Transcriptome-Wide Identification and Functional Characterization of CIPK Gene Family Members in Actinidia valvata under Salt Stress
Abstract
1. Introduction
2. Results
2.1. Identification and Phylogenetic Analysis of AvCIPK Genes in A. valvata
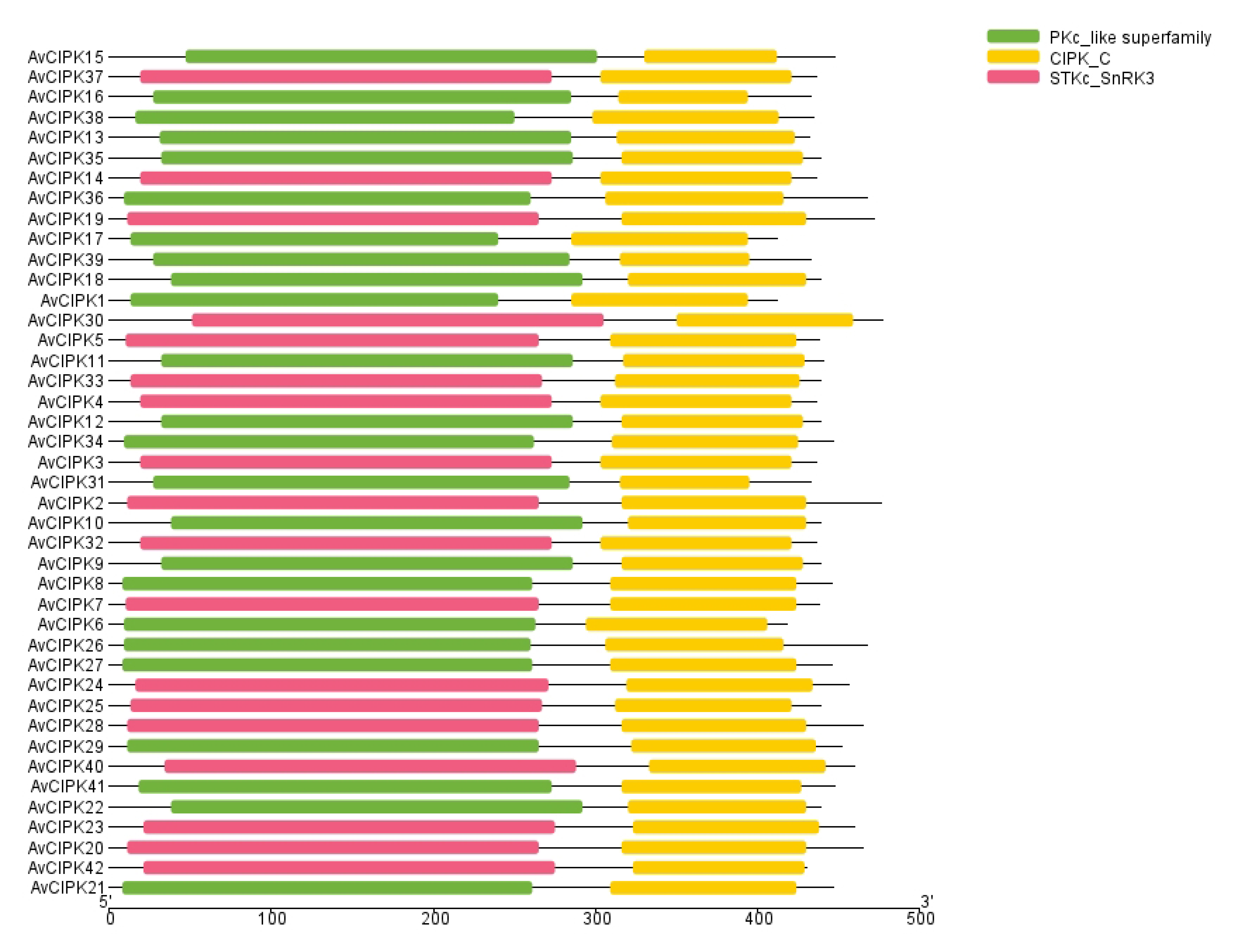
2.2. Analysis of Conserved Motif and Gene Structure for AvCIPKs
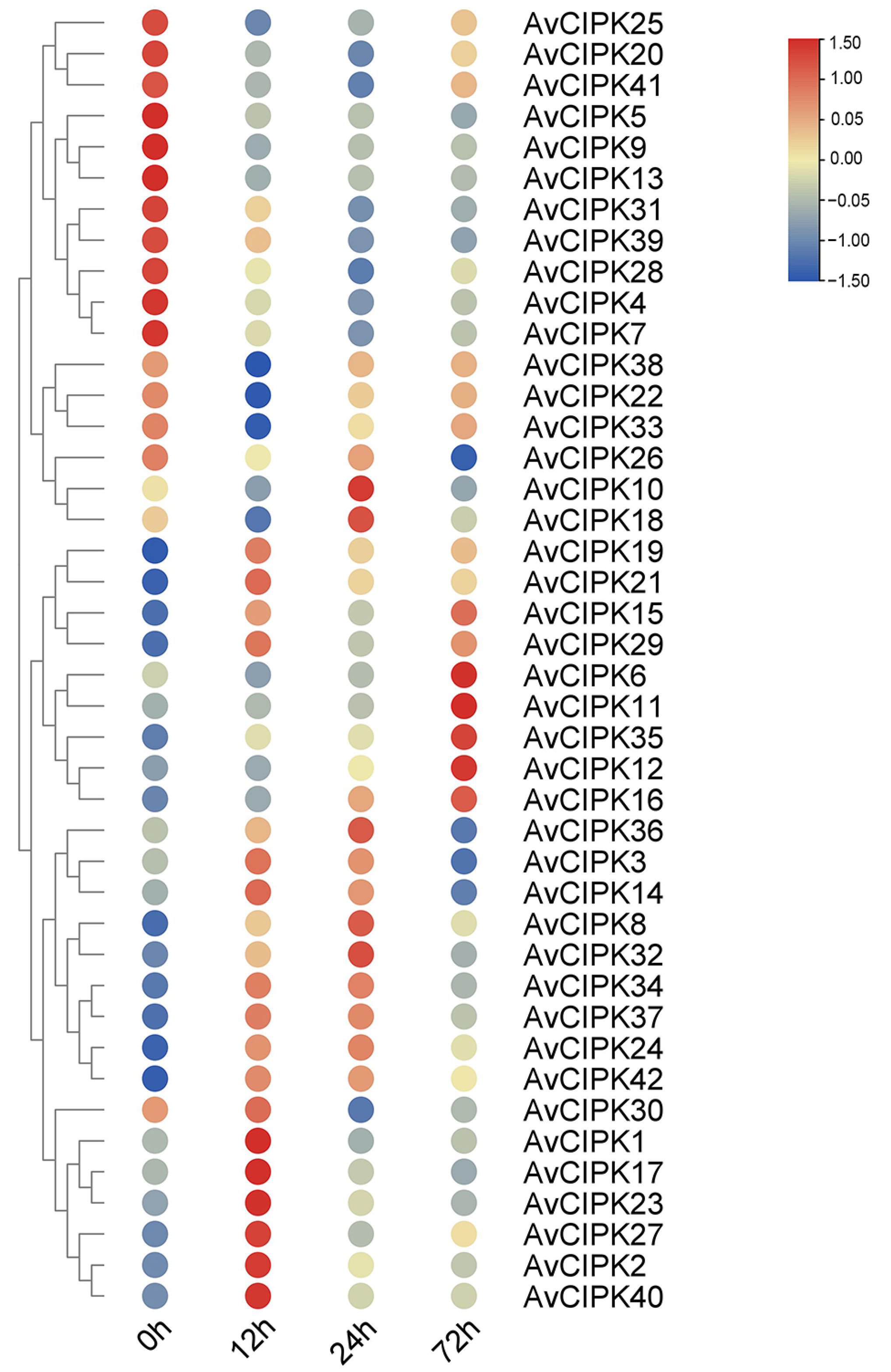
2.3. Expression Profiles of AvCIPK Genes
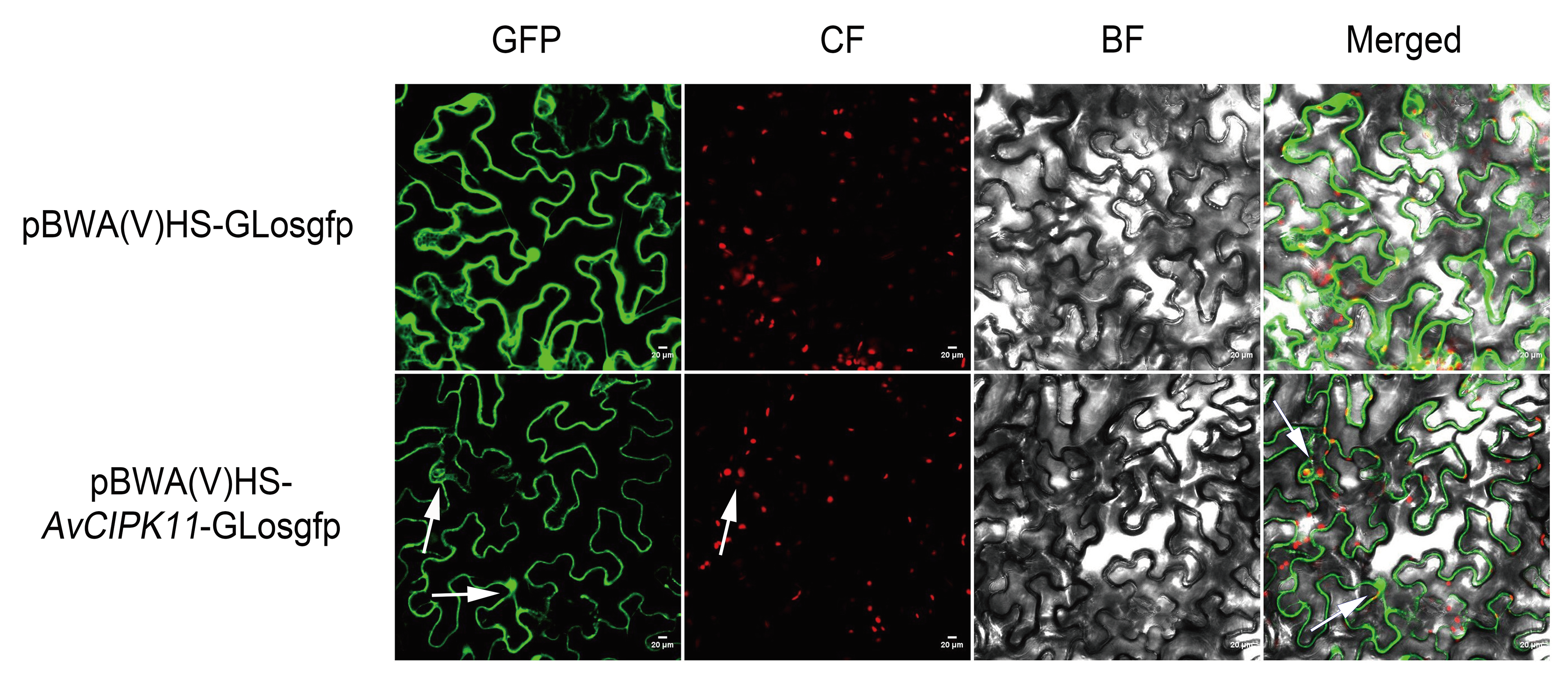
2.4. Subcellular Localization Analysis of AvCIPK11
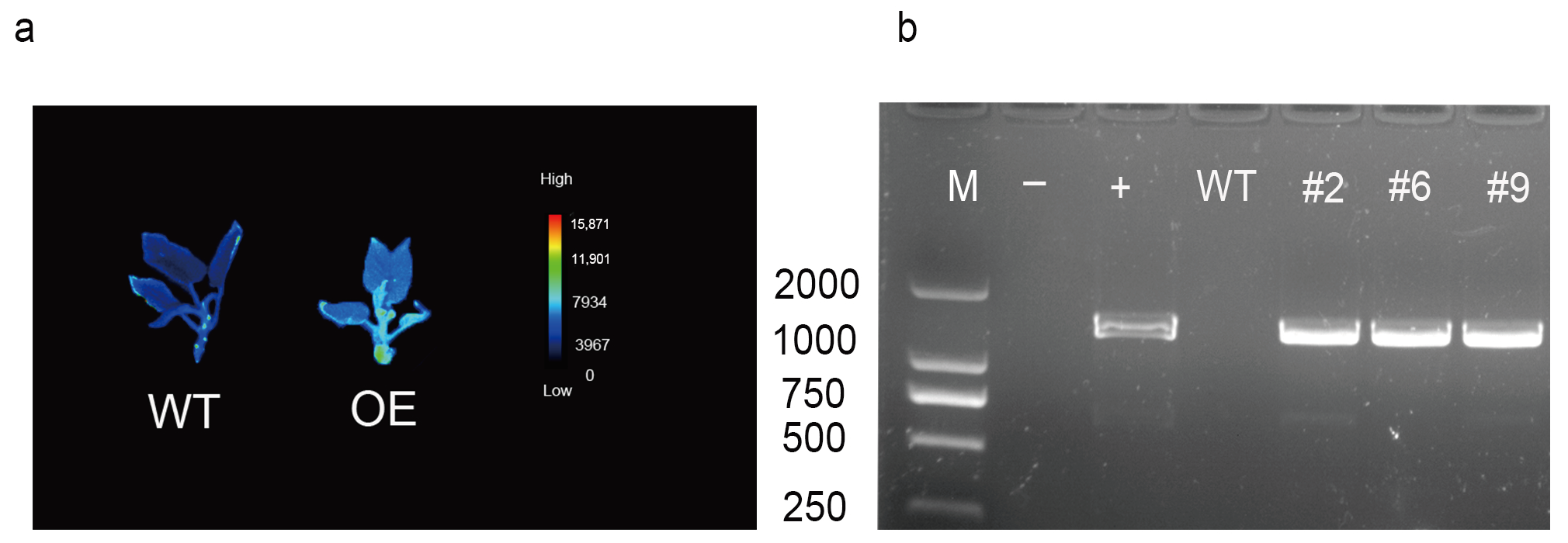
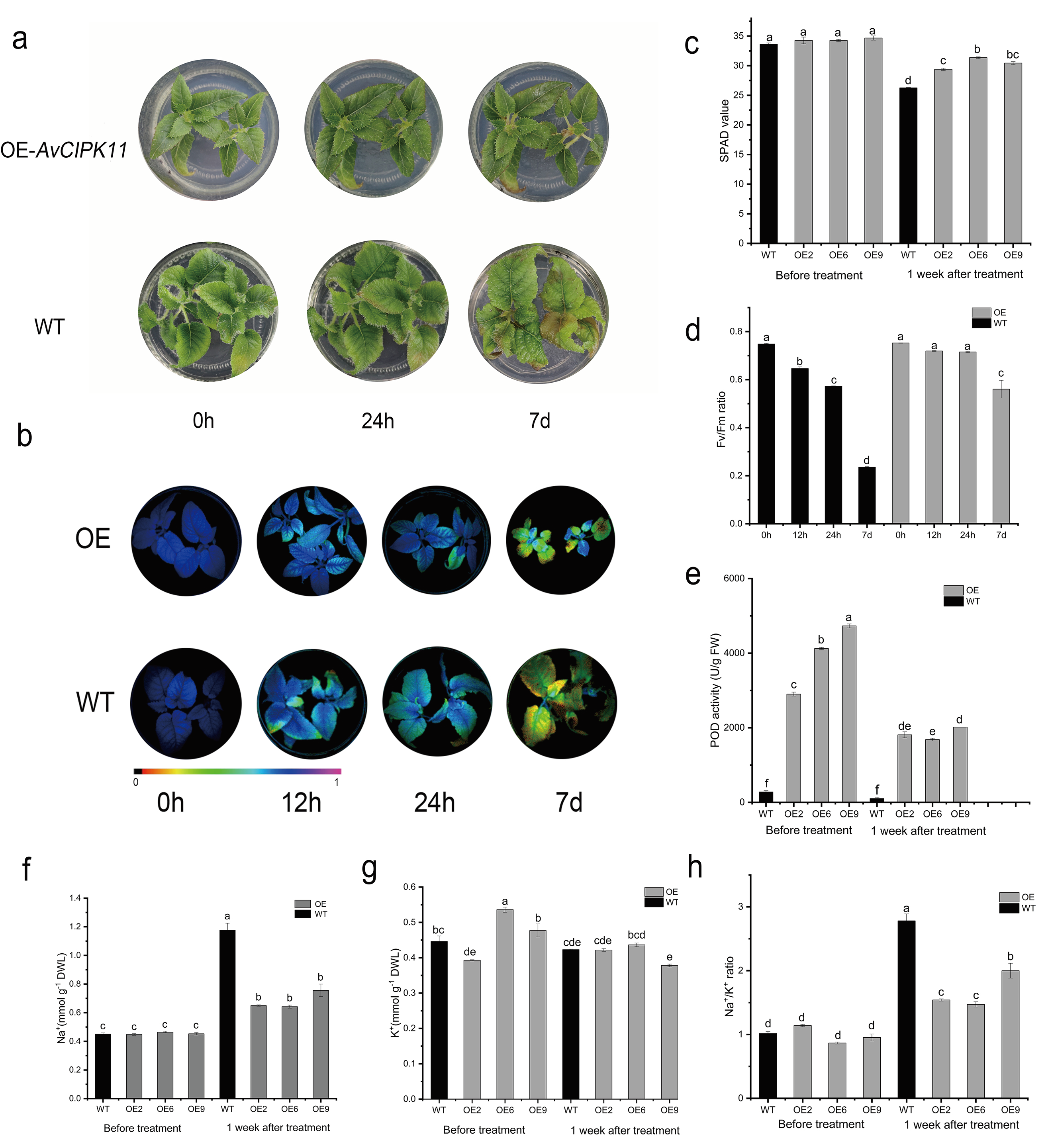
2.5. Overexpression of AvCIPK11 Enhances Salt Tolerance of A. chinensis
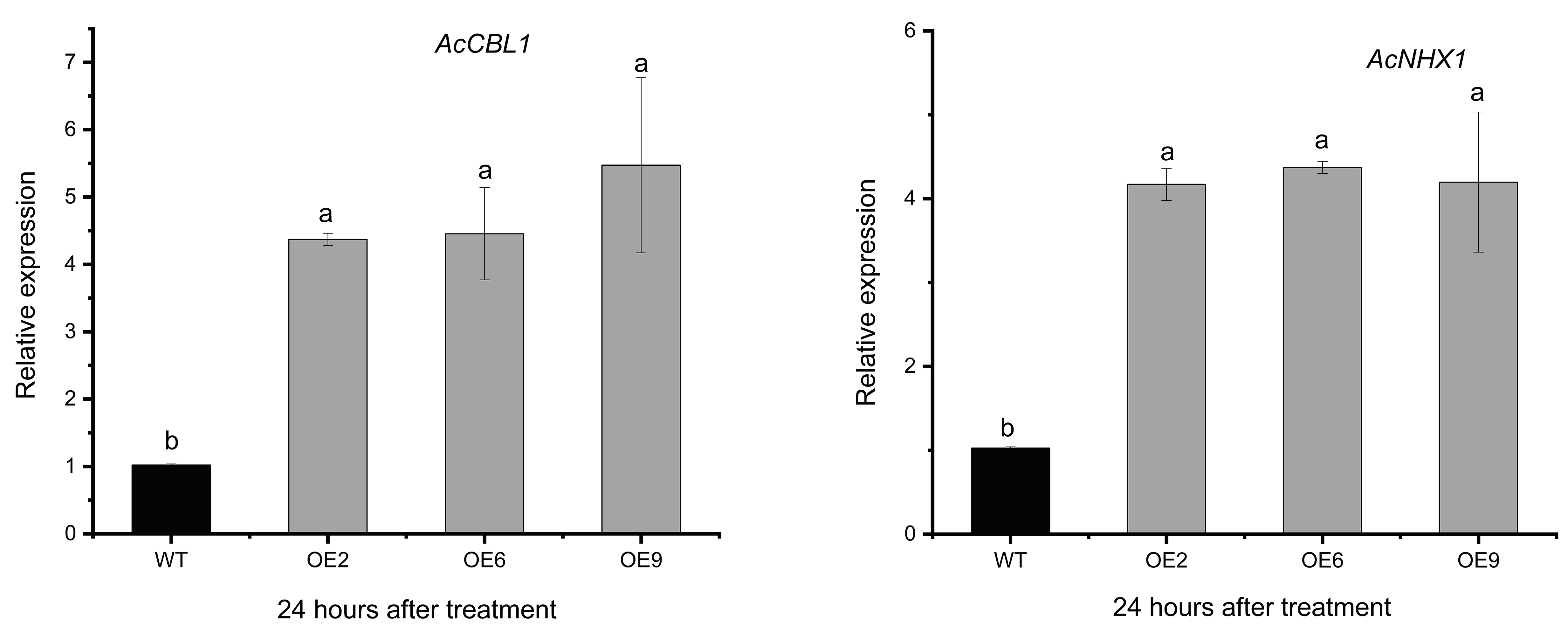
2.6. AvCIPK11 Activates the Expression of Salt Stress-Related Genes
3. Discussion
4. Materials and Methods
4.1. Identification of CIPK Family Members in A. valata and Arabidopsis
4.2. Phylogenetic Analysis and Conserved Motif Identification
4.3. Plant Materials Preparation and Application of Salt Treatment
4.4. RNA Isolation and RT-qPCR Analysis
4.5. Subcellular Localization Analysis
4.6. Construction of Overexpression Vector, Transformation, and Regeneration of Transgenic Kiwifruit Plants
4.7. Salt Tolerance Assay
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Majumder, A.L.; Sengupta, S.; Goswami, L. Osmolyte Regulation in Abiotic Stress; Springer: Berlin/Heidelberg, Germany, 2009; pp. 349–370. [Google Scholar]
- Maierhofer, T.; Diekmann, M.; Offenborn, J.N.; Lind, C.; Bauer, H.; Hashimoto, K.; KA, S.A.-R.; Luan, S.; Kudla, J.; Geiger, D.; et al. Site- and kinase-specific phosphorylation-mediated activation of SLAC1, a guard cell anion channel stimulated by abscisic acid. Sci. Signal. 2014, 7, ra86–ra97. [Google Scholar] [CrossRef]
- Luan, S. The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 2009, 14, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Kudla, J.; Becker, D.; Grill, E.; Hedrich, R.; Hippler, M.; Kummer, U.; Parniske, M.; Romeis, T.; Schumacher, K. Advances and current challenges in calcium signaling. New. Phytol. 2018, 218, 414–431. [Google Scholar] [CrossRef]
- Guo, Y.; Halfter, U.; Ishitani, M.; Zhu, J.K. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 2001, 13, 1383–1400. [Google Scholar] [CrossRef]
- Xiang, Y.; Huang, Y.; Xiong, L. Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 2007, 144, 1416–1428. [Google Scholar] [CrossRef]
- Hrabak, E.M.; Chan, C.W.; Gribskov, M.; Harper, J.F.; Choi, J.H.; Halford, N.; Kudla, J.; Luan, S.; Nimmo, H.G.; Sussman, M.R.; et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003, 132, 666–680. [Google Scholar] [CrossRef]
- Ishitani, M.; Liu, J.; Halfter, U.; Kim, C.S.; Shi, W.; Zhu, J.K. SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 2000, 12, 1667–1678. [Google Scholar] [CrossRef]
- Sánchez-Barrena, M.J.; Martínez-Ripoll, M.; Zhu, J.K.; Albert, A. The structure of the Arabidopsis thaliana SOS3: Molecular mechanism of sensing calcium for salt stress response. J. Mol. Biol. 2005, 345, 1253–1264. [Google Scholar] [CrossRef]
- Qiu, Q.S.; Guo, Y.; Dietrich, M.A.; Schumaker, K.S.; Zhu, J.K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 2002, 99, 8436–8441. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Ishitani, M.; Kim, C.; Zhu, J.K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901. [Google Scholar] [CrossRef] [PubMed]
- Batelli, G.; Verslues, P.E.; Agius, F.; Qiu, Q.; Fujii, H.; Pan, S.; Schumaker, K.S.; Grillo, S.; Zhu, J.K. SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity. Mol. Cell Biol. 2007, 27, 7781–7790. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, H.D.; Chen, L.Q.; Wang, Y.; Liu, L.L.; He, L.; Wu, W.H. A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 2006, 125, 1347–1360. [Google Scholar] [CrossRef]
- Ragel, P.; Ródenas, R.; García-Martín, E.; Andrés, Z.; Villalta, I.; Nieves-Cordones, M.; Rivero, R.M.; Martínez, V.; Pardo, J.M.; Quintero, F.J.; et al. The CBL-Interacting Protein Kinase CIPK23 Regulates HAK5-Mediated High-Affinity K+ Uptake in Arabidopsis Roots. Plant Physiol. 2015, 169, 2863–2873. [Google Scholar]
- Li, L.; Kim, B.G.; Cheong, Y.H.; Pandey, G.K.; Luan, S. A Ca(2)+ signaling pathway regulates a K(+) channel for low-K response in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 12625–12630. [Google Scholar] [CrossRef]
- Gierth, M.; Maser, P.; Schroeder, J.I. The potassium transporter AtHAK5 functions in K(+) deprivation-induced high-affinity K(+) uptake and AKT1 K(+) channel contribution to K(+) uptake kinetics in Arabidopsis roots. Plant Physiol. 2005, 137, 1105–1114. [Google Scholar] [CrossRef]
- Rubio, F.; Alemán, F.; Nieves-Cordones, M.; Martínez, V. Studies on Arabidopsis athak5, atakt1 double mutants disclose the range of concentrations at which AtHAK5, AtAKT1 and unknown systems mediate K uptake. Physiol. Plant 2010, 139, 220–228. [Google Scholar] [CrossRef]
- Tang, R.J.; Zhao, F.G.; Yang, Y.; Wang, C.; Li, K.; Kleist, T.J.; Lemaux, P.G.; Luan, S. A calcium signalling network activates vacuolar K(+) remobilization to enable plant adaptation to low-K environments. Nat. Plants 2020, 6, 384–393. [Google Scholar] [CrossRef]
- Yang, Y.; Qin, Y.; Xie, C.; Zhao, F.; Zhao, J.; Liu, D.; Chen, S.; Fuglsang, A.T.; Palmgren, M.G.; Schumaker, K.S.; et al. The Arabidopsis chaperone J3 regulates the plasma membrane H+-ATPase through interaction with the PKS5 kinase. Plant Cell 2010, 22, 1313–1332. [Google Scholar] [CrossRef]
- Tripathi, V.; Parasuraman, B.; Laxmi, A.; Chattopadhyay, D. CIPK6, a CBL-interacting protein kinase is required for development and salt tolerance in plants. Plant J. 2009, 58, 778–790. [Google Scholar] [CrossRef] [PubMed]
- Abdula, S.E.; Lee, H.-J.; Ryu, H.; Kang, K.K.; Nou, I.; Sorrells, M.E.; Cho, Y.-G. Overexpression of BrCIPK1 Gene Enhances Abiotic Stress Tolerance by Increasing Proline Biosynthesis in Rice. Plant Mol. Biol. Rep. 2015, 34, 501–511. [Google Scholar] [CrossRef]
- Deng, X.; Hu, W.; Wei, S.; Zhou, S.; Zhang, F.; Han, J.; Chen, L.; Li, Y.; Feng, J.; Fang, B.; et al. TaCIPK29, a CBL-Interacting Protein Kinase Gene from Wheat, Confers Salt Stress Tolerance in Transgenic Tobacco. PLoS ONE 2013, 8, e69881. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, T.; Li, T.; Wang, M.; Yang, G.; He, G. A CBL-Interacting Protein Kinase TaCIPK2 Confers Drought Tolerance in Transgenic Tobacco Plants through Regulating the Stomatal Movement. PLoS ONE 2016, 11, e0167962. [Google Scholar] [CrossRef] [PubMed]
- Luan, S.; Kudla, J.; Rodriguez-Concepcion, M.; Yalovsky, S.; Gruissem, W. Calmodulins and calcineurin B-like proteins: Calcium sensors for specific signal response coupling in plants. Plant Cell 2002, 14, S389–S400. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, B.; Liu, W.-Z.; Li, H.; Wang, L.; Wang, B.; Deng, M.; Liang, W.; Deyholos, M.K.; Jiang, Y.-Q. Identification and characterization of CBL and CIPK gene families in canola (Brassica napus L.). BMC Plant Biol. 2014, 14, 8. [Google Scholar] [CrossRef]
- Kolukisaoglu, Ü.; Weinl, S.; Blazevic, D.; Batistic, O.; Kudla, J. Calcium Sensors and Their Interacting Protein Kinases: Genomics of the Arabidopsis and Rice CBL-CIPK Signaling Networks. Plant Physiol. 2004, 134, 43–58. [Google Scholar] [CrossRef]
- Chen, X.; Gu, Z.; Xin, D.; Hao, L.; Liu, C.; Huang, J.; Ma, B.; Zhang, H. Identification and characterization of putative CIPK genes in maize. J. Genet. Genom. 2011, 38, 77–87. [Google Scholar] [CrossRef]
- Yu, Y.; Xia, X.; Yin, W.; Zhang, H. Comparative genomic analysis of CIPK gene family in Arabidopsis and Populus. Plant Growth Regul. 2007, 52, 101–110. [Google Scholar] [CrossRef]
- Xi, Y.; Liu, J.; Dong, C.; Cheng, Z.-M. The CBL and CIPK Gene Family in Grapevine (Vitis vinifera): Genome-Wide Analysis and Expression Profiles in Response to Various Abiotic Stresses. Front Plant Sci 2017, 8, 978–993. [Google Scholar] [CrossRef]
- Niu, L.; Dong, B.; Song, Z.; Meng, D.; Fu, Y. Genome-Wide Identification and Characterization of CIPK Family and Analysis Responses to Various Stresses in Apple (Malus domestica). Int. J. Mol. Sci. 2018, 19, 19072131. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Su, Y.; Wang, J.; Jia, B.; Wu, M.; Pei, W.; Zhang, J.; Yu, J. Genome-Wide Characterization and Analysis of CIPK Gene Family in Two Cultivated Allopolyploid Cotton Species: Sequence Variation, Association with Seed Oil Content, and the Role of GhCIPK6. Int. J. Mol. Sci. 2020, 21, 863. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Zhang, X.; Zhang, W.; Tian, Y.; Wang, X.; Hao, Z.; Chang, Y. Genome-wide identification, characterization and expression analysis of the MITF gene in Yesso scallops (Patinopecten yessoensis) with different shell colors. Gene 2019, 688, 155–162. [Google Scholar] [CrossRef]
- Abid, M.; Zhang, Y.J.; Li, Z.; Bai, D.F.; Zhong, Y.P.; Fang, J.B. Effect of Salt stress on growth, physiological and biochemical characters of Four kiwifruit genotypes. Sci. Hortic. 2020, 271, 109473. [Google Scholar] [CrossRef]
- Zhong, Y.-P.; Qi, X.-J.; Chen, J.-Y.; Li, Z.; Bai, D.-F.; Wei, C.-G.; Fang, J.-B. Growth and physiological responses of four kiwifruit genotypes to salt stress and resistance evaluation. J. Integr. Agric. 2019, 18, 83–95. [Google Scholar] [CrossRef]
- Abid, M.; Gu, S.; Zhang, Y.-J.; Sun, S.; Li, Z.; Bai, D.-F.; Sun, L.; Qi, X.-J.; Zhong, Y.-P.; Fang, J.-B. Comparative transcriptome and metabolome analysis reveal key regulatory defense networks and genes involved in enhanced salt tolerance of Actinidia (kiwifruit). Hortic. Res. 2022, 9, uhac189. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Wang, X.; Bai, J.; Wei, T.; Sun, M.; Zhu, L.; Wang, M.; Zhao, Y.; Wei, W. The kinase CIPK11 functions as a positive regulator in cadmium stress response in Arabidopsis. Gene 2020, 772, 145372. [Google Scholar] [CrossRef]
- Yang, Y.H.; Li, M.J.; Li, X.Y.; Chen, X.J.; Lin, W.X.; Zhang, Z.Y. Transcriptome-wide identification of the genes responding to replanting disease in Rehmannia glutinosa L. roots. Mol. Biol. Rep. 2014, 42, 881–892. [Google Scholar] [CrossRef]
- Chen, X.; Chen, G.; Li, J.; Hao, X.; Tuerxun, Z.; Chang, X.; Gao, S.; Huang, Q. A maize calcineurin B-like interacting protein kinase ZmCIPK42 confers salt stress tolerance. Physiol. Plant. 2020, 171, 161–172. [Google Scholar] [CrossRef]
- Plasencia, F.A.; Estrada, Y.; Flores, F.B.; Ortíz-Atienza, A.; Lozano, R.; Egea, I. The Ca(2+) Sensor Calcineurin B-Like Protein 10 in Plants: Emerging New Crucial Roles for Plant Abiotic Stress Tolerance. Front. Plant Sci. 2020, 11, 599944. [Google Scholar] [CrossRef]
- Ma, R.; Liu, W.; Li, S.; Zhu, X.; Yang, J.; Zhang, N.; Si, H. Genome-Wide Identification, Characterization and Expression Analysis of the CIPK Gene Family in Potato (Solanum tuberosum L.) and the Role of StCIPK10 in Response to Drought and Osmotic Stress. Int. J. Mol. Sci. 2021, 22, 13535. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Xia, Y.; Xie, Q.; Cao, Y.; Wang, Z.; Hao, G.; Song, J.; Zhou, Y.; Jiang, X. The protein kinase complex CBL10–CIPK8–SOS1 functions in Arabidopsis to regulate salt tolerance. J. Exp. Bot. 2019, 71, 1801–1814. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Cao, J.; Chen, Q.; He, J.; Liu, Z.; Wang, J.; Li, X.; Yang, Y. The Kinase CIPK11 Functions as a Negative Regulator in Drought Stress Response in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 2422. [Google Scholar] [CrossRef]
- Lu, L.; Chen, X.; Wang, P.; Lu, Y.; Zhang, J.; Yang, X.; Cheng, T.; Shi, J.; Chen, J. CIPK11: A calcineurin B-like protein-interacting protein kinase from Nitraria tangutorum, confers tolerance to salt and drought in Arabidopsis. BMC Plant Biol. 2021, 21, 123. [Google Scholar] [CrossRef]
- Fichman, Y.; Mittler, R. Integration of electric, calcium, reactive oxygen species and hydraulic signals during rapid systemic signaling in plants. Plant J. 2021, 107, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Seifikalhor, M.; Aliniaeifard, S.; Shomali, A.; Azad, N.; Hassani, B.; Lastochkina, O.; Li, T. Calcium signaling and salt tolerance are diversely entwined in plants. Plant Signal. Behav. 2019, 14, 1665455. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Zou, L.H.; Zheng, B.Q.; Wang, Y. Circadian Regulation of Alternative Splicing of Drought-Associated CIPK Genes in Dendrobium catenatum (Orchidaceae). Int. J. Mol. Sci. 2019, 20, 688. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, X.; Liu, S.; Yu, A.; Yang, C.; Chen, X.; Liu, J.; Wang, A. Identification and Functional Analysis of Tomato CIPK Gene Family. Int. J. Mol. Sci. 2019, 21, 110. [Google Scholar] [CrossRef]
- Ma, Q.-J.; Sun, M.-H.; Kang, H.; Lu, J.; You, C.-X.; Hao, Y.-J. A CIPK protein kinase targets sucrose transporter MdSUT2.2 at Ser254 for phosphorylation to enhance salt tolerance. Plant Cell Environ. 2019, 42, 918–930. [Google Scholar] [CrossRef]
- Su, Y.; Guo, A.; Huang, Y.; Wang, Y.; Hua, J. GhCIPK6a increases salt tolerance in transgenic upland cotton by involving in ROS scavenging and MAPK signaling pathways. BMC Plant Biol. 2020, 20, 421. [Google Scholar] [CrossRef]
- Ketehouli, T.; Zhou, Y.-G.; Dai, S.-Y.; Carther, K.F.I.; Sun, D.-Q.; Li, Y.; Nguyen, Q.V.H.; Xu, H.; Wang, F.-W.; Liu, W.-C.; et al. A soybean calcineurin B-like protein-interacting protein kinase, GmPKS4, regulates plant responses to salt and alkali stresses. J. Plant Physiol. 2021, 256, 153331. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.-G.; Ma, Q.-J.; Sun, C.-H.; Sun, M.-H.; You, C.-X.; Hao, Y.-J. Overexpression of MdSOS2L1, a CIPK protein kinase, increases the antioxidant metabolites to enhance salt tolerance in apple and tomato. Physiol. Plant 2016, 156, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Batistič, O.; Waadt, R.; Steinhorst, L.; Held, K.; Kudla, J. CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J. 2010, 61, 211–222. [Google Scholar] [CrossRef]
- Turan, S.; Tripathy, B.C. Salt-stress induced modulation of chlorophyll biosynthesis during de-etiolation of rice seedlings. Physiol. Plant 2015, 153, 477–491. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Zhou, S.; Hu, W.; Feng, J.; Zhang, F.; Chen, L.; Huang, C.; Luo, Q.; He, Y.; Yang, G.; et al. Ectopic expression of wheatTaCIPK14, encoding a calcineurin B-like protein-interacting protein kinase, confers salinity and cold tolerance in tobacco. Physiol. Plant. 2013, 149, 367–377. [Google Scholar] [CrossRef]
- Siddiqui, N.; Mostofa, M.G.; Akter, M.M.; Srivastava, A.K.; Sayed, A.; Hasan, M.S.; Tran, L.-S.P. Impact of salt-induced toxicity on growth and yield-potential of local wheat cultivars: Oxidative stress and ion toxicity are among the major determinants of salt-tolerant capacity. Chemosphere 2017, 187, 385–394. [Google Scholar] [CrossRef]
- Barragán, V.; Leidi, E.O.; Andrés, Z.; Rubio, L.; De Luca, A.; Fernández, J.A.; Cubero, B.; Pardo, J.M. Ion Exchangers NHX1 and NHX2 Mediate Active Potassium Uptake into Vacuoles to Regulate Cell Turgor and Stomatal Function in Arabidopsis. Plant Cell 2012, 24, 1127–1142. [Google Scholar] [CrossRef]
- Ho, C.H.; Lin, S.H.; Hu, H.C.; Tsay, Y.F. CHL1 functions as a nitrate sensor in plants. Cell 2009, 138, 1184–1194. [Google Scholar] [CrossRef]
- Liu, J.; Ishitani, M.; Halfter, U.; Kim, C.-S.; Zhu, J.-K. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. USA 2000, 97, 3730–3734. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, J.-K. A calcium sensor homolog required for plant salt tolerance. Science 1998, 280, 1943–1945. [Google Scholar] [CrossRef]
- Rubio, F.; Nieves-Cordones, M.; Horie, T.; Shabala, S. Doing ‘business as usual’ comes with a cost: Evaluating energy cost of maintaining plant intracellular K(+) homeostasis under saline conditions. New. Phytol. 2020, 225, 1097–1104. [Google Scholar] [CrossRef] [PubMed]
- Cuin, T.A.; Betts, S.A.; Chalmandrier, R.; Shabala, S. A root’s ability to retain K+ correlates with salt tolerance in wheat. J. Exp. Bot. 2008, 59, 2697–2706. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Halfter, U.; Ishitani, M.; Zhu, J.K. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 2000, 97, 3735–3740. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Wang, Y.; Wang, M.; Li, T.; Zhou, Y.; Wang, X.; Wei, S.; He, G.; Yang, G. Identification and comprehensive analyses of the CBL and CIPK gene families in wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 269. [Google Scholar] [CrossRef]
- Zhang, H.; Feng, H.; Zhang, J.; Ge, R.; Zhang, L.; Wang, Y.; Li, L.; Wei, J.; Li, R. Emerging crosstalk between two signaling pathways coordinates K+ and Na+ homeostasis in the halophyte Hordeum brevisubulatum. J. Exp. Bot. 2020, 71, 4345–4358. [Google Scholar] [CrossRef]
- Blumwald, E.; Poole, R.J. Na+/H+ Antiport in Isolated Tonoplast Vesicles from Storage Tissue of Beta vulgaris 1. Plant Physiol. 1985, 78, 163–167. [Google Scholar] [CrossRef]
- Bassil, E.; Coku, A.; Blumwald, E. Cellular ion homeostasis: Emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J. Exp. Bot. 2012, 63, 5727–5740. [Google Scholar] [CrossRef]
- Apse, M.P.; Aharon, G.S.; Snedden, W.A.; Blumwald, E.J.S. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 1999, 285, 1256–1258. [Google Scholar] [CrossRef]
- Liu, H.U.A.; Wang, Q.; Yu, M.; Zhang, Y.; Wu, Y.; Zhang, H. Transgenic salt-tolerant sugar beet (Beta vulgaris L.) constitutively expressing an Arabidopsis thaliana vacuolar Na+/H+ antiporter gene, AtNHX3, accumulates more soluble sugar but less salt in storage roots. Plant Cell Environ. 2008, 31, 1325–1334. [Google Scholar] [CrossRef]
- Ohta, M.; Hayashi, Y.; Nakashima, A.; Hamada, A.; Tanaka, A.; Nakamura, T.; Hayakawa, T. Introduction of a Na+/H+ antiporter gene from Atriplex gmelini confers salt tolerance to rice. FEBS Lett. 2002, 532, 279–282. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, Z.-N.; Li, Q.; Zhu, W.-L.; Wang, X.-H.; Xu, P.; Cao, X.; Cui, X.-Y. CBL-Interacting Protein Kinase 2 Improves Salt Tolerance in Soybean (Glycine max L.). Agronomy 2022, 12, 1595. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.K. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fang, J.; Qi, X.; Lin, M.; Zhong, Y.; Sun, L.; Cui, W. Combined Analysis of the Fruit Metabolome and Transcriptome Reveals Candidate Genes Involved in Flavonoid Biosynthesis in Actinidia arguta. Int. J. Mol. Sci. 2018, 1471–1488. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wang, T.; Atkinson, R.; Janssen, B. The choice of agrobacterium strain for transformation of kiwifruit. Acta Hortic. 2007, 753, 227–232. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Su, L.; Dai, Z.; Li, S.; Xin, H. A novel system for evaluating drought-cold tolerance of grapevines using chlorophyll fluorescence. BMC Plant Biol. 2015, 15, 82. [Google Scholar] [CrossRef]
- Abdelbaki, A.; Schlerf, M.; Retzlaff, R.; Machwitz, M.; Verrelst, J.; Udelhoven, T. Comparison of Crop Trait Retrieval Strategies Using UAV-Based VNIR Hyperspectral Imaging. Remote Sens. 2021, 13, 1748. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, S.; Abid, M.; Bai, D.; Chen, C.; Sun, L.; Qi, X.; Zhong, Y.; Fang, J. Transcriptome-Wide Identification and Functional Characterization of CIPK Gene Family Members in Actinidia valvata under Salt Stress. Int. J. Mol. Sci. 2023, 24, 805. https://doi.org/10.3390/ijms24010805
Gu S, Abid M, Bai D, Chen C, Sun L, Qi X, Zhong Y, Fang J. Transcriptome-Wide Identification and Functional Characterization of CIPK Gene Family Members in Actinidia valvata under Salt Stress. International Journal of Molecular Sciences. 2023; 24(1):805. https://doi.org/10.3390/ijms24010805
Chicago/Turabian StyleGu, Shichao, Muhammad Abid, Danfeng Bai, Chen Chen, Leiming Sun, Xiujuan Qi, Yunpeng Zhong, and Jinbao Fang. 2023. "Transcriptome-Wide Identification and Functional Characterization of CIPK Gene Family Members in Actinidia valvata under Salt Stress" International Journal of Molecular Sciences 24, no. 1: 805. https://doi.org/10.3390/ijms24010805
APA StyleGu, S., Abid, M., Bai, D., Chen, C., Sun, L., Qi, X., Zhong, Y., & Fang, J. (2023). Transcriptome-Wide Identification and Functional Characterization of CIPK Gene Family Members in Actinidia valvata under Salt Stress. International Journal of Molecular Sciences, 24(1), 805. https://doi.org/10.3390/ijms24010805






