G-Protein-Coupled Receptors Mediate Modulations of Cell Viability and Drug Sensitivity by Aberrantly Expressed Recoverin 3 within A549 Cells
Abstract
1. Introduction
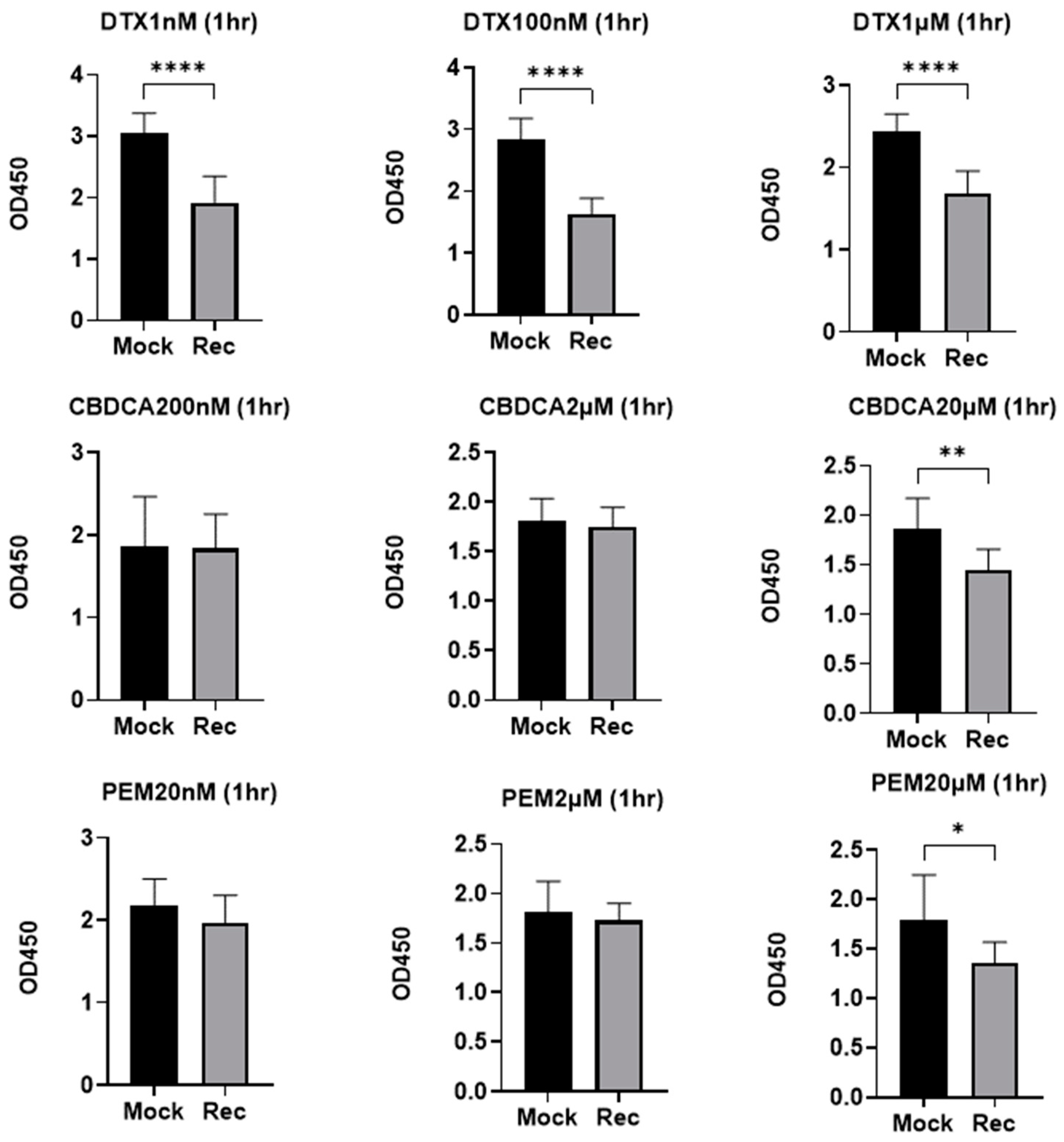
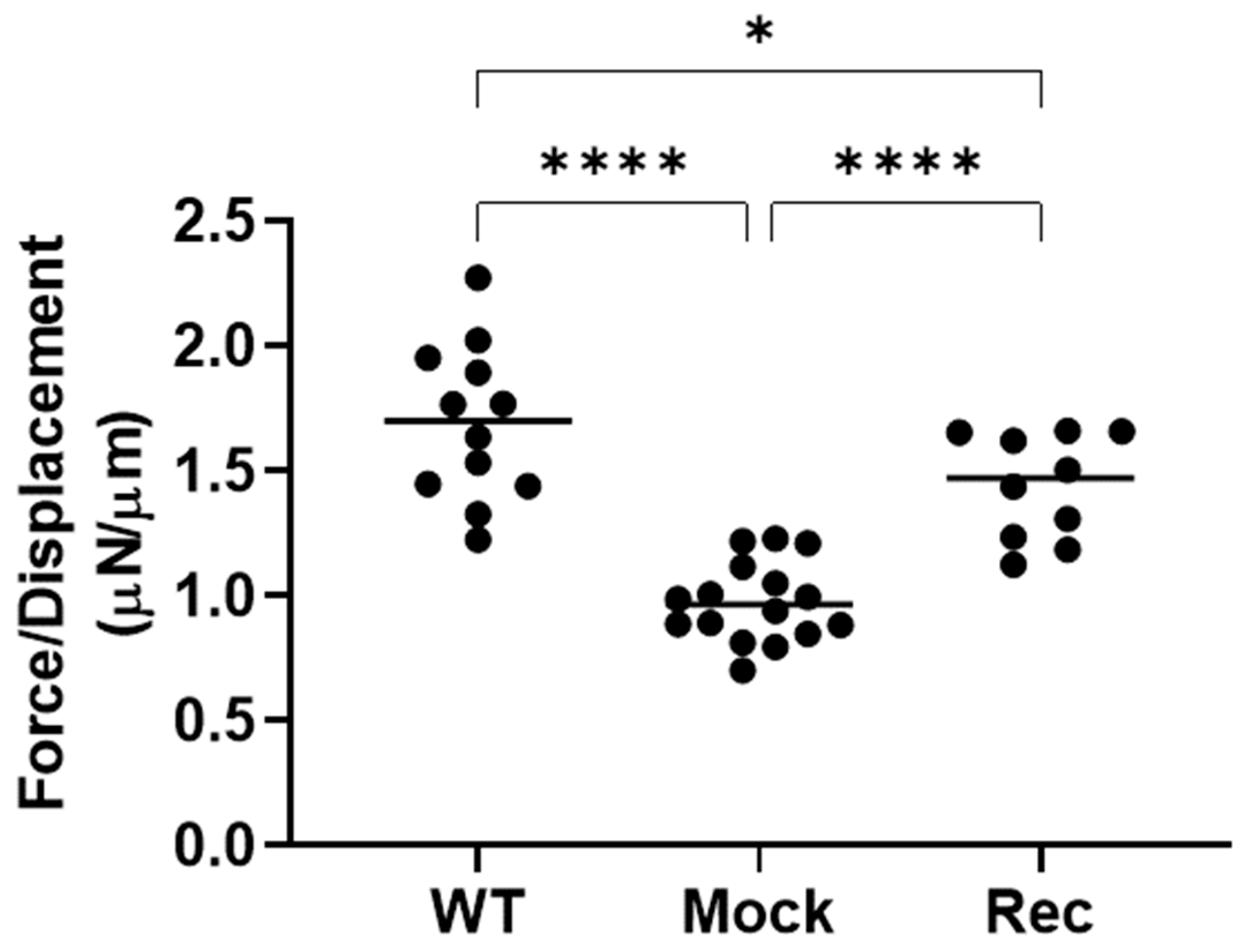
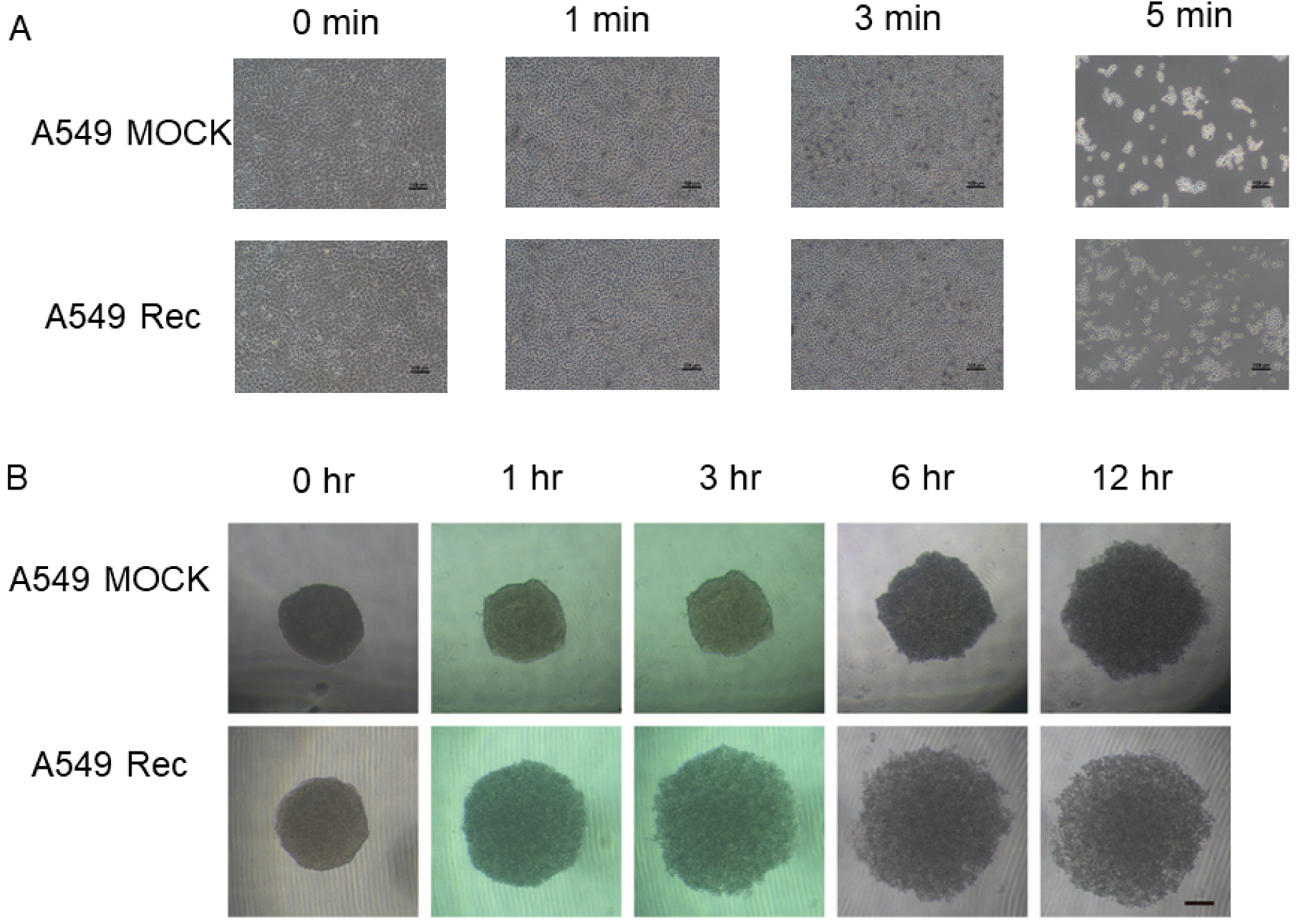
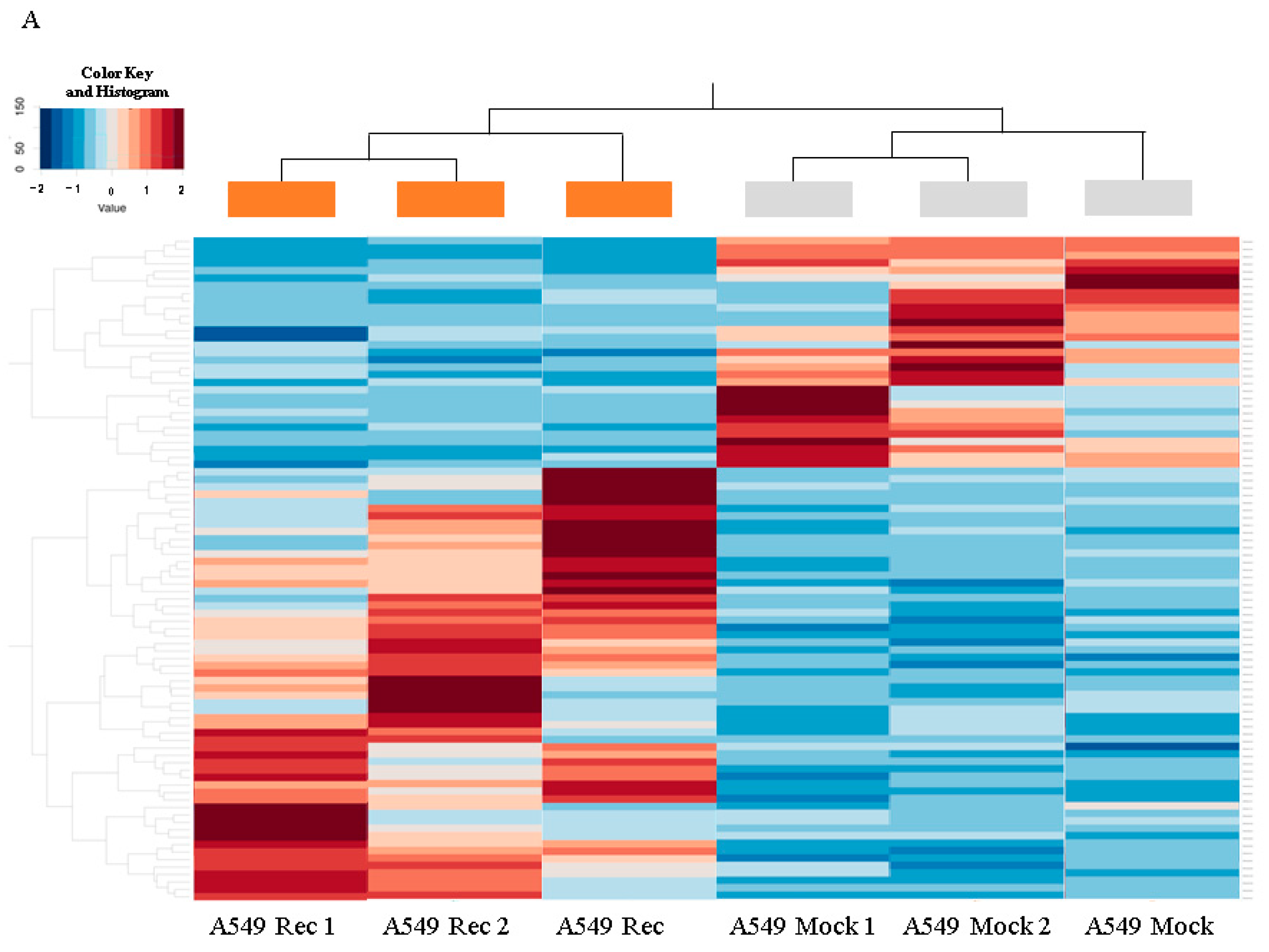
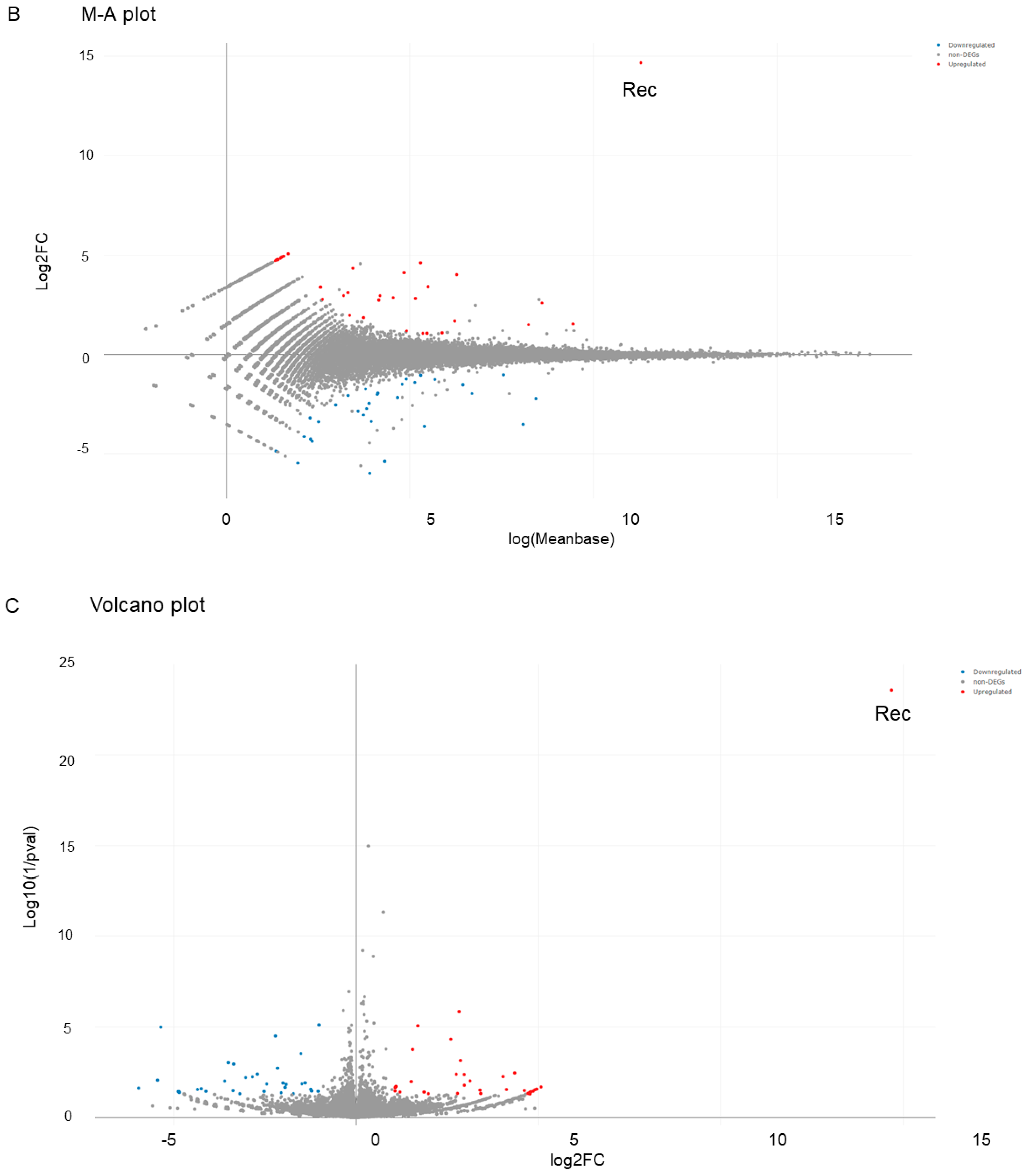
2. Results
3. Discussion
4. Materials and Methods
4.1. 2D and 3D Cultures of A549 Lung Adenoma Cells Transfected with or without Human Recoverin cDNA
4.2. Measurement of Mitochondrial and Glycolytic Functions of 2D-Cultured A549 Cells
4.3. Phase Contrast Microscopy of the 3D Spheroids
4.4. RNA Sequencing, Gene Function and Analysis of Pathways
4.5. Other Analytical Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lennon, V.A. Paraneoplastic autoantibodies: The case for a descriptive generic nomenclature. Neurology 1994, 44, 2236–2240. [Google Scholar] [CrossRef] [PubMed]
- Höftberger, R.; Rosenfeld, M.R.; Dalmau, J. Update on neurological paraneoplastic syndromes. Curr. Opin. Oncol. 2015, 27, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Ganaraja, V.H.; Rezk, M.; Dubey, D. Paraneoplastic neurological syndrome: Growing spectrum and relevance. Neurol. Sci. Off. J. Ital. Neurol. Soc. Ital. Soc. Clin. Neurophysiol. 2022, 43, 3583–3594. [Google Scholar] [CrossRef] [PubMed]
- Sawyer, R.A.; Selhorst, J.B.; Zimmerman, L.E.; Hoyt, W.F. Blindness caused by photoreceptor degeneration as a remote effect of cancer. Am. J. Ophthalmol. 1976, 81, 606–613. [Google Scholar] [CrossRef]
- Berson, E.L.; Lessell, S. Paraneoplastic night blindness with malignant melanoma. Am. J. Ophthalmol. 1988, 106, 307–311. [Google Scholar] [CrossRef]
- Grunwald, G.B.; Klein, R.; Simmonds, M.A.; Kornguth, S.E. Autoimmune basis for visual paraneoplastic syndrome in patients with small-cell lung carcinoma. Lancet 1985, 1, 658–661. [Google Scholar] [CrossRef]
- Kornguth, S.E.; Klein, R.; Appen, R.; Choate, J. Occurrence of anti-retinal ganglion cell antibodies in patients with small cell carcinoma of the lung. Cancer 1982, 50, 1289–1293. [Google Scholar] [CrossRef]
- Keltner, J.L.; Roth, A.M.; Chang, R.S. Photoreceptor degeneration. Possible autoimmune disorder. Arch. Ophthalmol. 1983, 101, 564–569. [Google Scholar] [CrossRef]
- Kornguth, S.E.; Kalinke, T.; Grunwald, G.B.; Schutta, H.; Dahl, D. Anti-neurofilament antibodies in the sera of patients with small cell carcinoma of the lung and with visual paraneoplastic syndrome. Cancer Res. 1986, 46, 2588–2595. [Google Scholar]
- Jacobson, D.M.; Thirkill, C.E.; Tipping, S.J. A clinical triad to diagnose paraneoplastic retinopathy. Ann. Neurol. 1990, 28, 162–167. [Google Scholar] [CrossRef]
- Yang, S.; Dizhoor, A.; Wilson, D.J.; Adamus, G. GCAP1, Rab6, and HSP27: Novel Autoantibody Targets in Cancer-Associated Retinopathy and Autoimmune Retinopathy. Transl. Vis. Sci. Technol. 2016, 5, 1. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Adamus, G. Are Anti-Retinal Autoantibodies a Cause or a Consequence of Retinal Degeneration in Autoimmune Retinopathies? Front. Immunol. 2018, 9, 765. [Google Scholar] [CrossRef] [PubMed]
- Polans, A.S.; Buczyłko, J.; Crabb, J.; Palczewski, K. A photoreceptor calcium binding protein is recognized by autoantibodies obtained from patients with cancer-associated retinopathy. J. Cell Biol. 1991, 112, 981–989. [Google Scholar] [CrossRef] [PubMed]
- Polans, A.S.; Burton, M.D.; Haley, T.L.; Crabb, J.W.; Palczewski, K. Recoverin, but not visinin, is an autoantigen in the human retina identified with a cancer-associated retinopathy. Investig. Ophthalmol. Vis. Sci. 1993, 34, 81–90. [Google Scholar]
- Kawamura, S. Rhodopsin phosphorylation as a mechanism of cyclic GMP phosphodiesterase regulation by S-modulin. Nature 1993, 362, 855–857. [Google Scholar] [CrossRef]
- Ohguro, H.; Ogawa, K.; Maeda, T.; Maeda, A.; Maruyama, I. Cancer-associated retinopathy induced by both anti-recoverin and anti-hsc70 antibodies in vivo. Investig. Ophthalmol. Vis. Sci. 1999, 40, 3160–3167. [Google Scholar]
- Adamus, G.; Machnicki, M.; Elerding, H.; Sugden, B.; Blocker, Y.S.; Fox, D.A. Antibodies to recoverin induce apoptosis of photoreceptor and bipolar cells in vivo. J. Autoimmun. 1998, 11, 523–533. [Google Scholar] [CrossRef]
- Maeda, T.; Maeda, A.; Maruyama, I.; Ogawa, K.I.; Kuroki, Y.; Sahara, H.; Sato, N.; Ohguro, H. Mechanisms of photoreceptor cell death in cancer-associated retinopathy. Investig. Ophthalmol. Vis. Sci. 2001, 42, 705–712. [Google Scholar]
- Polans, A.S.; Witkowska, D.; Haley, T.L.; Amundson, D.; Baizer, L.; Adamus, G. Recoverin, a photoreceptor-specific calcium-binding protein, is expressed by the tumor of a patient with cancer-associated retinopathy. Proc. Natl. Acad. Sci. USA 1995, 92, 9176–9180. [Google Scholar] [CrossRef]
- Yamaji, Y.; Matsubara, S.; Yamadori, I.; Sato, M.; Fujita, T.; Fujita, J.; Takahara, J. Characterization of a small-cell-lung-carcinoma cell line from a patient with cancer-associated retinopathy. Int. J. Cancer 1996, 65, 671–676. [Google Scholar] [CrossRef]
- Matsubara, S.; Yamaji, Y.; Sato, M.; Fujita, J.; Takahara, J. Expression of a photoreceptor protein, recoverin, as a cancer-associated retinopathy autoantigen in human lung cancer cell lines. Br. J. Cancer 1996, 74, 1419–1422. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Matsuo, S.; Ohguro, H.; Ohguro, I.; Nakazawa, M. Clinicopathological roles of aberrantly expressed recoverin in malignant tumor cells. Ophthalmic Res. 2010, 43, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Maeda, A.; Ohguro, H.; Maeda, T.; Wada, I.; Sato, N.; Kuroki, Y.; Nakagawa, T. Aberrant expression of photoreceptor-specific calcium-binding protein (recoverin) in cancer cell lines. Cancer Res. 2000, 60, 1914–1920. [Google Scholar]
- Verweij, J.; Clavel, M.; Chevalier, B. Paclitaxel (Taxol) and docetaxel (Taxotere): Not simply two of a kind. Annals Oncol. Off. J. Eur. Soc. Med. Oncol. 1994, 5, 495–505. [Google Scholar] [CrossRef]
- Szefler, B.; Czeleń, P.; Krawczyk, P. The Affinity of Carboplatin to B-Vitamins and Nucleobases. Int. J. Mol. Sci. 2021, 22, 3634. [Google Scholar] [CrossRef]
- Liang, J.; Lu, T.; Chen, Z.; Zhan, C.; Wang, Q. Mechanisms of resistance to pemetrexed in non-small cell lung cancer. Transl. Lung Cancer Res. 2019, 8, 1107–1118. [Google Scholar] [CrossRef]
- Hikage, F.; Atkins, S.; Kahana, A.; Smith, T.J.; Chun, T.H. HIF2A-LOX Pathway Promotes Fibrotic Tissue Remodeling in Thyroid-Associated Orbitopathy. Endocrinology 2019, 160, 20–35. [Google Scholar] [CrossRef] [PubMed]
- Ida, Y.; Hikage, F.; Itoh, K.; Ida, H.; Ohguro, H. Prostaglandin F2α agonist-induced suppression of 3T3-L1 cell adipogenesis affects spatial formation of extra-cellular matrix. Sci. Rep. 2020, 10, 7958. [Google Scholar] [CrossRef]
- Watanabe, M.; Ida, Y.; Ohguro, H.; Ota, C.; Hikage, F. Establishment of appropriate glaucoma models using dexamethasone or TGFβ2 treated three-dimension (3D) cultured human trabecular meshwork (HTM) cells. Sci. Rep. 2021, 11, 19369. [Google Scholar] [CrossRef]
- Miyagawa, Y.; Ohguro, H.; Odagiri, H.; Maruyama, I.; Maeda, T.; Maeda, A.; Sasaki, M.; Nakazawa, M. Aberrantly expressed recoverin is functionally associated with G-protein-coupled receptor kinases in cancer cell lines. Biochem. Biophys. Res. Commun. 2003, 300, 669–673. [Google Scholar] [CrossRef]
- Couet, J.; Li, S.; Okamoto, T.; Ikezu, T.; Lisanti, M.P. Identification of peptide and protein ligands for the caveolin-scaffolding domain. Implications for the interaction of caveolin with caveolae-associated proteins. J. Biol. Chem. 1997, 272, 6525–6533. [Google Scholar] [CrossRef] [PubMed]
- Pitcher, J.A.; Freedman, N.J.; Lefkowitz, R.J. G protein-coupled receptor kinases. Annu. Rev. Biochem. 1998, 67, 653–692. [Google Scholar] [CrossRef] [PubMed]
- Carman, C.V.; Lisanti, M.P.; Benovic, J.L. Regulation of G protein-coupled receptor kinases by caveolin. J. Biol. Chem. 1999, 274, 8858–8864. [Google Scholar] [CrossRef] [PubMed]
- Machado, F.S.; Rodriguez, N.E.; Adesse, D.; Garzoni, L.R.; Esper, L.; Lisanti, M.P.; Burk, R.D.; Albanese, C.; Van Doorslaer, K.; Weiss, L.M.; et al. Recent developments in the interactions between caveolin and pathogens. Adv. Exp. Med. Biol. 2012, 729, 65–82. [Google Scholar] [PubMed]
- Fridolfsson, H.N.; Roth, D.M.; Insel, P.A.; Patel, H.H. Regulation of intracellular signaling and function by caveolin. FASEB J. 2014, 28, 3823–3831. [Google Scholar] [CrossRef] [PubMed]
- Van Krieken, R.; Krepinsky, J.C. Caveolin-1 in the Pathogenesis of Diabetic Nephropathy: Potential Therapeutic Target? Curr. Diabetes Rep. 2017, 17, 19. [Google Scholar] [CrossRef]
- Couet, J.; Sargiacomo, M.; Lisanti, M.P. Interaction of a receptor tyrosine kinase, EGF-R, with caveolins. Caveolin binding negatively regulates tyrosine and serine/threonine kinase activities. J. Biol. Chem. 1997, 272, 30429–30438. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Ying, Y.; Anderson, R.G. Platelet-derived growth factor activates mitogen-activated protein kinase in isolated caveolae. Proc. Natl. Acad. Sci. USA 1997, 94, 13666–13670. [Google Scholar] [CrossRef]
- Wary, K.K.; Mariotti, A.; Zurzolo, C.; Giancotti, F.G. A requirement for caveolin-1 and associated kinase Fyn in integrin signaling and anchorage-dependent cell growth. Cell 1998, 94, 625–634. [Google Scholar] [CrossRef]
- Dupree, P.; Parton, R.G.; Raposo, G.; Kurzchalia, T.V.; Simons, K. Caveolae and sorting in the trans-Golgi network of epithelial cells. Embo J. 1993, 12, 1597–1605. [Google Scholar] [CrossRef]
- Feron, O.; Smith, T.W.; Michel, T.; Kelly, R.A. Dynamic targeting of the agonist-stimulated m2 muscarinic acetylcholine receptor to caveolae in cardiac myocytes. J. Biol. Chem. 1997, 272, 17744–17748. [Google Scholar] [CrossRef]
- Ishizaka, N.; Griendling, K.K.; Lassègue, B.; Alexander, R.W. Angiotensin II type 1 receptor: Relationship with caveolae and caveolin after initial agonist stimulation. Hypertension 1998, 32, 459–466. [Google Scholar] [CrossRef]
- Venema, V.J.; Ju, H.; Zou, R.; Venema, R.C. Interaction of neuronal nitric-oxide synthase with caveolin-3 in skeletal muscle. Identification of a novel caveolin scaffolding/inhibitory domain. J. Biol. Chem. 1997, 272, 28187–28190. [Google Scholar] [CrossRef]
- Ohguro, H.; Odagiri, H.; Miyagawa, Y.; Ohguro, I.; Sasaki, M.; Nakazawa, M. Clinicopathological features of gastric cancer cases and aberrantly expressed recoverin. Tohoku J. Exp. Med. 2004, 202, 213–219. [Google Scholar] [CrossRef]
- Doerner, A.; Pauschinger, M.; Badorff, A.; Noutsias, M.; Giessen, S.; Schulze, K.; Bilger, J.; Rauch, U.; Schultheiss, H.P. Tissue-specific transcription pattern of the adenine nucleotide translocase isoforms in humans. FEBS Lett. 1997, 414, 258–262. [Google Scholar] [PubMed]
- Stepien, G.; Torroni, A.; Chung, A.B.; Hodge, J.A.; Wallace, D.C. Differential expression of adenine nucleotide translocator isoforms in mammalian tissues and during muscle cell differentiation. J. Biol. Chem. 1992, 267, 14592–14597. [Google Scholar] [CrossRef] [PubMed]
- Graham, B.H.; Waymire, K.G.; Cottrell, B.; Trounce, I.A.; MacGregor, G.R.; Wallace, D.C. A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/muscle isoform of the adenine nucleotide translocator. Nat. Genet. 1997, 16, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.J.; Webb-Wood, S.; Faulkner, A.E.; Jabbar, S.B.; Biousse, V.; Newman, N.J.; Do, V.T.; Boatright, J.H.; Wallace, D.C.; Pardue, M.T. Retinal function and structure in Ant1-deficient mice. Investig. Ophthalmol. Vis. Sci. 2010, 51, 6744–6752. [Google Scholar] [CrossRef] [PubMed]
- Ohguro, H.; Ida, Y.; Hikage, F.; Umetsu, A.; Ichioka, H.; Watanabe, M.; Furuhashi, M. STAT3 Is the Master Regulator for the Forming of 3D Spheroids of 3T3-L1 Preadipocytes. Cells 2022, 11, 300. [Google Scholar] [CrossRef] [PubMed]
- Akama, T.; Leung, B.M.; Labuz, J.; Takayama, S.; Chun, T.H. Designing 3-D Adipospheres for Quantitative Metabolic Study. Methods Mol. Biol. 2017, 1566, 177–183. [Google Scholar]
- Ida, Y.; Hikage, F.; Umetsu, A.; Ida, H.; Ohguro, H. Omidenepag, a non-prostanoid EP2 receptor agonist, induces enlargement of the 3D organoid of 3T3-L1 cells. Sci. Rep. 2020, 10, 16018. [Google Scholar] [CrossRef] [PubMed]
- Itoh, K.; Hikage, F.; Ida, Y.; Ohguro, H. Prostaglandin F2α Agonists Negatively Modulate the Size of 3D Organoids from Primary Human Orbital Fibroblasts. Investig. Ophthalmol. Vis. Sci. 2020, 61, 13. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhou, Y.; Jiang, H.; Chen, Z.; Lu, B. Analysis of core genes for colorectal cancer prognosis based on immune and stromal scores. PeerJ 2021, 9, e12452. [Google Scholar] [CrossRef] [PubMed]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ichioka, H.; Hirohashi, Y.; Sato, T.; Furuhashi, M.; Watanabe, M.; Ida, Y.; Hikage, F.; Torigoe, T.; Ohguro, H. G-Protein-Coupled Receptors Mediate Modulations of Cell Viability and Drug Sensitivity by Aberrantly Expressed Recoverin 3 within A549 Cells. Int. J. Mol. Sci. 2023, 24, 771. https://doi.org/10.3390/ijms24010771
Ichioka H, Hirohashi Y, Sato T, Furuhashi M, Watanabe M, Ida Y, Hikage F, Torigoe T, Ohguro H. G-Protein-Coupled Receptors Mediate Modulations of Cell Viability and Drug Sensitivity by Aberrantly Expressed Recoverin 3 within A549 Cells. International Journal of Molecular Sciences. 2023; 24(1):771. https://doi.org/10.3390/ijms24010771
Chicago/Turabian StyleIchioka, Hanae, Yoshihiko Hirohashi, Tatsuya Sato, Masato Furuhashi, Megumi Watanabe, Yosuke Ida, Fumihito Hikage, Toshihiko Torigoe, and Hiroshi Ohguro. 2023. "G-Protein-Coupled Receptors Mediate Modulations of Cell Viability and Drug Sensitivity by Aberrantly Expressed Recoverin 3 within A549 Cells" International Journal of Molecular Sciences 24, no. 1: 771. https://doi.org/10.3390/ijms24010771
APA StyleIchioka, H., Hirohashi, Y., Sato, T., Furuhashi, M., Watanabe, M., Ida, Y., Hikage, F., Torigoe, T., & Ohguro, H. (2023). G-Protein-Coupled Receptors Mediate Modulations of Cell Viability and Drug Sensitivity by Aberrantly Expressed Recoverin 3 within A549 Cells. International Journal of Molecular Sciences, 24(1), 771. https://doi.org/10.3390/ijms24010771










