Triggering of Polymer-Degrading Enzymes from Layered Double Hydroxides for Recycling Strategies
Abstract
1. Introduction
2. Experimental
2.1. Materials
2.2. Film Weight Loss
2.3. Immobilization of Cutinase on LDH
2.4. Triggering of Enzyme from LDH
2.5. Characterization
3. Results and Discussion
3.1. Effect of Medium on the Release of Enzyme from the LDH Structure
3.2. Effect of Sodium Phosphate Buffer Concentration and pH on the Triggering of Enzyme from the LDH Structure
4. Conclusions
5. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rosato, A.; Romano, A.; Totaro, G.; Celli, A.; Fava, F.; Zanaroli, G.; Sisti, L. Enzymatic Degradation of the Most Common Aliphatic Bio-Polyesters and Evaluation of the Mechanisms Involved: An Extended Study. Polymers 2022, 14, 1850. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Su, L.; Chen, J.; Wu, J. Cutinase: Characteristics, preparation, and application. Biotechnol. Adv. 2013, 31, 1754–1767. [Google Scholar] [CrossRef] [PubMed]
- Gamerith, C.; Vastano, M.; Ghorbanpour, S.M.; Zitzenbacher, S.; Ribitsch, D.; Zumstein, M.T.; Sander, M.; Acero, E.H.; Pellis, A.; Guebitz, G.M. Enzymatic Degradation of Aromatic and Aliphatic Polyesters by P. pastoris Expressed Cutinase 1 from Thermobifida cellulosilytica. Front. Microbiol. 2017, 8, 938. [Google Scholar] [CrossRef] [PubMed]
- Romano, A.; Rosato, A.; Totaro, G.; Zanaroli, G.; Celli, A.; Sisti, L. Recycling by-design of plastic through formulation with thermally protected enzymes in layered double hydroxide structures. J. Clean. Prod. 2023, 384, 135517. [Google Scholar] [CrossRef]
- Marek, A.A.; Verney, V.; Totaro, G.; Sisti, L.; Celli, A.; Leroux, F. Composites for «white and green» solutions: Coupling UV resistance and chain extension effect from poly(butylene succinate) and layered double hydroxides composites. J. Solid State Chem. 2018, 268, 9–15. [Google Scholar] [CrossRef]
- Marek, A.A.; Verney, V.; Totaro, G.; Sisti, L.; Celli, A.; Bozzi Cionci, N.; Di Gioia, D.; Massacrier, L.; Leroux, F. Organo-modified LDH fillers endowing multi-functionality to bio-based poly(butylene succinate): An extended study from the laboratory to possible market. Appl. Clay Sci. 2020, 188, 105502. [Google Scholar] [CrossRef]
- Dong, L.; Ge, C.; Qin, P.; Chen, Y.; Xu, Q. Immobilization and catalytic properties of Candida lipolytic lipase on surface of organic intercalated and modified MgAl-LDHs. Solid State Sci. 2014, 31, 8–15. [Google Scholar] [CrossRef]
- Zou, N.; Plank, J. Intercalation of papain enzyme into hydrotalcite type layered double hydroxide. J. Phys. Chem. Solids 2012, 73, 1127–1130. [Google Scholar] [CrossRef]
- Zou, N.; Plank, J. Intercalation of cellulase enzyme into a hydrotalcite layer structure. J. Phys. Chem. Solids 2015, 76, 34–39. [Google Scholar] [CrossRef]
- Yang, Q.-Z.; Chang, Y.-Y.; Zhao, H.-Z. Preparation and antibacterial activity of lysozyme and layered double hydroxide nanocomposites. Water Res. 2013, 47, 6712–6718. [Google Scholar] [CrossRef]
- Bouaziz, Z.; Soussan, L.; Janot, J.-M.; Lepoitevin, M.; Bechelany, M.; Djebbi, M.A.; Amara, A.B.H.; Balme, S. Structure and antibacterial activity relationships of native andamyloid fibril lysozyme loaded on layered double hydroxide. Colloids Surf. B Biointerfaces 2017, 157, 10–17. [Google Scholar] [CrossRef]
- Bruna, F.; Mousty, C.; Besse-Hoggan, P.; Batisson, I.; Prevot, V. Assembly of nitroreductase and layered double hydroxides toward functional biohybrid materials. J. Colloid Interface Sci. 2019, 533, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Djebbi, M.A.; Braiek, M.; Hidouri, S.; Namour, P.; Jaffrezic-Renault, N.; Amara, A.B.H. Novel biohybrids of layered double hydroxide and lactate dehydrogenase enzyme: Synthesis, characterization and catalytic activity studies. J. Mol. Struct. 2016, 1105, 381–388. [Google Scholar] [CrossRef]
- Ding, Y.; Liu, L.; Fang, Y.; Zhang, X.; Lyu, M.; Wang, S. The adsorption of dextranase onto Mg/Fe-Layered Double Hydroxide: Insight into the immobilization. Nanomaterials 2018, 8, 173. [Google Scholar] [CrossRef] [PubMed]
- Soussou, A.; Gammoudi, I.; Moroté, F.; Kalboussi, A.; Cohen-Bouhacina, T.; Grauby-Heywang, C.; Baccar, Z.M. Efficient immobilization of Tyrosinase enzyme on layered double hydroxide hybrid nanomaterials for electrochemical detection of polyphenols. IEEE Sens. J. 2017, 17, 4340–4348. [Google Scholar] [CrossRef]
- Rives, V.; del Arco, M.; Martín, C. Intercalation of drugs in layered double hydroxides and their controlled release: A review. Appl. Clay Sci. 2014, 88, 239–269. [Google Scholar] [CrossRef]
- Allou, N.B.; Saikia, P.; Borah, A.; Goswamee, R.L. Hybrid nanocomposites of layered double hydroxides: An update of their biological applications and future prospects. Colloid Polym. Sci. 2017, 295, 725–747. [Google Scholar] [CrossRef]
- Bernard, E.; Zucha, W.J.; Lothenbach, B.; Mäder, U. Stability of hydrotalcite (Mg-Al layered double hydroxide) in presence of different anions. Cem. Concr. Res. 2022, 152, 106674. [Google Scholar] [CrossRef]
- Ralla, K.; Sohling, U.; Suck, K.; Sander, F.; Kasper, C.; Ruf, F.; Scheper, T. Adsorption and separation of proteins by a synthetic hydrotalcite. Colloids Surf. B 2011, 87, 217–225. [Google Scholar] [CrossRef]
- Nakayama, H.; Wada, N.; Tsuhako, M. Intercalation of amino acids and peptides into Mg–Al layered double hydroxide by reconstruction method. Int. J. Pharm. 2004, 269, 469–478. [Google Scholar] [CrossRef]
- Dias, G.S.; Bandeira, P.T.; Jaerger, S.; Piovan, L.; Mitchell, D.A.; Wypych, F.; Krieger, N. Immobilization of Pseudomonas cepacia lipase on layered double hydroxide of Zn/Al-Cl for kinetic resolution of rac-1-phenylethanol. Enzyme Microb. Technol. 2019, 130, 109365. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, D. Synthesis, characterization, and controlled release antibacterial behavior of antibiotic intercalated Mg–Al layered double hydroxides. Mater. Res. Bull. 2012, 47, 3185–3194. [Google Scholar] [CrossRef]
- Pavlovic, M.; Szerlauth, A.; Muráth, S.; Varga, G.; Szilagyi, I. Surface modification of two-dimensional layered double hydroxide nanoparticles with biopolymers for biomedical applications. Adv. Drug Deliv. Rev. 2022, 191, 114590. [Google Scholar] [CrossRef] [PubMed]
- Violante, A.; Pucci, M.; Cozzolino, V.; Zhu, J.; Pigna, M. Sorption/desorption of arsenate on/from Mg–Al layered double hydroxides: Influence of phosphate. J. Colloid Interface Sci. 2009, 333, 63–70. [Google Scholar] [CrossRef]
- Yang, W.; Kim, Y.; Liu, P.K.T.; Sahimi, M.; Tsotsis, T.T. A study by in situ techniques of the thermal evolution of the structure of a Mg–Al–CO3 layered double hydroxide. Chem. Eng. Sci. 2002, 57, 2945–2953. [Google Scholar] [CrossRef]
- Zaak, H.; Fernandez-Lopez, L.; Velasco-Lozano, S.; Alcaraz-Fructuoso, M.T.; Sassi, M.; Lopez-Gallego, F.; Fernandez-Lafuente, R. Effect of high salt concentrations on the stability of immobilized lipases: Dramatic deleterious effects of phosphate anions. Process Biochem. 2017, 62, 128–134. [Google Scholar] [CrossRef]
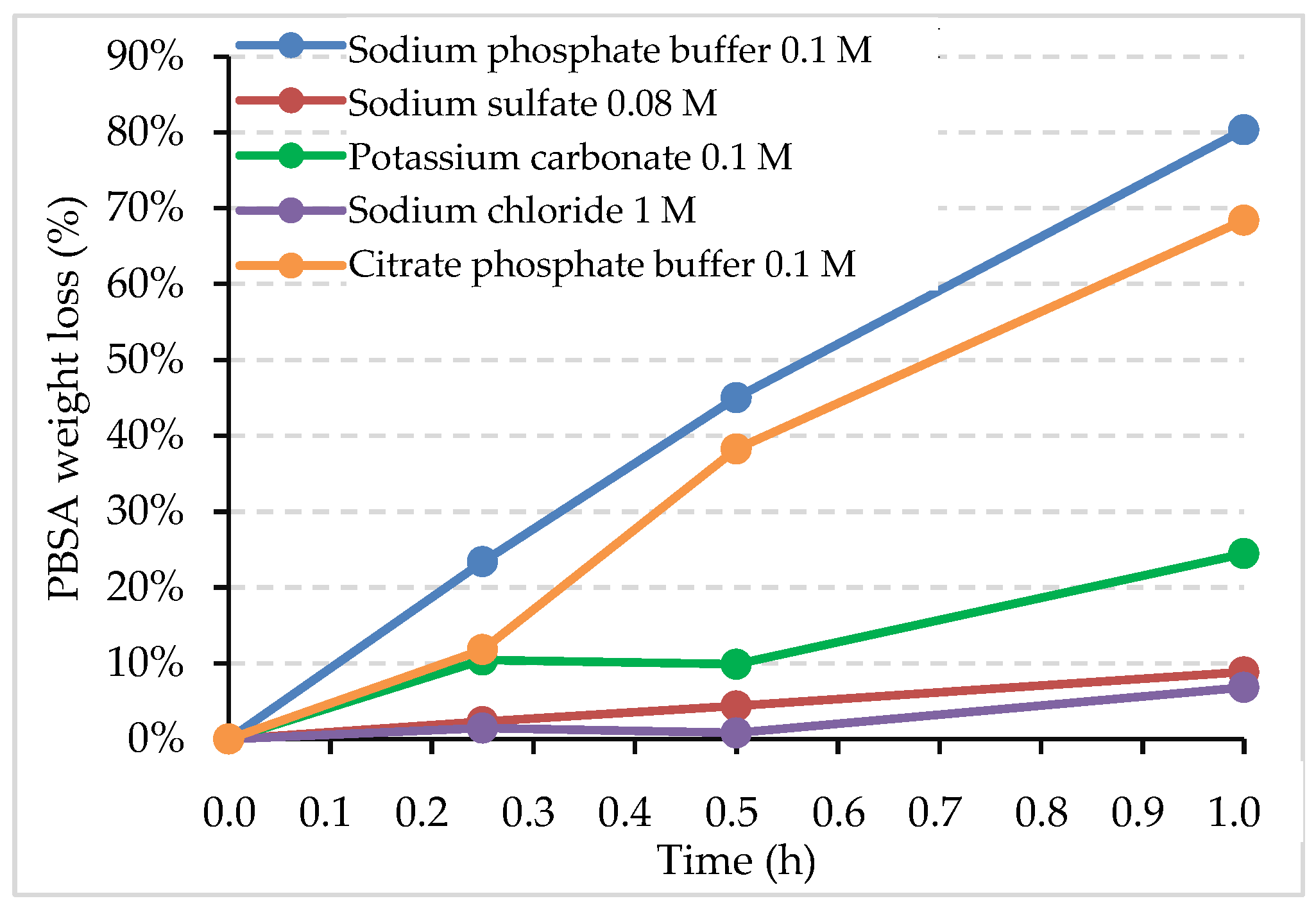
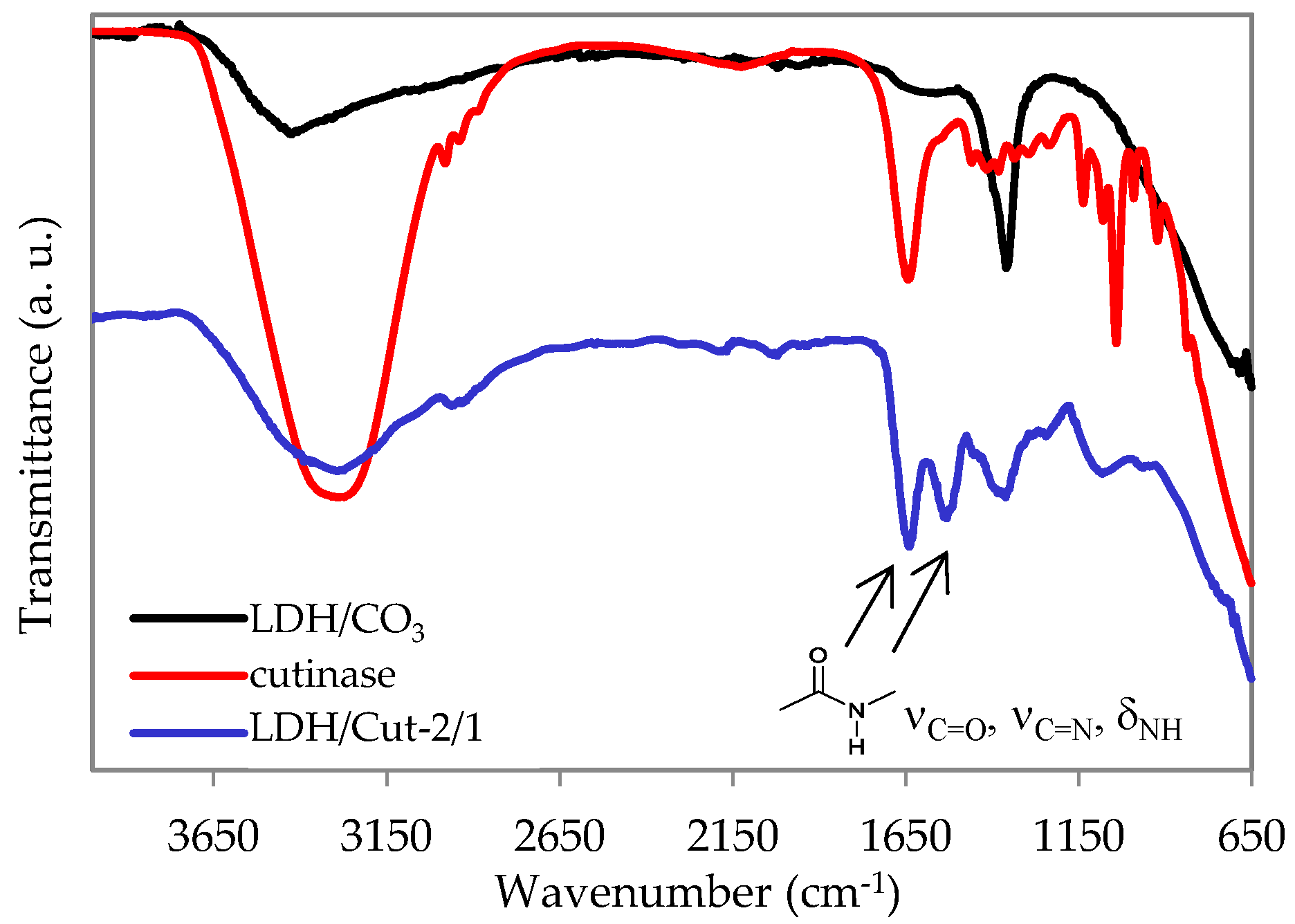
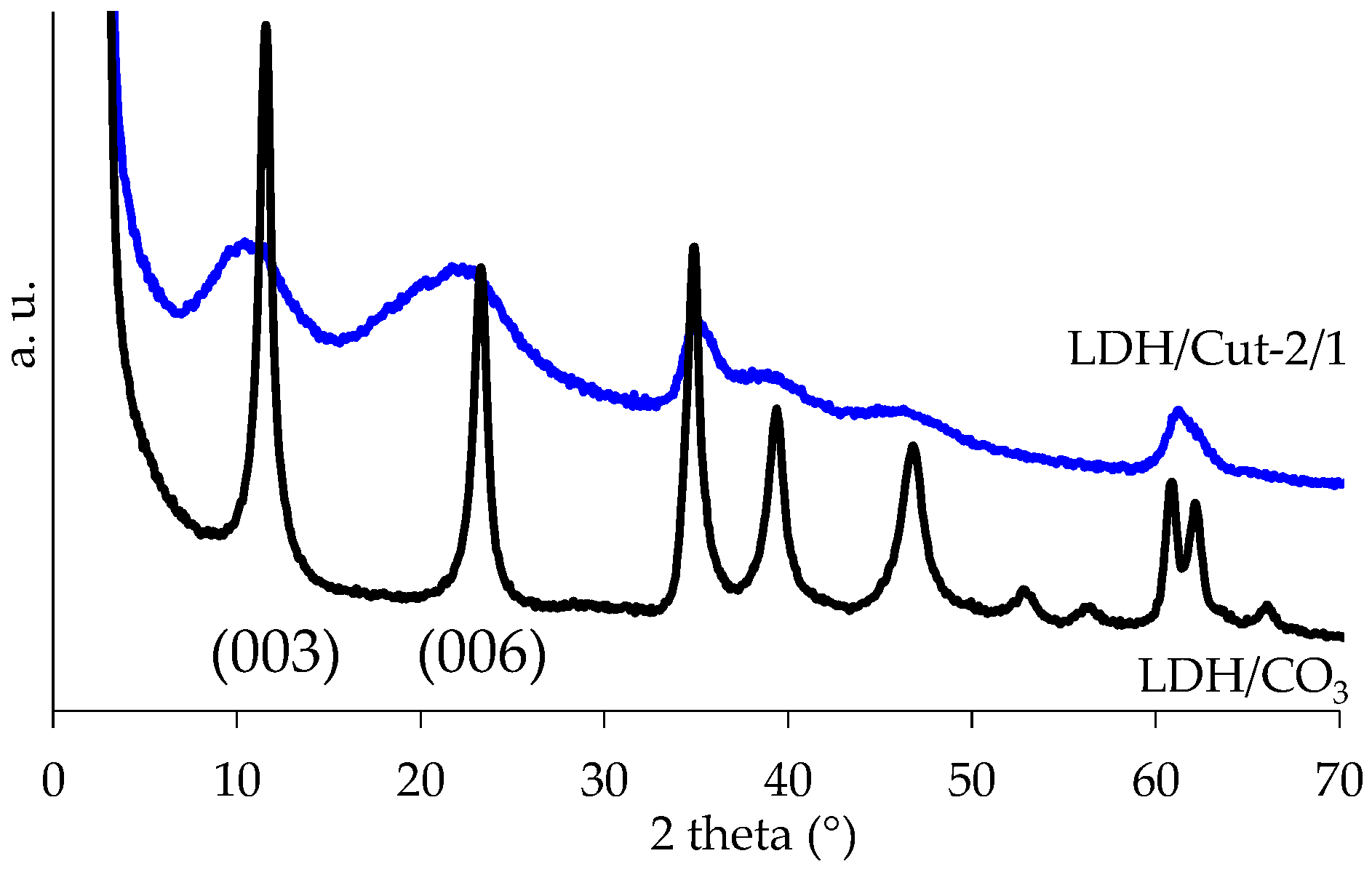
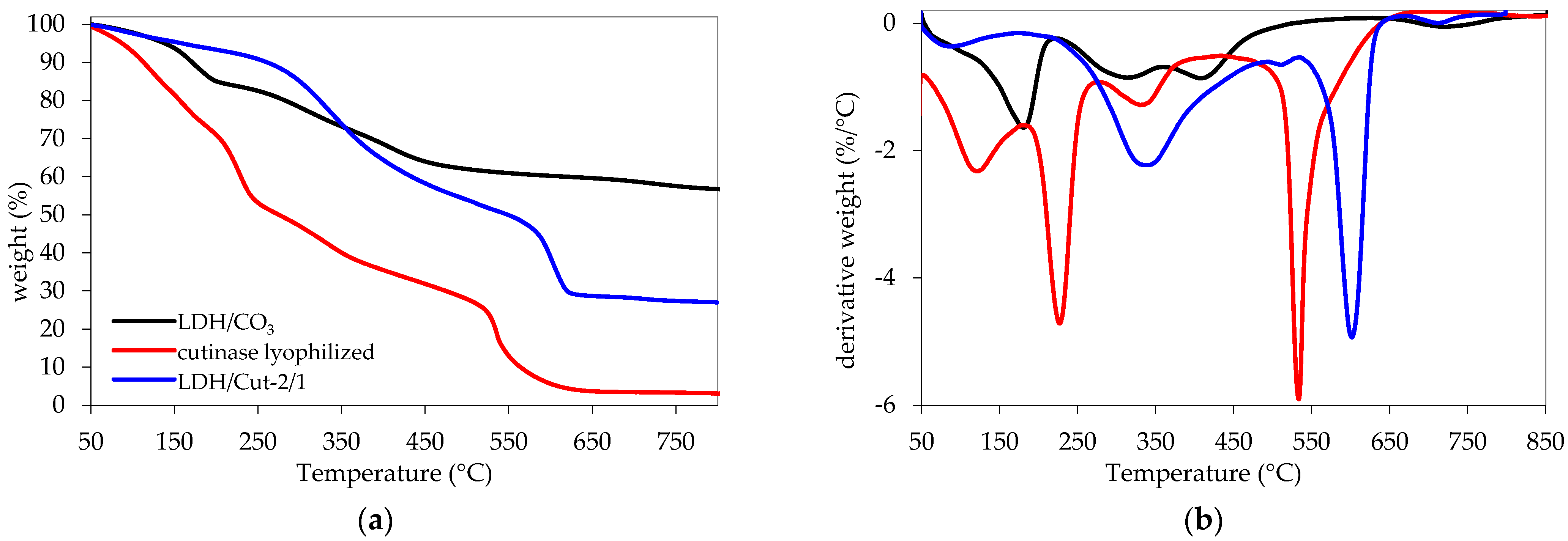


| Triggering Solution | Concentration (M) | pH |
|---|---|---|
| Sodium phosphate buffer | 0.10 | 8.0 |
| Citrate phosphate buffer | 0.10 | 8.0 |
| Sodium sulfate (Na2SO4) | 0.08 | 7.5 |
| Potassium carbonate (K2CO3) | 0.10 | 11.5 |
| Sodium chloride (NaCl) | 1.00 | 7.4 |
| Water (H2O) | - | 6.5 |
| Triggering Solution | Degradation Rate (mgPBSA/h/cm2) | Activity (U/mL) | Relative Activity (%) |
|---|---|---|---|
| Sodium phosphate buffer | 6.4 | 33,975 ± 531 | 100 ± 3 |
| Citrate phosphate buffer | 5.4 | 33,891 ± 1237 | 100 ± 7 |
| Sodium sulfate | 0.7 | 19,546 ± 1096 | 58 ± 6 |
| Potassium carbonate | 2.8 | n.a | n.a |
| Sodium chloride | 0.5 | 4247 ± 163 | 12 ± 1 |
| Code | T20D (°C) 1 | Residue (%) 1 | Step | |||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |||
| Cutinase | 153 | 3 | 28% 50–190 °C | 22% 190–280 °C | 18% 280–450 °C | 29% 450–850 °C |
| LDH/CO3 | 283 | 56 | 15.9% 50–220 °C | 12.6% 220–370 °C | 10.5% 370–550 °C | 4.7% 550–850 °C |
| LDH/Cut-2/1 | 322 | 27 | 6% 50–190 °C | 42% 190–520 °C | 23% 520–640 °C | 2% 640–850 °C |
| Triggering Solution | Protein Recovery (%) | Activity Recovery (%) | Retention Activity of Released Proteins (%) |
|---|---|---|---|
| Sodium phosphate buffer | 59 ± 1 | 59 ± 2 | 100 |
| Citrate phosphate buffer | 69 ± 1 | 70 ± 1 | 100 |
| Sodium sulfate | 57 ± 3 | 59 ± 0 | 100 |
| Potassium carbonate | 69 ± 1 | 69 ± 0 | 100 |
| Sodium chloride | 4 ± 0 | 2 ± 0 | 50 |
| Water | 10 ± 0 | 10 ± 1 | 100 |
| Triggering Solution | Protein Recovery (%) | Activity Recovery (%) | Retention Activity of Released Proteins (%) |
|---|---|---|---|
| Sodium phosphate buffer | 63 ± 1 | 59 ± 5 | 94 |
| Citrate phosphate buffer | 63 ± 1 | 57 ± 3 | 90 |
| Potassium carbonate | 60 ± 4 | 56 ± 3 | 92 |
| Concentration (M) | pH | Protein Release (%) | Activity Release (%) | Retention Activity of Released Proteins (%) |
|---|---|---|---|---|
| 0.1 | 8 | 55 ± 4 | 49 ± 3 | 89 |
| 0.3 | 8 | 57 ± 2 | 47 ± 4 | 83 |
| 0.5 | 8 | 59 ± 1 | 50 ± 3 | 85 |
| Concentration (M) | pH | Protein Release (%) | Activity Release (%) | Retention Activity of Released Proteins (%) |
|---|---|---|---|---|
| 0.1 | 8 | 55 ± 4 | 49 ± 3 | 89 |
| 0.1 | 6 | 50 ± 3 | 43 ± 3 | 86 |
| 0.1 | 4 | 52 ± 4 | 47 ± 3 | 90 |
| Solution | Degradation Rate (mgPBSA/h/cm2) |
|---|---|
| Sodium phosphate buffer 0.3 M pH 8 | 6.0 |
| Sodium phosphate buffer 0.5 M pH 8 | 6.2 |
| Sodium phosphate buffer 0.1 M pH 8 | 6.4 |
| Sodium phosphate buffer 0.1 M pH 4 | 5.6 |
| Sodium phosphate buffer 0.1 M pH 6 | 6.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romano, A.; Rosato, A.; Bianchi, S.; Zanaroli, G.; Celli, A.; Totaro, G.; Sisti, L. Triggering of Polymer-Degrading Enzymes from Layered Double Hydroxides for Recycling Strategies. Int. J. Mol. Sci. 2023, 24, 831. https://doi.org/10.3390/ijms24010831
Romano A, Rosato A, Bianchi S, Zanaroli G, Celli A, Totaro G, Sisti L. Triggering of Polymer-Degrading Enzymes from Layered Double Hydroxides for Recycling Strategies. International Journal of Molecular Sciences. 2023; 24(1):831. https://doi.org/10.3390/ijms24010831
Chicago/Turabian StyleRomano, Angela, Antonella Rosato, Stefano Bianchi, Giulio Zanaroli, Annamaria Celli, Grazia Totaro, and Laura Sisti. 2023. "Triggering of Polymer-Degrading Enzymes from Layered Double Hydroxides for Recycling Strategies" International Journal of Molecular Sciences 24, no. 1: 831. https://doi.org/10.3390/ijms24010831
APA StyleRomano, A., Rosato, A., Bianchi, S., Zanaroli, G., Celli, A., Totaro, G., & Sisti, L. (2023). Triggering of Polymer-Degrading Enzymes from Layered Double Hydroxides for Recycling Strategies. International Journal of Molecular Sciences, 24(1), 831. https://doi.org/10.3390/ijms24010831








