi, Y.; Ueno, R.; Kondo, H.; Hirono, I.; et al. Differential Gene Expression in Black Tiger Shrimp, Penaeus Monodon, Following Administration of Oxytetracycline and Oxolinic Acid. Dev. Comp. Immunol. 2009, 33, 1088–1092. [Google Scholar] [CrossRef]
- Du, J.; Liu, Q.; Fu, L. Metabolic and Transcriptional Disruption of American Shad (Alosa sapidissima) by Enrofloxacin in Commercial Aquaculture. Environ. Sci. Pollut. Res. 2022, 29, 2052–2062. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.; Liu, X.; Yang, F.; Li, R.; Xiong, Y.; Fu, C.; Li, G.; Liu, S.; Zheng, C. Single and Joint Toxic Effects of Four Antibiotics on Some Metabolic Pathways of Zebrafish (Danio rerio) Larvae. Sci. Total Environ. 2020, 716, 137062. [Google Scholar] [CrossRef] [PubMed]
- Luan, Y.; Zhao, J.; Han, H.; Shen, J.; Tang, S.; Cheng, L. Toxicologic Effect and Transcriptome Analysis for Short-Term Orally Dosed Enrofloxacin Combined with Two Veterinary Antimicrobials on Rat Liver. Ecotoxicol. Environ. Saf. 2021, 220, 112398. [Google Scholar] [CrossRef]
- Trombini, C.; Kazakova, J.; Montilla-López, A.; Fernández-Cisnal, R.; Hampel, M.; Fernández-Torres, R.; Bello-López, M.Á.; Abril, N.; Blasco, J. Assessment of Pharmaceutical Mixture (Ibuprofen, Ciprofloxacin and Flumequine) Effects to the Crayfish Procambarus clarkii: A Multilevel Analysis (Biochemical, Transcriptional and Proteomic Approaches). Environ. Res. 2021, 200, 111396. [Google Scholar] [CrossRef]
- Xu, R.; Zheng, R.; Wang, Y.; Ma, R.; Tong, G.; Wei, X.; Feng, D.; Hu, K. Transcriptome Analysis to Elucidate the Toxicity Mechanisms of Fenvalerate, Sulfide Gatifloxacin, and Ridomil on the Hepatopancreas of Procambarus clarkii. Fish Shellfish Immunol. 2021, 116, 140–149. [Google Scholar] [CrossRef]
- Hou, L.; Fu, Y.; Zhao, C.; Fan, L.; Hu, H.; Yin, S. Ciprofloxacin and Enrofloxacin Can Cause Reproductive Toxicity via Endocrine Signaling Pathways. Ecotoxicol. Environ. Saf. 2022, 244, 114049. [Google Scholar] [CrossRef]
- Rico, A.; Vighi, M.; Van den Brink, P.J.; ter Horst, M.; Macken, A.; Lillicrap, A.; Falconer, L.; Telfer, T.C. Use of Models for the Environmental Risk Assessment of Veterinary Medicines in European Aquaculture: Current Situation and Future Perspectives. In Reviews in Aquaculture; Wiley-Blackwell: Hoboken, NJ, USA, 2019; pp. 969–988. [Google Scholar] [CrossRef]
- Samuelsen, O.B.; Lunestadb, B.T.; Ervik, A.; Fjelde, S. Stability of Antibacterial Agents in an Artificial Marine Aquaculture Sediment Studied under Laboratory Conditions. Aquaculture 1994, 126, 283–290. [Google Scholar] [CrossRef]
- Santos, M.A.P.F.; Vicensotti, J.; Monteiro, R.T.R. Sensitivity of Four Test Organisms (Chironomus xanthus, Daphnia magna, Hydra attenuata and Pseudokirchneriella subcapitata) to NaCl: An Alternative Reference Toxicant. J. Braz. Soc. Ecotoxicol. 2007, 2, 229–236. [Google Scholar] [CrossRef]
- De Liguoro, M.; Di Leva, V.; Dalla Bona, M.; Merlanti, R.; Caporale, G.; Radaelli, G. Sublethal Effects of Trimethoprim on Four Freshwater Organisms. Ecotoxicol. Environ. Saf. 2012, 82, 114–121. [Google Scholar] [CrossRef] [PubMed]
- ISO 6341; BSI Standards Publication Water Quality-Determination of the Inhibition of the Mobility of Daphnia magna Straus (Cladocera, Crustacea)-Acute Toxicity Test. International Organization for Standardization: Geneva, Switzerland, 2012.
- Organisation for Economic Co-operation and Development (OECD). Test No. 211: Daphnia magna Reproduction Test. In OECD Guideline for The Testing of Chemicals; Section 2 Effects on Biotic Systems; OECD: Paris, France, 2012. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 21 February 2022).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kopylova, E.; Noé, L.; Touzet, H.; Nó, L. SortMeRNA: Fast and Accurate Filtering of Ribosomal RNAs in Metatranscriptomic Data. SortMeRNA: Fast and Accurate Filtering of Ribosomal RNAs in Metatranscriptomic Data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Törönen, P.; Medlar, A.; Holm, L. PANNZER2: A Rapid Functional Annotation Web Server. Nucleic Acids Res. 2018, 46, W84–W88. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. ClusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an Enhanced Web Interface to Primer3. Nucleic Acids Res. 2007, 35 (Suppl. S2), W71–W74. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA Primers for Amplification of Mitochondrial Cytochrome c Oxidase Subunit I from Diverse Metazoan Invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Prosser, S.; Martínez-Arce, A.; Elías-Gutiérrez, M. A New Set of Primers for COI Amplification from Freshwater Microcrustaceans. Mol. Ecol. Resour. 2013, 13, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The Barcode of Life Data System: Barcoding. Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef]
- Wei, X.; Li, X.; Liu, H.; Lei, H.; Sun, W.; Li, D.; Dong, W.; Chen, H.; Xie, L. Altered life history traits and transcripts of molting- and reproduction-related genes by cadmium in Daphnia magna. Ecotoxicology 2022, 31, 735–745. [Google Scholar] [CrossRef]
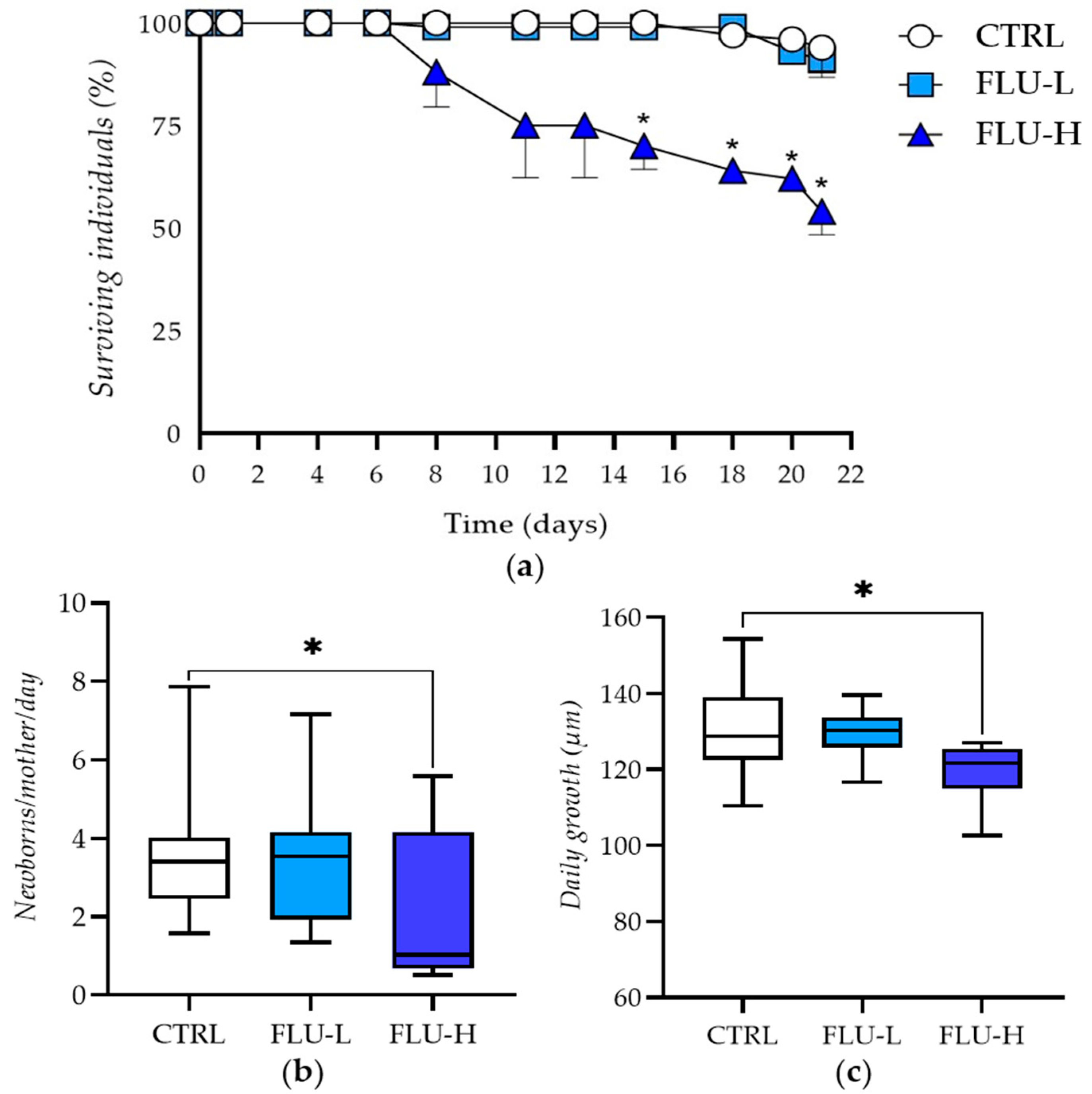
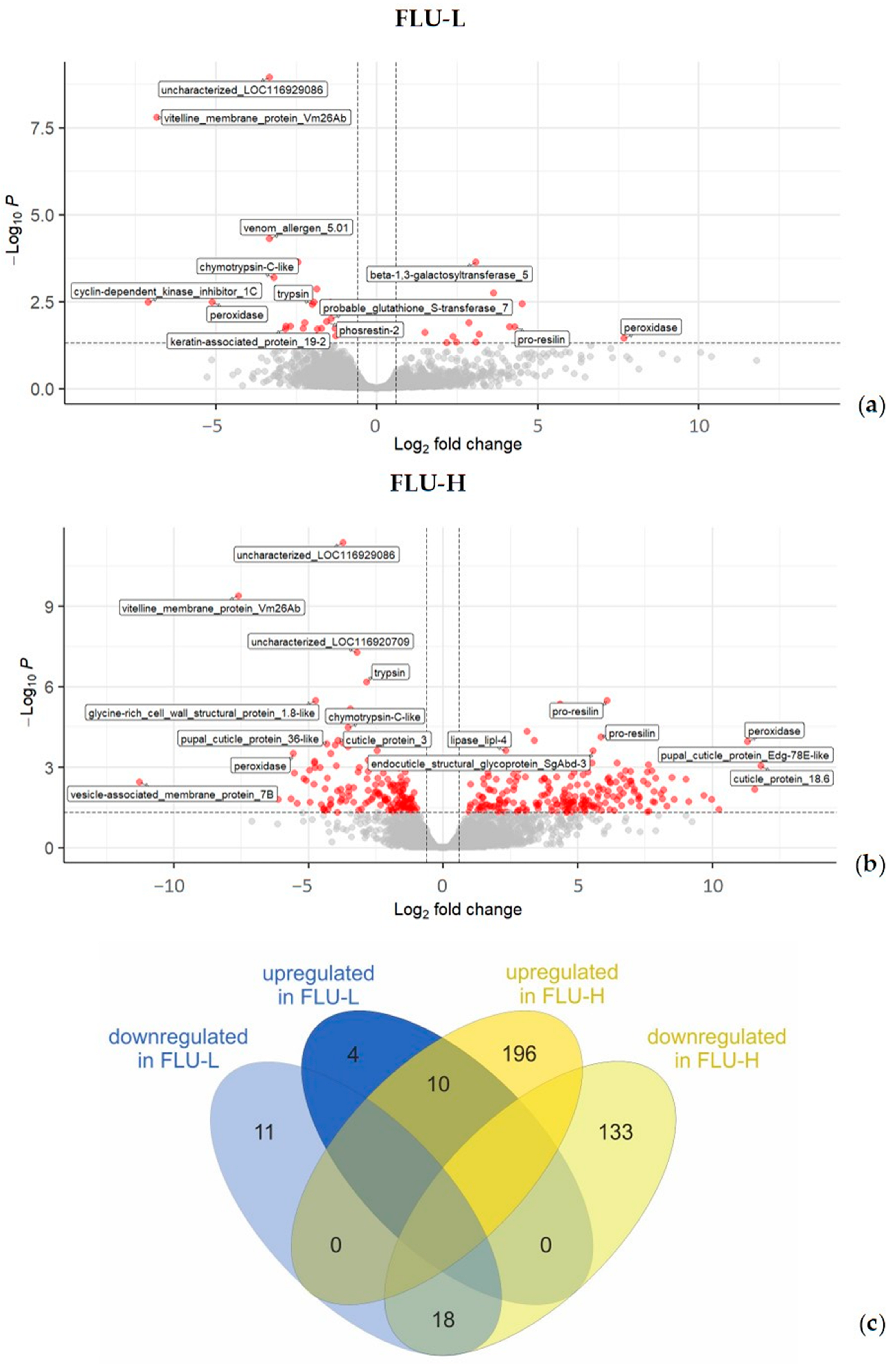
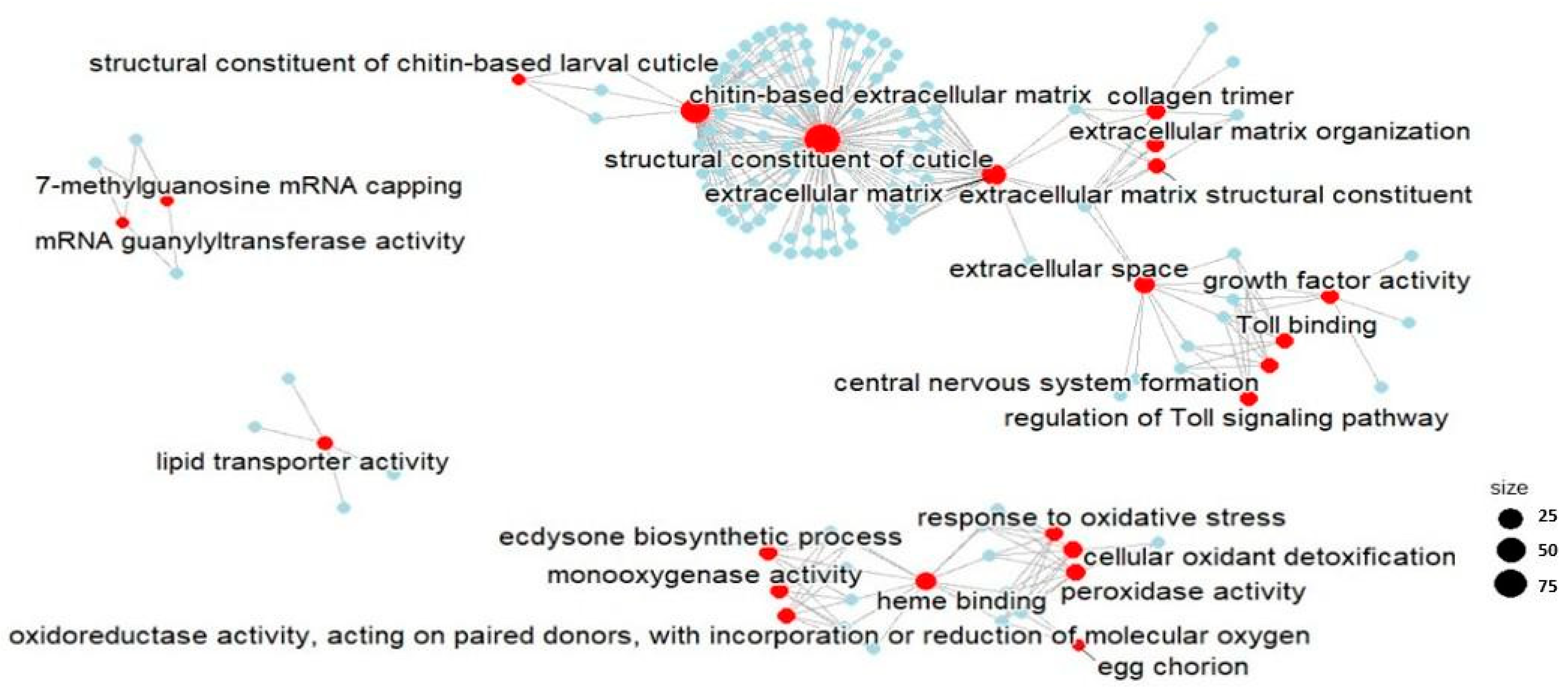
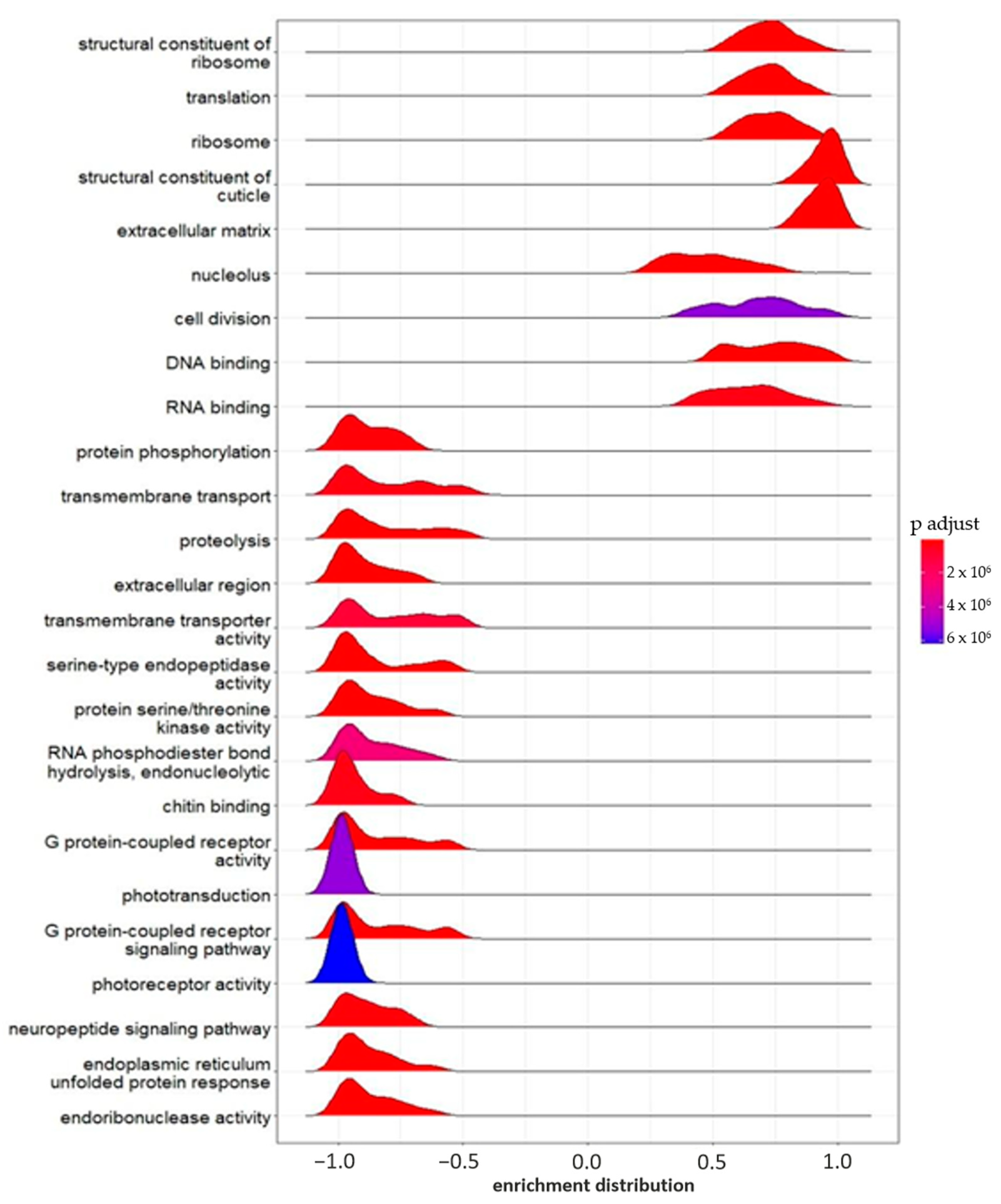
| Group | Mortality Rate (%) N = 100 | Daily Newborns per Parent Animal at the Start of the Test | Reproduction Inhibition Rate (%) | Daily Growth (Length, µm) | Daily Growth Inhibition (%) |
|---|---|---|---|---|---|
| CTRL | 6 | 3.62 ± 1.86 | 130.41 ± 12.98 | ||
| FLU-L | 9 | 3.45 ± 1.73 | 4.56 | 129.12 ± 6.52 | 1.00 |
| FLU-H | 46 * | 1.94 ± 1.79 | 46.50 * | 118.89 ± 8.43 | 8.82 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pietropoli, E.; Pauletto, M.; Tolosi, R.; Iori, S.; Lopparelli, R.M.; Montanucci, L.; Giantin, M.; Dacasto, M.; De Liguoro, M. An In Vivo Whole-Transcriptomic Approach to Assess Developmental and Reproductive Impairments Caused by Flumequine in Daphnia magna. Int. J. Mol. Sci. 2023, 24, 9396. https://doi.org/10.3390/ijms24119396
Pietropoli E, Pauletto M, Tolosi R, Iori S, Lopparelli RM, Montanucci L, Giantin M, Dacasto M, De Liguoro M. An In Vivo Whole-Transcriptomic Approach to Assess Developmental and Reproductive Impairments Caused by Flumequine in Daphnia magna. International Journal of Molecular Sciences. 2023; 24(11):9396. https://doi.org/10.3390/ijms24119396
Chicago/Turabian StylePietropoli, Edoardo, Marianna Pauletto, Roberta Tolosi, Silvia Iori, Rosa Maria Lopparelli, Ludovica Montanucci, Mery Giantin, Mauro Dacasto, and Marco De Liguoro. 2023. "An In Vivo Whole-Transcriptomic Approach to Assess Developmental and Reproductive Impairments Caused by Flumequine in Daphnia magna" International Journal of Molecular Sciences 24, no. 11: 9396. https://doi.org/10.3390/ijms24119396
APA StylePietropoli, E., Pauletto, M., Tolosi, R., Iori, S., Lopparelli, R. M., Montanucci, L., Giantin, M., Dacasto, M., & De Liguoro, M. (2023). An In Vivo Whole-Transcriptomic Approach to Assess Developmental and Reproductive Impairments Caused by Flumequine in Daphnia magna. International Journal of Molecular Sciences, 24(11), 9396. https://doi.org/10.3390/ijms24119396








