E3 Ubiquitin Ligase Midline 1 Regulates Endothelial Cell ICAM-1 Expression and Neutrophil Adhesion in Abdominal Sepsis
Abstract
1. Introduction
2. Results
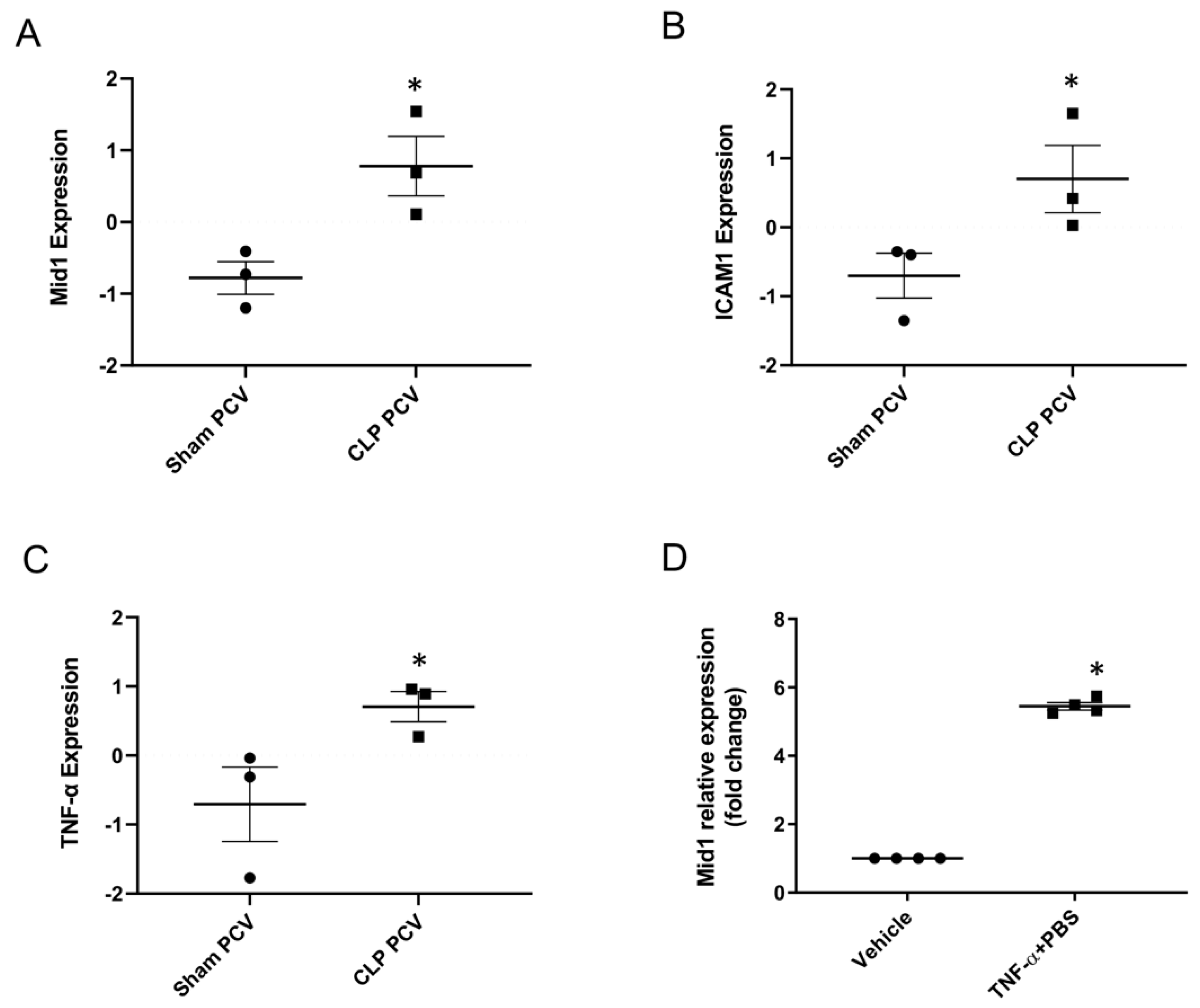
2.1. Mid1 Expression in Lung Post-Capillary Venules of Cecal Ligation Puncture (CLP) Mice and Mouse Endothelial Cell Line
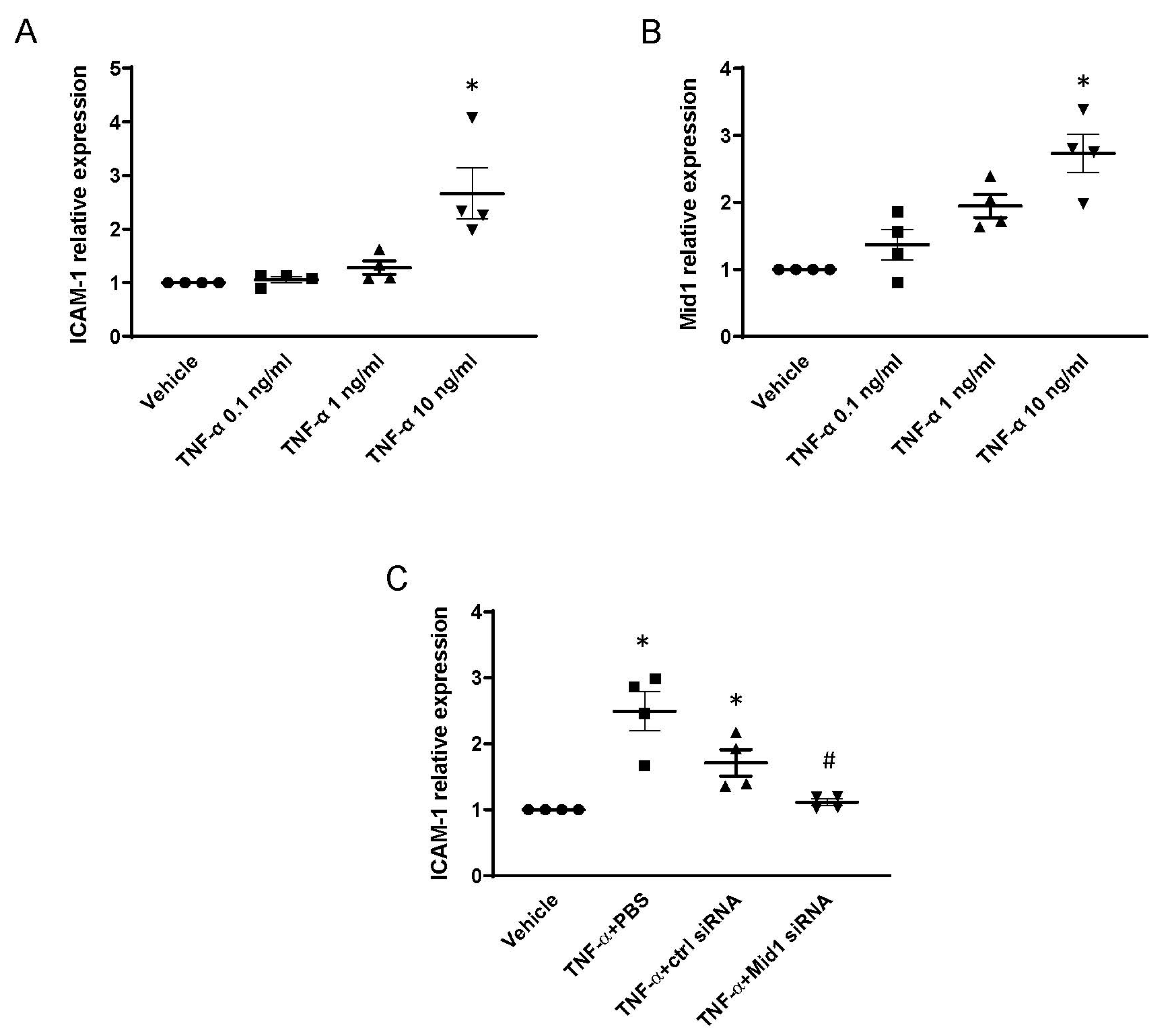
2.2. Mid1 Silencing Reduces ICAM-1 Expression and Neutrophil Adhesion In Vitro
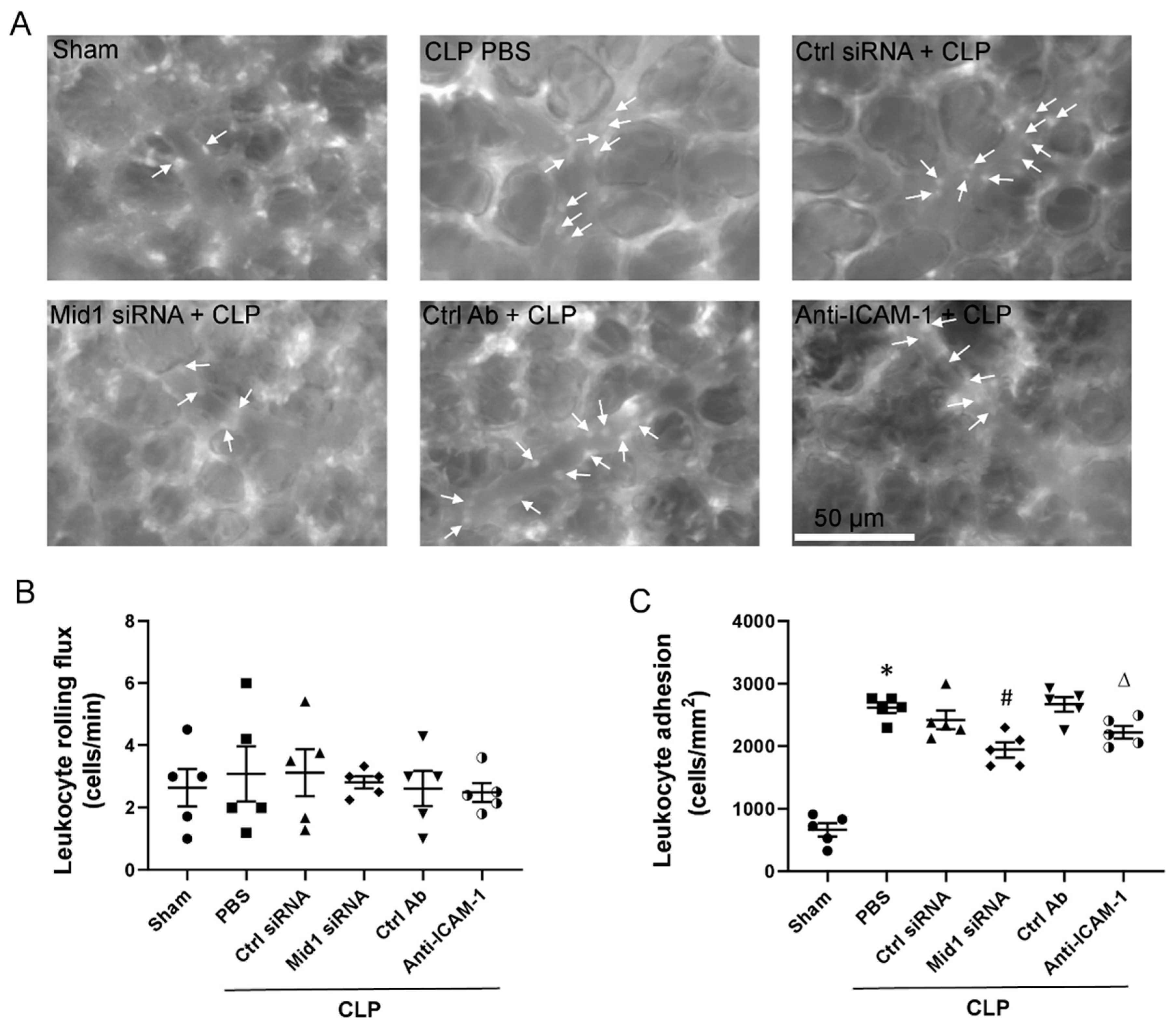
2.3. Mid1 Silencing Reduces Leukocyte Adhesion In Vivo
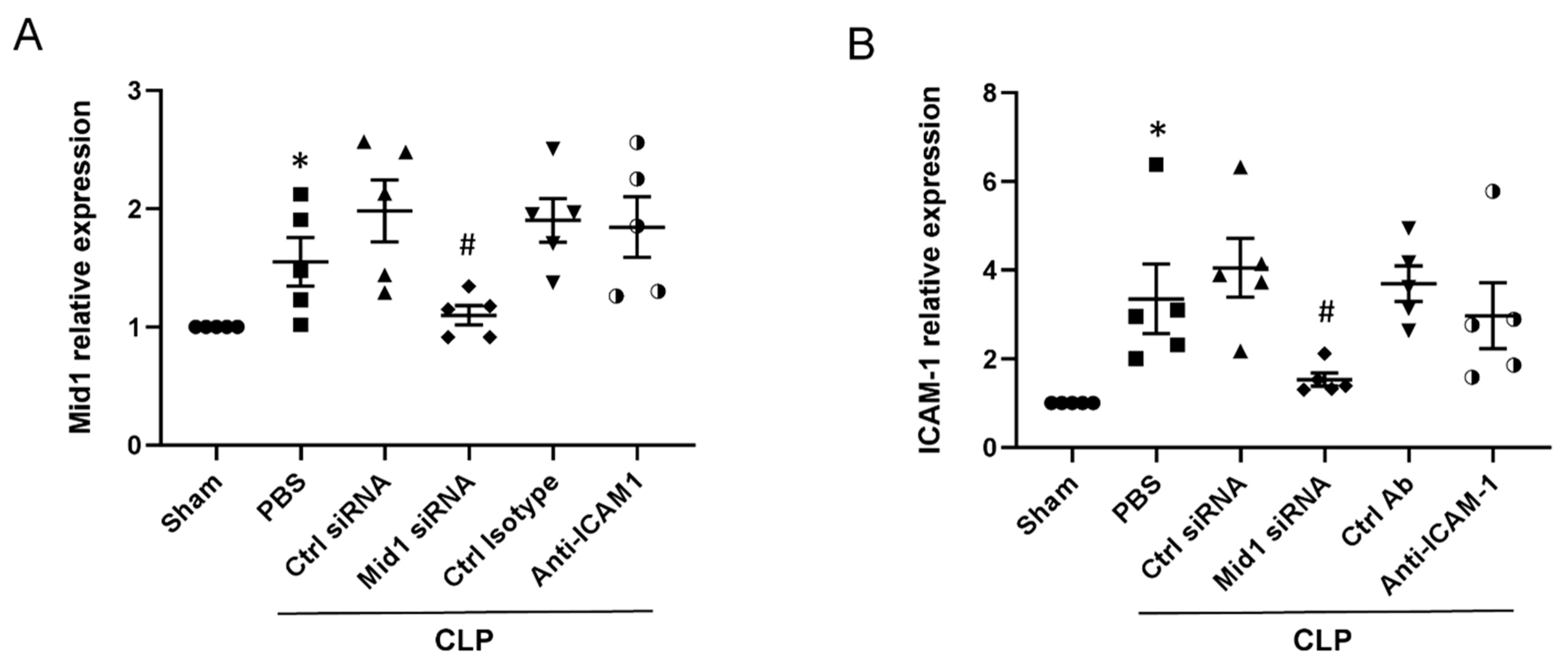
2.4. Mid1 Silencing Reduces ICAM-1 Expression In Vivo
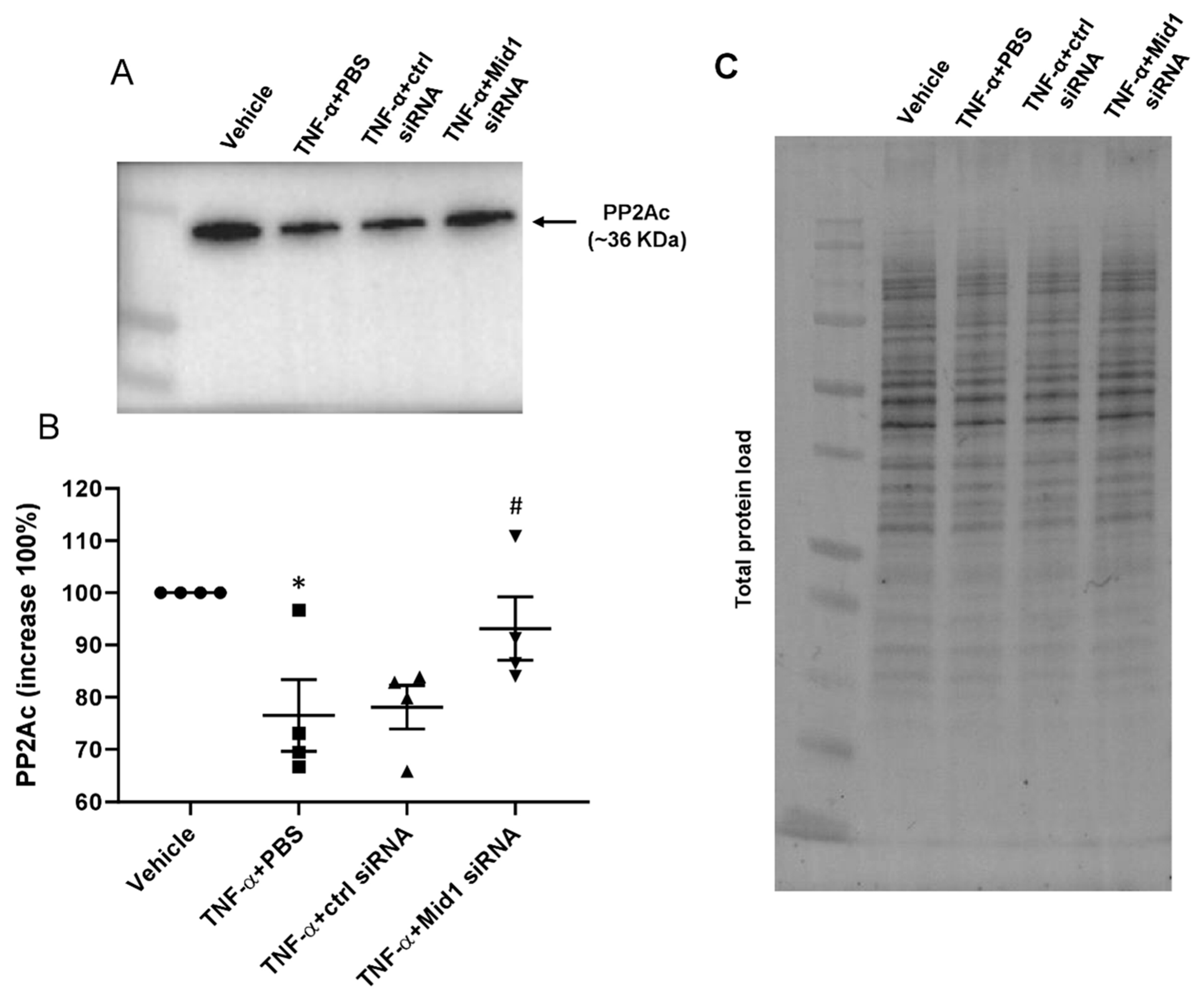
2.5. Mid1 Regulates PP2Ac Expression
2.6. Mid1 Regulates ICAM-1 Expression in Human Lung Microvascular Endothelial Cells (HLMVEC) In Vitro
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Experimental Protocol of Sepsis
4.3. Lung Intravital Fluorescence Microscopy
4.4. RNA Sequence Data Analysis
4.5. Endothelial Cells Activation
4.6. Transfection
4.7. Quantitative Real-Time Polymerase Chain Reaction
4.8. Flow Cytometry
4.9. Adhesion Assay
4.10. Confocal Microscopy
4.11. Western Blot
4.12. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Asaduzzaman, M.; Zhang, S.; Lavasani, S.; Wang, Y.; Thorlacius, H. LFA-1 and MAC-1 mediate pulmonary recruitment of neutrophils and tissue damage in abdominal sepsis. Shock 2008, 30, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Kamochi, M.; Kamochi, F.; Kim, Y.B.; Sawh, S.; Sanders, J.M.; Sarembock, I.; Green, S.; Young, J.S.; Ley, K.; Fu, S.; et al. P-selectin and ICAM-1 mediate endotoxin-induced neutrophil recruitment and injury to the lung and liver. Am. J. Physiol. 1999, 277, L310–L319. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Roller, J.; Slotta, J.E.; Zhang, S.; Luo, L.; Rahman, M.; Syk, I.; Menger, M.D.; Thorlacius, H. Distinct patterns of leukocyte recruitment in the pulmonary microvasculature in response to local and systemic inflammation. Am. J. Physiol. Lung Cell. Mol. Physiol. 2013, 304, L298–L305. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Roller, J.; Menger, M.; Thorlacius, H. Sepsis-induced leukocyte adhesion in the pulmonary microvasculature in vivo is mediated by CD11a and CD11b. Eur. J. Pharmacol. 2013, 702, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Mayadas, T.N.; Johnson, R.C.; Rayburn, H.; Hynes, R.O.; Wagner, D.D. Leukocyte rolling and extravasation are severely compromised in P selectin-deficient mice. Cell 1993, 74, 541–554. [Google Scholar] [CrossRef]
- Patel, K.D.; Cuvelier, S.L.; Wiehler, S. Selectins: Critical mediators of leukocyte recruitment. Semin. Immunol. 2002, 14, 73–81. [Google Scholar] [CrossRef]
- Carlos, T.M.; Harlan, J.M. Leukocyte-endothelial adhesion molecules. Blood 1994, 84, 2068–2101. [Google Scholar] [CrossRef]
- Albelda, S.M.; Smith, C.W.; Ward, P.A. Adhesion molecules and inflammatory injury. FASEB J. 1994, 8, 504–512. [Google Scholar] [CrossRef]
- Schmidt, S.; Moser, M.; Sperandio, M. The molecular basis of leukocyte recruitment and its deficiencies. Mol. Immunol. 2013, 55, 49–58. [Google Scholar] [CrossRef]
- Yan, J.; Nunn, A.D.; Thomas, R. Selective induction of cell adhesion molecules by proinflammatory mediators in human cardiac microvascular endothelial cells in culture. Int. J. Clin. Exp. Med. 2010, 3, 315–331. [Google Scholar]
- Xue, M.; Qiqige, C.; Zhang, Q.; Zhao, H.; Su, L.; Sun, P.; Zhao, P. Effects of Tumor Necrosis Factor alpha (TNF-alpha) and Interleukina 10 (IL-10) on Intercellular Cell Adhesion Molecule-1 (ICAM-1) and Cluster of Differentiation 31 (CD31) in Human Coronary Artery Endothelial Cells. Med. Sci. Monit. 2018, 24, 4433–4439. [Google Scholar] [CrossRef] [PubMed]
- Doerschuk, C.M.; Quinlan, W.M.; Doyle, N.A.; Bullard, D.C.; Vestweber, D.; Jones, M.L.; Takei, F.; Ward, P.A.; Beaudet, A.L. The role of P-selectin and ICAM-1 in acute lung injury as determined using blocking antibodies and mutant mice. J. Immunol. 1996, 157, 4609–4614. [Google Scholar] [PubMed]
- Kumasaka, T.; Quinlan, W.M.; Doyle, N.A.; Condon, T.P.; Sligh, J.; Takei, F.; Beaudet, A.; Bennett, C.F.; Doerschuk, C.M. Role of the intercellular adhesion molecule-1(ICAM-1) in endotoxin-induced pneumonia evaluated using ICAM-1 antisense oligonucleotides, anti-ICAM-1 monoclonal antibodies, and ICAM-1 mutant mice. J. Clin. Investig. 1996, 97, 2362–2369. [Google Scholar] [CrossRef] [PubMed]
- Collison, A.M.; Li, J.; de Siqueira, A.P.; Lv, X.; Toop, H.D.; Morris, J.C.; Starkey, M.R.; Hansbro, P.M.; Zhang, J.; Mattes, J. TRAIL signals through the ubiquitin ligase MID1 to promote pulmonary fibrosis. BMC Pulm. Med. 2019, 19, 31. [Google Scholar] [CrossRef]
- Winter, J.; Basilicata, M.F.; Stemmler, M.P.; Krauss, S. The MID1 protein is a central player during development and in disease. Front. Biosci. (Landmark Ed.) 2016, 21, 664–682. [Google Scholar]
- Demir, U.; Koehler, A.; Schneider, R.; Schweiger, S.; Klocker, H. Metformin anti-tumor effect via disruption of the MID1 translational regulator complex and AR downregulation in prostate cancer cells. BMC Cancer 2014, 14, 52. [Google Scholar] [CrossRef]
- Chen, Q.; Gao, C.; Wang, M.; Fei, X.; Zhao, N. TRIM18-Regulated STAT3 Signaling Pathway via PTP1B Promotes Renal Epithelial-Mesenchymal Transition, Inflammation, and Fibrosis in Diabetic Kidney Disease. Front. Physiol. 2021, 12, 709506. [Google Scholar] [CrossRef]
- Collison, A.; Hatchwell, L.; Verrills, N.; Wark, P.A.; de Siqueira, A.P.; Tooze, M.; Carpenter, H.; Don, A.S.; Morris, J.C.; Zimmermann, N.; et al. The E3 ubiquitin ligase midline 1 promotes allergen and rhinovirus-induced asthma by inhibiting protein phosphatase 2A activity. Nat. Med. 2013, 19, 232–237. [Google Scholar] [CrossRef]
- Pober, J.S. Activation and injury of endothelial cells by cytokines. Pathologie-Biologie 1998, 46, 159–163. [Google Scholar]
- Kempe, S.; Kestler, H.; Lasar, A.; Wirth, T. NF-kappaB controls the global pro-inflammatory response in endothelial cells: Evidence for the regulation of a pro-atherogenic program. Nucleic Acids Res. 2005, 33, 5308–5319. [Google Scholar] [CrossRef]
- Curtis, A.M.; Wilkinson, P.F.; Gui, M.; Gales, T.L.; Hu, E.; Edelberg, J.M. p38 mitogen-activated protein kinase targets the production of proinflammatory endothelial microparticles. J. Thromb. Haemost. 2009, 7, 701–709. [Google Scholar] [CrossRef] [PubMed]
- Uz, Y.H.; Murk, W.; Bozkurt, I.; Kizilay, G.; Arici, A.; Kayisli, U.A. Increased c-Jun N-terminal kinase activation in human endometriotic endothelial cells. Histochem. Cell Biol. 2011, 135, 83–91. [Google Scholar] [CrossRef]
- Grech, G.; Baldacchino, S.; Saliba, C.; Grixti, M.P.; Gauci, R.; Petroni, V.; Fenech, A.G.; Scerri, C. Deregulation of the protein phosphatase 2A, PP2A in cancer: Complexity and therapeutic options. Tumour Biol. 2016, 37, 11691–11700. [Google Scholar] [CrossRef] [PubMed]
- Trockenbacher, A.; Suckow, V.; Foerster, J.; Winter, J.; Krauss, S.; Ropers, H.H.; Schneider, R.; Schweiger, S. MID1, mutated in Opitz syndrome, encodes an ubiquitin ligase that targets phosphatase 2A for degradation. Nat. Genet. 2001, 29, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Granger, D.N.; Kubes, P. The microcirculation and inflammation: Modulation of leukocyte-endothelial cell adhesion. J. Leukoc. Biol. 1994, 55, 662–675. [Google Scholar] [CrossRef]
- Park, I.; Kim, M.; Choe, K.; Song, E.; Seo, H.; Hwang, Y.; Ahn, J.; Lee, S.H.; Lee, J.H.; Jo, Y.H.; et al. Neutrophils disturb pulmonary microcirculation in sepsis-induced acute lung injury. Eur. Respir. J. 2019, 53, 1800786. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xia, Y.; Kuang, D.; Duan, Y.; Wang, G. PP2A regulates SCF-induced cardiac stem cell migration through interaction with p38 MAPK. Life Sci. 2017, 191, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Shanley, T.P.; Vasi, N.; Denenberg, A.; Wong, H.R. The serine/threonine phosphatase, PP2A: Endogenous regulator of inflammatory cell signaling. J. Immunol. 2001, 166, 966–972. [Google Scholar] [CrossRef]
- Zhang, L.; Li, J.; Lv, X.; Guo, T.; Li, W.; Zhang, J. MID1-PP2A complex functions as new insights in human lung adenocarcinoma. J. Cancer Res. Clin. Oncol. 2018, 144, 855–864. [Google Scholar] [CrossRef]
- Monteiro, O.; Chen, C.; Bingham, R.; Argyrou, A.; Buxton, R.; Jonsson, C.P.; Jones, E.; Bridges, A.; Gatfield, K.; Krauss, S.; et al. Pharmacological disruption of the MID1/alpha4 interaction reduces mutant Huntingtin levels in primary neuronal cultures. Neurosci. Lett. 2018, 673, 44–50. [Google Scholar] [CrossRef]
- Mackay, F.; Loetscher, H.; Stueber, D.; Gehr, G.; Lesslauer, W. Tumor necrosis factor alpha (TNF-alpha)-induced cell adhesion to human endothelial cells is under dominant control of one TNF receptor type, TNF-R55. J. Exp. Med. 1993, 177, 1277–1286. [Google Scholar] [CrossRef]
- Chang, Y.L.; Chen, C.L.; Kuo, C.L.; Chen, B.C.; You, J.S. Glycyrrhetinic acid inhibits ICAM-1 expression via blocking JNK and NF-kappaB pathways in TNF-alpha-activated endothelial cells. Acta Pharmacol. Sin. 2010, 31, 546–553. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Connell, M.C.; MacEwan, D.J. TNFR1-induced NF-kappaB, but not ERK, p38MAPK or JNK activation, mediates TNF-induced ICAM-1 and VCAM-1 expression on endothelial cells. Cell Signal 2007, 19, 1238–1248. [Google Scholar] [CrossRef] [PubMed]
- Muller, W.A. Leukocyte-endothelial-cell interactions in leukocyte transmigration and the inflammatory response. Trends Immunol. 2003, 24, 327–334. [Google Scholar] [CrossRef]
- Alon, R.; Feigelson, S. From rolling to arrest on blood vessels: Leukocyte tap dancing on endothelial integrin ligands and chemokines at sub-second contacts. Semin. Immunol. 2002, 14, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Long, E.O. ICAM-1: Getting a grip on leukocyte adhesion. J. Immunol. 2011, 186, 5021–5023. [Google Scholar] [CrossRef] [PubMed]
- Osborn, L. Leukocyte adhesion to endothelium in inflammation. Cell 1990, 62, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Roller, J.; Zhang, S.; Syk, I.; Menger, M.D.; Jeppsson, B.; Thorlacius, H. Metalloproteinases regulate CD40L shedding from platelets and pulmonary recruitment of neutrophils in abdominal sepsis. Inflamm. Res. 2012, 61, 571–579. [Google Scholar] [CrossRef]
- Benjamim, C.F.; Silva, J.S.; Fortes, Z.B.; Oliveira, M.A.; Ferreira, S.H.; Cunha, F.Q. Inhibition of leukocyte rolling by nitric oxide during sepsis leads to reduced migration of active microbicidal neutrophils. Infect. Immun. 2002, 70, 3602–3610. [Google Scholar] [CrossRef]
- Rittirsch, D.; Huber-Lang, M.S.; Flierl, M.A.; Ward, P.A. Immunodesign of experimental sepsis by cecal ligation and puncture. Nat. Protoc. 2009, 4, 31–36. [Google Scholar] [CrossRef]
- Rahman, M.; Ding, Z.; Rönnow, C.-F.; Thorlacius, H. Transcriptomic Analysis Reveals Differential Expression of Genes between Lung Capillary and Post Capillary Venules in Abdominal Sepsis. Int. J. Mol. Sci. 2021, 22, 10181. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, F.; Hawez, A.; Ding, Z.; Wang, Y.; Rönnow, C.-F.; Rahman, M.; Thorlacius, H. E3 Ubiquitin Ligase Midline 1 Regulates Endothelial Cell ICAM-1 Expression and Neutrophil Adhesion in Abdominal Sepsis. Int. J. Mol. Sci. 2023, 24, 705. https://doi.org/10.3390/ijms24010705
Du F, Hawez A, Ding Z, Wang Y, Rönnow C-F, Rahman M, Thorlacius H. E3 Ubiquitin Ligase Midline 1 Regulates Endothelial Cell ICAM-1 Expression and Neutrophil Adhesion in Abdominal Sepsis. International Journal of Molecular Sciences. 2023; 24(1):705. https://doi.org/10.3390/ijms24010705
Chicago/Turabian StyleDu, Feifei, Avin Hawez, Zhiyi Ding, Yongzhi Wang, Carl-Fredrik Rönnow, Milladur Rahman, and Henrik Thorlacius. 2023. "E3 Ubiquitin Ligase Midline 1 Regulates Endothelial Cell ICAM-1 Expression and Neutrophil Adhesion in Abdominal Sepsis" International Journal of Molecular Sciences 24, no. 1: 705. https://doi.org/10.3390/ijms24010705
APA StyleDu, F., Hawez, A., Ding, Z., Wang, Y., Rönnow, C.-F., Rahman, M., & Thorlacius, H. (2023). E3 Ubiquitin Ligase Midline 1 Regulates Endothelial Cell ICAM-1 Expression and Neutrophil Adhesion in Abdominal Sepsis. International Journal of Molecular Sciences, 24(1), 705. https://doi.org/10.3390/ijms24010705






