The Role of Proteolysis in Amyloidosis
Abstract
1. Introduction
1.1. Clinical Features of Amyloid Diseases
1.2. Amyloid Fibrils Composition
2. Proteolysis-Driven Amyloidosis
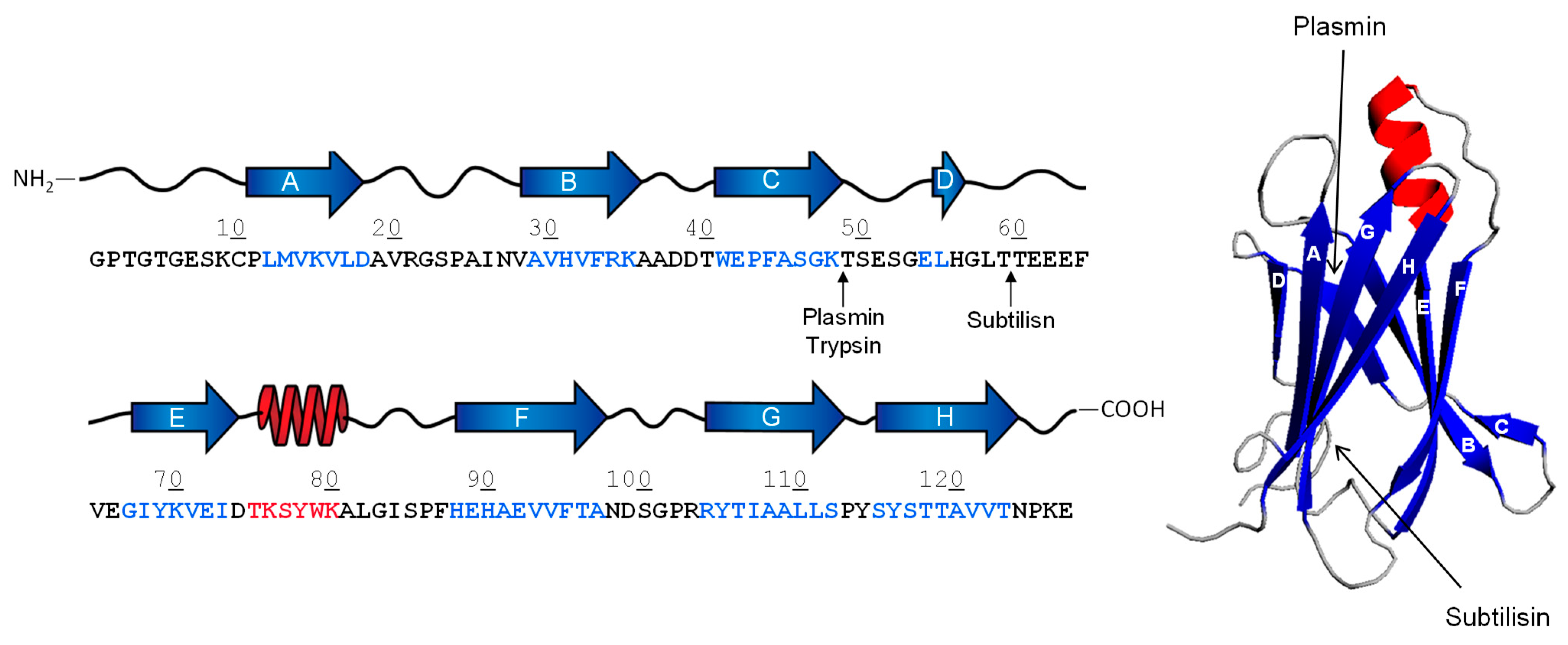
2.1. Proteolysis of Transthyretin
2.2. Proteolysis of β-Amyloid Precursor Protein
2.3. Proteolysis of α-Synuclein
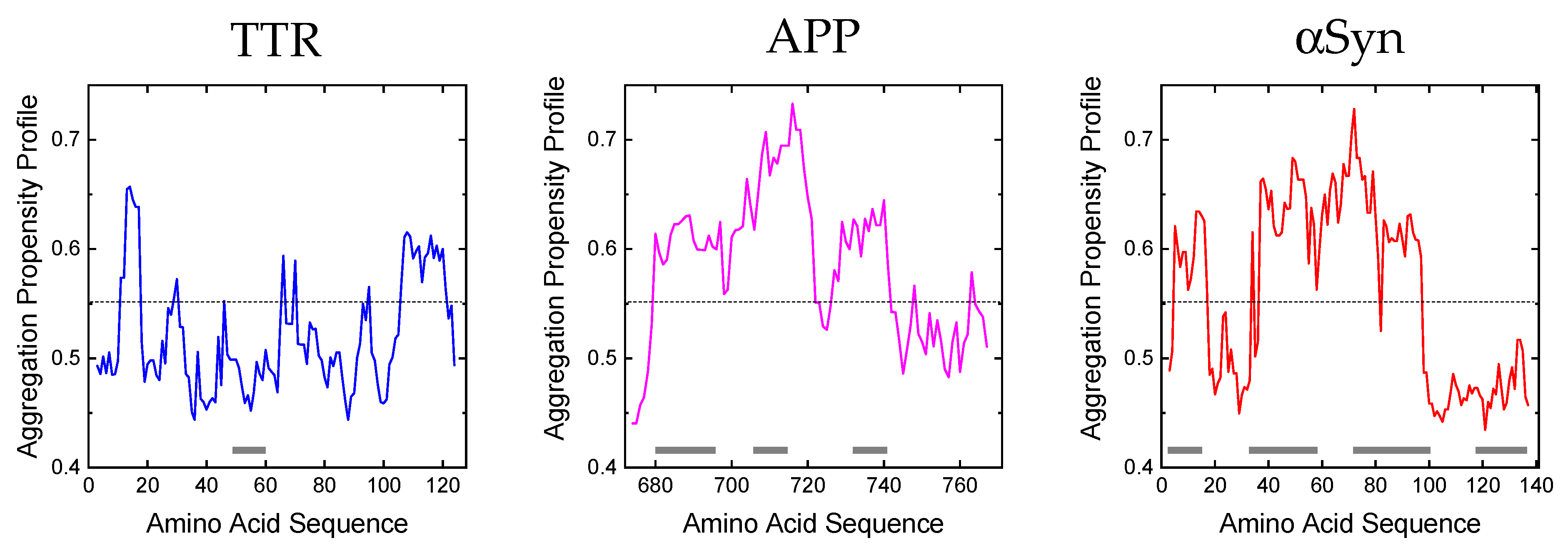
3. Generation and Amyloid-Forming Properties of Proteolytic Fragments
4. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gillmore, J.D.; Hawkins, P.N. Pathophysiology and treatment of systemic amyloidosis. Nat. Rev. Nephrol. 2013, 9, 574–586. [Google Scholar] [CrossRef] [PubMed]
- Merlini, G.; Bellotti, V. Molecular Mechanisms of Amyloidosis. N. Engl. J. Med. 2003, 349, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Functional Amyloid, and Human Disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [PubMed]
- Benson, M.D.; Buxbaum, J.N.; Eisenberg, D.S.; Merlini, G.; Saraiva, M.J.M.; Sekijima, Y.; Sipe, J.D.; Westermark, P. Amyloid nomenclature 2020: Update and recommendations by the International Society of Amyloidosis (ISA) nomenclature committee. Amyloid 2020, 27, 217–222. [Google Scholar] [CrossRef]
- Falk, R.H. Diagnosis and Management of the Cardiac Amyloidoses. Circulation 2005, 112, 2047–2060. [Google Scholar] [CrossRef]
- Tsai, S.B.; Seldin, D.C.; Wu, H.; O’Hara, C.; Ruberg, F.; Sanchorawala, V. Myocardial infarction with “clean coronaries” caused by amyloid light-chain AL amyloidosis: A case report and literature review. Amyloid 2011, 18, 160–164. [Google Scholar] [CrossRef]
- Dember, L.M. Amyloidosis-Associated Kidney Disease. J. Am. Soc. Nephrol. 2006, 17, 3458–3471. [Google Scholar] [CrossRef]
- Wang, A.K.; Fealey, R.D.; Gehrking, T.L.; Low, P.A. Patterns of Neuropathy and Autonomic Failure in Patients with Amyloidosis. Mayo Clin. Proc. 2008, 83, 1226–1230. [Google Scholar] [CrossRef]
- Berk, J.L.; O’Regan, A.; Skinner, M. Pulmonary and Tracheobronchial Amyloidosis. Semin. Respir. Crit. Care Med. 2002, 23, 155–166. [Google Scholar] [CrossRef]
- O’regan, A.; Fenlon, H.M.; Beamis, J.F.; Steele, M.P.; Skinner, M.; Berk, J. Tracheobronchial Amyloidosis: The Boston University Experience from 1984 to 1999. Medicine 2000, 79, 69–79. [Google Scholar] [CrossRef]
- Wechalekar, A.D.; Gillmore, J.D.; Hawkins, P.N. Systemic amyloidosis. Lancet 2016, 387, 2641–2654. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, D.; Jucker, M. The Amyloid State of Proteins in Human Diseases. Cell 2012, 148, 1188–1203. [Google Scholar] [CrossRef]
- Ni, W.; Jagust, W.; Wang, D. Multiplex Mass Spectrometry Analysis of Amyloid Proteins in Human Plasma for Alzheimer’s Disease Diagnosis. J. Proteome Res. 2021, 20, 4106–4112. [Google Scholar] [CrossRef] [PubMed]
- Bleiholder, C.; Dupuis, N.F.; Wyttenbach, T.; Bowers, M.T. Ion mobility-mass spectrometry reveals a conformational conversion from random assembly to β-sheet in amyloid fibril formation. Nat. Chem. 2011, 3, 172–177. [Google Scholar] [CrossRef]
- Bonar, L.; Cohen, A.S.; Skinner, M.M. Characterization of the Amyloid Fibril as a Cross-beta Protein. Proc. Soc. Exp. Biol. Med. 1969, 131, 1373–1375. [Google Scholar] [CrossRef]
- Paravastu, A.K.; Leapman, R.D.; Yau, W.-M.; Tycko, R. Molecular structural basis for polymorphism in Alzheimer’s β-amyloid fibrils. Proc. Natl. Acad. Sci. USA 2008, 105, 18349–18354. [Google Scholar] [CrossRef]
- Wasmer, C.; Lange, A.; Van Melckebeke, H.; Siemer, A.B.; Riek, R.; Meier, B.H. Amyloid Fibrils of the HET-s(218–289) Prion Form a β Solenoid with a Triangular Hydrophobic Core. Science 2008, 319, 1523–1526. [Google Scholar] [CrossRef]
- Nilsson, M.R. Techniques to study amyloid fibril formation in vitro. Methods 2004, 34, 151–160. [Google Scholar] [CrossRef]
- Kushnirov, V.V.; Dergalev, A.A.; Alexandrov, A.I. Proteinase K resistant cores of prions and amyloids. Prion 2020, 14, 11–19. [Google Scholar] [CrossRef]
- Pepys, M.B.; Rademacher, T.W.; Amatayakul-Chantler, S.; Williams, P.; Noble, G.E.; Hutchinson, W.L.; Hawkins, P.N.; Nelson, S.R.; Gallimore, J.R.; Herbert, J. Human serum amyloid P component is an invariant constituent of amyloid deposits and has a uniquely homogeneous glycostructure. Proc. Natl. Acad. Sci. USA 1994, 91, 5602–5606. [Google Scholar] [CrossRef] [PubMed]
- Coker, A.R.; Purvis, A.; Baker, D.; Pepys, M.B.; Wood, S.P. Molecular chaperone properties of serum amyloid P component. FEBS Lett. 2000, 473, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Tennent, G.A.; Lovat, L.B.; Pepys, M.B. Serum amyloid P component prevents proteolysis of the amyloid fibrils of Alzheimer disease and systemic amyloidosis. Proc. Natl. Acad. Sci. USA 1995, 92, 4299–4303. [Google Scholar] [CrossRef] [PubMed]
- Kisilevsky, R.; Kyle, R.A. Proteoglycans, glycosaminoglycans, amyloid-enhancing factor, and amyloid deposition. J. Intern. Med. 1992, 232, 515–516. [Google Scholar] [CrossRef]
- Knowles, T.P.J.; Waudby, C.A.; Devlin, G.L.; Cohen, S.I.A.; Aguzzi, A.; Vendruscolo, M.; Terentjev, E.M.; Welland, M.E.; Dobson, C.M. An Analytical Solution to the Kinetics of Breakable Filament Assembly. Science 2009, 326, 1533–1537. [Google Scholar] [CrossRef] [PubMed]
- Arosio, P.; Knowles, T.P.J.; Linse, S. On the lag phase in amyloid fibril formation. Phys. Chem. Chem. Phys. 2015, 17, 7606–7618. [Google Scholar] [CrossRef]
- Morris, A.M.; Watzky, M.A.; Finke, R.G. Protein aggregation kinetics, mechanism, and curve-fitting: A review of the literature. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2009, 1794, 375–397. [Google Scholar] [CrossRef] [PubMed]
- Bucciantini, M.; Giannoni, E.; Chiti, F.; Baroni, F.; Formigli, L.; Zurdo, J.; Taddei, N.; Ramponi, G.; Dobson, C.M.; Stefani, M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 2002, 416, 507–511. [Google Scholar] [CrossRef]
- Okamoto, A.; Hosoda, N.; Hoshino, S.-I. Proteolysis: A double-edged sword for the development of amyloidoses. Prion 2018, 12, 273–279. [Google Scholar] [CrossRef]
- Marcoux, J.; Mangione, P.P.; Porcari, R.; Degiacomi, M.; Verona, G.; Taylor, G.W.; Giorgetti, S.; Raimondi, S.; Cianferani, S.; Benesch, J.; et al. A novel mechano-enzymatic cleavage mechanism underlies transthyretin amyloidogenesis. EMBO Mol. Med. 2015, 7, 1337–1349. [Google Scholar] [CrossRef]
- Enqvist, S.; Sletten, K.; Westermark, P. Fibril protein fragmentation pattern in systemic AL-amyloidosis. J. Pathol. 2009, 219, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Höhn, A.; Tramutola, A.; Cascella, R. Proteostasis Failure in Neurodegenerative Diseases: Focus on Oxidative Stress. Oxidative Med. Cell. Longev. 2020, 2020, 5497046. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, G.; Richardson, S.J. The Evolution of Gene Expression, Structure and Function of Transthyretin. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 1997, 116, 137–160. [Google Scholar] [CrossRef]
- Foss, T.R.; Wiseman, A.R.L.; Kelly, J.W. The Pathway by Which the Tetrameric Protein Transthyretin Dissociates. Biochemistry 2005, 44, 15525–15533. [Google Scholar] [CrossRef]
- Blake, C.; Swan, I.; Rerat, C.; Berthou, J.; Laurent, A.; Rerat, B. An X-ray study of the subunit structure of prealbumin. J. Mol. Biol. 1971, 61, 217–224. [Google Scholar] [CrossRef]
- Hamilton, J.A.; Benson, M.D. Transthyretin: A review from a structural perspective. Cell Mol. Life Sci. 2001, 58, 1491–1521. [Google Scholar] [CrossRef]
- Schmidt, H.H.; Waddington-Cruz, M.; Botteman, M.F.; Carter, J.; Chopra, A.S.; Hopps, M.; Stewart, M.; Fallet, S.; Amass, L. Estimating the global prevalence of transthyretin familial amyloid polyneuropathy. Muscle Nerve 2018, 57, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.; Koike, H.; Slama, M.; Coelho, T. Hereditary transthyretin amyloidosis: A model of medical progress for a fatal disease. Nat. Rev. Neurol. 2019, 15, 387–404. [Google Scholar] [CrossRef]
- Mangione, P.P.; Porcari, R.; Gillmore, J.D.; Pucci, P.; Monti, M.; Porcari, M.; Giorgetti, S.; Marchese, L.; Raimondi, S.; Serpell, L.C.; et al. Proteolytic cleavage of Ser52Pro variant transthyretin triggers its amyloid fibrillogenesis. Proc. Natl. Acad. Sci. USA 2014, 111, 1539–1544. [Google Scholar] [CrossRef]
- Sipe, J.D.; Cohen, A.S. Amyloidosis. Crit. Rev. Clin. Lab. Sci. 1994, 31, 325–354. [Google Scholar] [CrossRef]
- Jiang, X.; Buxbaum, J.N.; Kelly, J.W. The V122I cardiomyopathy variant of transthyretin increases the velocity of rate-limiting tetramer dissociation, resulting in accelerated amyloidosis. Proc. Natl. Acad. Sci. USA 2001, 98, 14943–14948. [Google Scholar] [CrossRef]
- Hanson, J.L.; Arvanitis, M.; Koch, C.M.; Berk, J.L.; Ruberg, F.L.; Prokaeva, T.; Connors, L.H. Use of Serum Transthyretin as a Prognostic Indicator and Predictor of Outcome in Cardiac Amyloid Disease Associated With Wild-Type Transthyretin. Circ. Heart Fail. 2018, 11, e004000. [Google Scholar] [CrossRef] [PubMed]
- Saelices, L.; Johnson, L.M.; Liang, W.Y.; Sawaya, M.R.; Cascio, D.; Ruchala, P.; Whitelegge, J.; Jiang, L.; Riek, R.; Eisenberg, D.S. Uncovering the Mechanism of Aggregation of Human Transthyretin. J. Biol. Chem. 2015, 290, 28932–28943. [Google Scholar] [CrossRef] [PubMed]
- Ihse, E.; Rapezzi, C.; Merlini, G.; Benson, M.D.; Ando, Y.; Suhr, O.B.; Ikeda, S.-I.; Lavatelli, F.; Obici, L.; Quarta, C.C.; et al. Amyloid fibrils containing fragmented ATTR may be the standard fibril composition in ATTR amyloidosis. Amyloid 2013, 20, 142–150. [Google Scholar] [CrossRef]
- Bergström, J.; Gustavsson, A.; Hellman, U.; Sletten, K.; Murphy, C.L.; Weiss, D.T.; Solomon, A.; Olofsson, B.-O.; Westermark, P. Amyloid deposits in transthyretin-derived amyloidosis: Cleaved transthyretin is associated with distinct amyloid morphology. J. Pathol. 2005, 206, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Liepnieks, J.J.; Wilson, D.L.; Benson, M.D. Biochemical characterization of vitreous and cardiac amyloid in Ile84Ser transthyretin amyloidosis. Amyloid 2006, 13, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Wiese, S.; Adak, V.; Engler, J.; Agarwal, S.; Fritz, G.; Westermark, P.; Zacharias, M.; Fändrich, M. Cryo-EM structure of a transthyretin-derived amyloid fibril from a patient with hereditary ATTR amyloidosis. Nat. Commun. 2019, 10, 5008. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, S.; Ihse, E.; Henein, M.Y.; Westermark, P.; Lindqvist, P.; Suhr, O.B. Amyloid Fibril Composition as a Predictor of Development of Cardiomyopathy After Liver Transplantation for Hereditary Transthyretin Amyloidosis. Transplantation 2012, 93, 1017–1023. [Google Scholar] [CrossRef]
- Mangione, P.P.; Verona, G.; Corazza, A.; Marcoux, J.; Canetti, D.; Giorgetti, S.; Raimondi, S.; Stoppini, M.; Esposito, M.; Relini, A.; et al. Plasminogen activation triggers transthyretin amyloidogenesis in vitro. J. Biol. Chem. 2018, 293, 14192–14199. [Google Scholar] [CrossRef]
- Thylén, C.; Wahlqvist, J.; Haettner, E.; Sandgren, O.; Holmgren, G.; Lundgren, E. Modifications of transthyretin in amyloid fibrils: Analysis of amyloid from homozygous and heterozygous individuals with the Met30 mutation. EMBO J. 1993, 12, 743–748. [Google Scholar] [CrossRef]
- Hermansen, L.F.; Bergman, T.; Jornvall, H.; Husby, G.; Ranlov, I.; Sletten, K. Purification and Characterization of Amyloid-Related Transthyretin Associated with Familial Amyloidotic Cardiomyopathy. Eur. J. Biochem. 1995, 227, 772–779. [Google Scholar] [CrossRef] [PubMed]
- Pontarollo, G.; Acquasaliente, L.; Peterle, D.; Frasson, R.; Artusi, I.; De Filippis, V. Non-canonical proteolytic activation of human prothrombin by subtilisin from Bacillus subtilis may shift the procoagulant-anticoagulant equilibrium toward thrombosis. J. Biol. Chem. 2017, 292, 15161–15179. [Google Scholar] [CrossRef]
- Peterle, D.; Pontarollo, G.; Spada, S.; Brun, P.; Palazzi, L.; Sokolov, A.V.; Spolaore, B.; de Laureto, P.P.; Vasilyev, V.B.; Castagliuolo, I.; et al. A serine protease secreted from Bacillus subtilis cleaves human plasma transthyretin to generate an amyloidogenic fragment. Commun. Biol. 2020, 3, 764. [Google Scholar] [CrossRef] [PubMed]
- Claesson, M.J.; Cusack, S.; O’Sullivan, O.; Greene-Diniz, R.; De Weerd, H.; Flannery, E.; Marchesi, J.R.; Falush, D.; Dinan, T.G.; Fitzgerald, G.F.; et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4586–4591. [Google Scholar] [CrossRef] [PubMed]
- Branca, J.J.; Gulisano, M.; Nicoletti, C. Intestinal epithelial barrier functions in ageing. Ageing Res. Rev. 2019, 54, 100938. [Google Scholar] [CrossRef]
- Da Costa, G.; Ribeiro-Silva, C.; Ribeiro, R.; Gilberto, S.; Gomes, R.A.; Ferreira, A.; Mateus, É.; Barroso, E.; Coelho, A.V.; Freire, A.P.; et al. Transthyretin Amyloidosis: Chaperone Concentration Changes and Increased Proteolysis in the Pathway to Disease. PLoS ONE 2015, 10, e0125392. [Google Scholar] [CrossRef]
- Lake-Bakaar, G.; Rubio, C.E.; McKavanagh, S.; Potter, B.J.; Summerfield, J.A. Metabolism of 125I-labelled trypsin in man: Evidence of recirculation. Gut 1980, 21, 580–586. [Google Scholar] [CrossRef][Green Version]
- Midtvedt, T.; Zabarovsky, E.; Norin, E.; Bark, J.; Gizatullin, R.; Kashuba, V.; Ljungqvist, O.; Zabarovska, V.; Möllby, R.; Ernberg, I. Increase of Faecal Tryptic Activity Relates to Changes in the Intestinal Microbiome: Analysis of Crohn’s Disease with a Multidisciplinary Platform. PLoS ONE 2013, 8, e66074. [Google Scholar] [CrossRef]
- Constantinescu, P.; Brown, R.A.; Wyatt, A.R.; Ranson, M.; Wilson, M.R. Amorphous protein aggregates stimulate plasminogen activation, leading to release of cytotoxic fragments that are clients for extracellular chaperones. J. Biol. Chem. 2017, 292, 14425–14437. [Google Scholar] [CrossRef]
- Tucker, H.M.; Kihiko-Ehmann, M.; Wright, S.; Rydel, R.E.; Estus, S. Tissue Plasminogen Activator Requires Plasminogen to Modulate Amyloid-β Neurotoxicity and Deposition. J. Neurochem. 2000, 75, 2172–2177. [Google Scholar] [CrossRef]
- Mumford, A.D.; O’Donnell, J.; Gillmore, J.D.; Manning, R.A.; Hawkins, P.N.; Laffan, M. Bleeding symptoms and coagulation abnormalities in 337 patients with AL-amyloidosis. Br. J. Haematol. 2000, 110, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Bouma, B.; Maas, C.; Hazenberg, B.P.C.; Lokhorst, H.M.; Gebbink, M.F.B.G. Increased plasmin-α2-antiplasmin levels indicate activation of the fibrinolytic system in systemic amyloidoses. J. Thromb. Haemost. 2007, 5, 1139–1142. [Google Scholar] [CrossRef] [PubMed]
- Uchiba, M.; Imamura, T.; Hata, H.; Tatetsu, H.; Yonemura, Y.; Ueda, M.; Wada, Y.; Mitsuya, H.; Ando, Y. Excessive fibrinolysis in AL-amyloidosis is induced by urokinae-type plasminogen activator from bone marrow plasma cells. Amyloid 2009, 16, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Priller, C.; Bauer, T.; Mitteregger, G.; Krebs, B.; Kretzschmar, H.A.; Herms, J. Synapse Formation and Function Is Modulated by the Amyloid Precursor Protein. J. Neurosci. 2006, 26, 7212–7221. [Google Scholar] [CrossRef] [PubMed]
- Turner, P.R.; O’Connor, K.; Tate, W.P.; Abraham, W.C. Roles of amyloid precursor protein and its fragments in regulating neural activity, plasticity and memory. Prog. Neurobiol. 2003, 70, 1–32. [Google Scholar] [CrossRef]
- Duce, J.A.; Tsatsanis, A.; Cater, M.A.; James, S.A.; Robb, E.; Wikhe, K.; Leong, S.L.; Perez, K.; Johanssen, T.; Greenough, M.A.; et al. Iron-Export Ferroxidase Activity of β-Amyloid Precursor Protein Is Inhibited by Zinc in Alzheimer’s Disease. Cell 2010, 142, 857–867. [Google Scholar] [CrossRef]
- Bayer, T.A.; Cappai, R.; Masters, C.L.; Beyreuther, K.; Multhaup, G. It all sticks together—The APP-related family of proteins and Alzheimer’s disease. Mol. Psychiatry 1999, 4, 524–528. [Google Scholar] [CrossRef]
- Guerreiro, R.J.; Gustafson, D.R.; Hardy, J. The genetic architecture of Alzheimer’s disease: Beyond APP, PSENs and APOE. Neurobiol. Aging 2012, 33, 437–456. [Google Scholar] [CrossRef]
- Kayed, R.; Head, E.; Thompson, J.L.; McIntire, T.M.; Milton, S.C.; Cotman, C.W.; Glabe, C.G. Common Structure of Soluble Amyloid Oligomers Implies Common Mechanism of Pathogenesis. Science 2003, 300, 486–489. [Google Scholar] [CrossRef]
- Iwatsubo, T. The γ-secretase complex: Machinery for intramembrane proteolysis. Curr. Opin. Neurobiol. 2004, 14, 379–383. [Google Scholar] [CrossRef]
- Joshi, G.; Wang, Y. Golgi defects enhance APP amyloidogenic processing in Alzheimer’s disease. BioEssays 2015, 37, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Olsson, F.; Schmidt, S.; Althoff, V.; Munter, L.; Jin, S.; Rosqvist, S.; Lendahl, U.; Multhaup, G.; Lundkvist, J. Characterization of Intermediate Steps in Amyloid Beta (Aβ) Production under Near-native Conditions. J. Biol. Chem. 2014, 289, 1540–1550. [Google Scholar] [CrossRef] [PubMed]
- Takami, M.; Nagashima, Y.; Sano, Y.; Ishihara, S.; Morishima-Kawashima, M.; Funamoto, S.; Ihara, Y. Gamma-Secretase: Successive Tripeptide and Tetrapeptide Release from the Transmembrane Domain of beta-Carboxyl Terminal Fragment. J. Neurosci. 2009, 29, 13042–13052. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.C.; Rabizadeh, S.; Chandra, S.; Shayya, R.F.; Ellerby, L.M.; Ye, X.; Salvesen, G.S.; Koo, E.H.; Bredesen, D.E. A second cytotoxic proteolytic peptide derived from amyloid β-protein precursor. Nat. Med. 2000, 6, 397–404. [Google Scholar] [CrossRef]
- Zhang, H.; Ma, Q.; Zhang, Y.-W.; Xu, H. Proteolytic processing of Alzheimer’s β-amyloid precursor protein. J. Neurochem. 2012, 120 (Suppl. S1), 9–21. [Google Scholar] [CrossRef]
- El-Agnaf, O.M.; Mahil, D.S.; Patel, B.P.; Austen, B.M. Oligomerization and Toxicity of β-Amyloid-42 Implicated in Alzheimer’s Disease. Biochem. Biophys. Res. Commun. 2000, 273, 1003–1007. [Google Scholar] [CrossRef]
- Hébert, S.S.; Serneels, L.; Dejaegere, T.; Horré, K.; Dabrowski, M.; Baert, V.; Annaert, W.; Hartmann, D.; De Strooper, B. Coordinated and widespread expression of γ-secretase in vivo: Evidence for size and molecular heterogeneity. Neurobiol. Dis. 2004, 17, 260–272. [Google Scholar] [CrossRef]
- Saito, S.; Araki, W. Expression profiles of two human APH-1 genes and their roles in formation of presenilin complexes. Biochem. Biophys. Res. Commun. 2005, 327, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Esler, W.P.; Kimberly, W.T.; Ostaszewski, B.L.; Ye, W.; Diehl, T.S.; Selkoe, D.J.; Wolfe, M.S. Activity-dependent isolation of the presenilin–γ-secretase complex reveals nicastrin and a γ-secretase substrate. Proc. Natl. Acad. Sci. USA 2002, 99, 2720–2725. [Google Scholar] [CrossRef]
- Li, Y.-M.; Xu, M.; Lai, M.-T.; Huang, Q.; Castro, J.L.; DiMuzio-Mower, J.; Harrison, T.; Lellis, C.; Nadin, A.; Neduvelil, J.G.; et al. Photoactivated γ-secretase inhibitors directed to the active site covalently label presenilin 1. Nature 2000, 405, 689–694. [Google Scholar] [CrossRef]
- Esler, W.P.; Kimberly, W.T.; Ostaszewski, B.L.; Diehl, T.S.; Moore, C.L.; Tsai, J.-Y.; Rahmati, T.; Xia, W.; Selkoe, D.J.; Wolfe, M.S. Transition-state analogue inhibitors of γ-secretase bind directly to presenilin-1. Nature 2000, 2, 428–434. [Google Scholar] [CrossRef] [PubMed]
- Ertekin-Taner, N. Genetics of Alzheimer’s Disease: A Centennial Review. Neurol. Clin. 2007, 25, 611–667. [Google Scholar] [CrossRef] [PubMed]
- Mirza, Z.; Pillai, V.; Kamal, M. Protein Interactions Between the C-Terminus of Aβ-Peptide and Phospholipase A2—A Structure Biology Based Approach to Identify Novel Alzheimer’s Therapeutics. CNS Neurol. Disord.—Drug Targets 2014, 13, 1224–1231. [Google Scholar] [CrossRef]
- Goedert, M. Alpha-synuclein and neurodegenerative diseases. Nat. Rev. Neurosci. 2001, 2, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Lautenschläger, J.; Kaminski, C.F.; Schierle, G.S.K. α-Synuclein—Regulator of Exocytosis, Endocytosis, or Both? Trends Cell Biol. 2017, 27, 468–479. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Li, J.; Souillac, P.; Millett, I.S.; Doniach, S.; Jakes, R.; Goedert, M.; Fink, A.L. Biophysical Properties of the Synucleins and Their Propensities to Fibrillate: Inhibition of alpha-synuclein assembly by beta- and gamma-synucleins. J. Biol. Chem. 2002, 277, 11970–11978. [Google Scholar] [CrossRef] [PubMed]
- Theillet, F.-X.; Binolfi, A.; Bekei, B.; Martorana, A.; Rose, H.M.; Stuiver, M.; Verzini, S.; Lorenz, D.; van Rossum, M.; Goldfarb, D.; et al. Structural disorder of monomeric α-synuclein persists in mammalian cells. Nature 2016, 530, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Mollenhauer, B.; Locascio, J.J.; Schulz-Schaeffer, W.; Sixel-Döring, F.; Trenkwalder, C.; Schlossmacher, M.G. α-Synuclein and tau concentrations in cerebrospinal fluid of patients presenting with parkinsonism: A cohort study. Lancet Neurol. 2011, 10, 230–240. [Google Scholar] [CrossRef]
- El-Agnaf, O.M.A.; Salem, S.A.; Paleologou, K.E.; Curran, M.D.; Gibson, M.J.; Court, J.A.; Schlossmacher, M.G.; Allsop, D. Detection of oligomeric forms of α-synuclein protein in human plasma as a potential biomarker for Parkinson’s disease. FASEB J. 2006, 20, 419–425. [Google Scholar] [CrossRef]
- Acquasaliente, L.; Pontarollo, G.; Radu, C.M.; Peterle, D.; Artusi, I.; Pagotto, A.; Uliana, F.; Negro, A.; Simioni, P.; De Filippis, V. Exogenous human α-Synuclein acts in vitro as a mild platelet antiaggregant inhibiting α-thrombin-induced platelet activation. Sci. Rep. 2022, 12, 9880. [Google Scholar] [CrossRef]
- Davidson, W.S.; Jonas, A.; Clayton, D.F.; George, J.M. Stabilization of α-Synuclein Secondary Structure upon Binding to Synthetic Membranes. J. Biol. Chem. 1998, 273, 9443–9449. [Google Scholar] [CrossRef]
- Dettmer, U.; Newman, A.J.; von Saucken, V.; Bartels, T.; Selkoe, D. KTKEGV repeat motifs are key mediators of normal α-synuclein tetramerization: Their mutation causes excess monomers and neurotoxicity. Proc. Natl. Acad. Sci. USA 2015, 112, 9596–9601. [Google Scholar] [CrossRef]
- Qin, Z.; Hu, D.; Han, S.; Hong, D.-P.; Fink, A.L. Role of Different Regions of α-Synuclein in the Assembly of Fibrils. Biochemistry 2007, 46, 13322–13330. [Google Scholar] [CrossRef] [PubMed]
- Eliezer, D.; Kutluay, E.; Bussell, R., Jr.; Browne, G. Conformational properties of α-synuclein in its free and lipid-associated states. J. Mol. Biol. 2001, 307, 1061–1073. [Google Scholar] [CrossRef] [PubMed]
- Ueda, K.; Fukushima, H.; Masliah, E.; Xia, Y.; Iwai, A.; Yoshimoto, M.; Otero, D.A.; Kondo, J.; Ihara, Y.; Saitoh, T. Molecular cloning of cDNA encoding an unrecognized component of amyloid in Alzheimer disease. Proc. Natl. Acad. Sci. USA 1993, 90, 11282–11286. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Weinreb, P.; Lansbury, P.T. The core Alzheimer’s peptide NAC forms amyloid fibrils which seed and are seeded by β-amyloid: Is NAC a common trigger or target in neurodegenerative disease? Chem. Biol. 1995, 2, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Iwai, A.; Masliah, E.; Yoshimoto, M.; Ge, N.; Flanagan, L.; de Silva, H.R.; Kittel, A.; Saitoh, T. The precursor protein of non-Aβ component of Alzheimer’s disease amyloid is a presynaptic protein of the central nervous system. Neuron 1995, 14, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Wood, S.J.; Wypych, J.; Steavenson, S.; Louis, J.-C.; Citron, M.; Biere, A.L. α-Synuclein Fibrillogenesis Is Nucleation-dependent. Implications for the pathogenesis of Parkinson’s disease. J. Biol. Chem. 1999, 274, 19509–19512. [Google Scholar] [CrossRef]
- Li, B.; Ge, P.; Murray, K.A.; Sheth, P.; Zhang, M.; Nair, G.; Sawaya, M.R.; Shin, W.S.; Boyer, D.R.; Ye, S.; et al. Cryo-EM of full-length α-synuclein reveals fibril polymorphs with a common structural kernel. Nat. Commun. 2018, 9, 3609. [Google Scholar] [CrossRef]
- Recchia, A.; Debetto, P.; Negro, A.; Guidolin, D.; Skaper, S.D.; Giusti, P. α-Synuclein and Parkinson’s disease. FASEB J. 2004, 18, 617–626. [Google Scholar] [CrossRef]
- Lashuel, H.A.; Overk, C.R.; Oueslati, A.; Masliah, E. The many faces of α-synuclein: From structure and toxicity to therapeutic target. Nat. Rev. Neurosci. 2013, 14, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, Z.A.; Giasson, B.I. The emerging role of α-synuclein truncation in aggregation and disease. J. Biol. Chem. 2020, 295, 10224–10244. [Google Scholar] [CrossRef]
- Schmidt, M.F.; Gan, Z.Y.; Komander, D.; Dewson, G. Ubiquitin signalling in neurodegeneration: Mechanisms and therapeutic opportunities. Cell Death Differ. 2021, 28, 570–590. [Google Scholar] [CrossRef] [PubMed]
- Tayebi, N.; Lopez, G.; Do, J.; Sidransky, E. Pro-cathepsin D, Prosaposin, and Progranulin: Lysosomal Networks in Parkinsonism. Trends Mol. Med. 2020, 26, 913–923. [Google Scholar] [CrossRef]
- Kim, K.S.; Choi, Y.R.; Park, J.-Y.; Lee, J.-H.; Kim, D.K.; Lee, S.-J.; Paik, S.R.; Jou, I.; Park, S.M. Proteolytic Cleavage of Extracellular α-Synuclein by Plasmin: Implications for Parkinson disease. J. Biol. Chem. 2012, 287, 24862–24872. [Google Scholar] [CrossRef] [PubMed]
- Iwata, A.; Maruyama, M.; Akagi, T.; Hashikawa, T.; Kanazawa, I.; Tsuji, S.; Nukina, N. Alpha-synuclein degradation by serine protease neurosin: Implication for pathogenesis of synucleinopathies. Hum. Mol. Genet. 2003, 12, 2625–2635. [Google Scholar] [CrossRef]
- Choi, D.-H.; Kim, Y.-J.; Kim, Y.-G.; Joh, T.H.; Beal, M.F.; Kim, Y.-S. Role of Matrix Metalloproteinase 3-mediated α-Synuclein Cleavage in Dopaminergic Cell Death. J. Biol. Chem. 2011, 286, 14168–14177. [Google Scholar] [CrossRef]
- Sung, J.Y.; Park, S.M.; Lee, C.-H.; Um, J.W.; Lee, H.J.; Kim, J.; Oh, Y.J.; Lee, S.-T.; Paik, S.R.; Chung, K.C. Proteolytic Cleavage of Extracellular Secreted α-Synuclein via Matrix Metalloproteinases. J. Biol. Chem. 2005, 280, 25216–25224. [Google Scholar] [CrossRef]
- Mishizen-Eberz, A.J.; Guttmann, R.P.; Giasson, B.I.; Day, G.A., 3rd; Hodara, R.; Ischiropoulos, H.; Lee, V.M.-Y.; Trojanowski, J.Q.; Lynch, D.R. Distinct cleavage patterns of normal and pathologic forms of α-synuclein by calpain I in vitro. J. Neurochem. 2003, 86, 836–847. [Google Scholar] [CrossRef]
- Shinkai-Ouchi, F.; Koyama, S.; Ono, Y.; Hata, S.; Ojima, K.; Shindo, M.; Duverle, D.; Ueno, M.; Kitamura, F.; Doi, N.; et al. Predictions of Cleavability of Calpain Proteolysis by Quantitative Structure-Activity Relationship Analysis Using Newly Determined Cleavage Sites and Catalytic Efficiencies of an Oligopeptide Array. Mol. Cell. Proteom. 2016, 15, 1262–1280. [Google Scholar] [CrossRef]
- Takahashi, M.; Ko, L.-W.; Kulathingal, J.; Jiang, P.; Sevlever, D.; Yen, S.-H.C. Oxidative stress-induced phosphorylation, degradation and aggregation of α-synuclein are linked to upregulated CK2 and cathepsin D. Eur. J. Neurosci. 2007, 26, 863–874. [Google Scholar] [CrossRef]
- McGlinchey, R.P.; Lee, J.C. Cysteine cathepsins are essential in lysosomal degradation of α-synuclein. Proc. Natl. Acad. Sci. USA 2015, 112, 9322–9327. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sheng, R.; Qin, Z. The lysosome and neurodegenerative diseases. Acta Biochim. Biophys. Sin. 2009, 41, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Calamai, M.; Taddei, N.; Stefani, M.; Ramponi, G.; Dobson, C.M. Studies of the aggregation of mutant proteins in vitro provide insights into the genetics of amyloid diseases. Proc. Natl. Acad. Sci. USA 2002, 99 (Suppl. S4), 16419–16426. [Google Scholar] [CrossRef] [PubMed]
- de la Paz, M.L.; Goldie, K.; Zurdo, J.; Lacroix, E.; Dobson, C.M.; Hoenger, A.; Serrano, L. De novo designed peptide-based amyloid fibrils. Proc. Natl. Acad. Sci. USA 2002, 99, 16052–16057. [Google Scholar] [CrossRef]
- Chiti, F.; Taddei, N.; Baroni, F.; Capanni, C.; Stefani, M.; Ramponi, G.; Dobson, C.M. Kinetic partitioning of protein folding and aggregation. Nat. Struct. Biol. 2002, 9, 137–143. [Google Scholar] [CrossRef]
- Otzen, D.E.; Kristensen, O.; Oliveberg, M. Designed protein tetramer zipped together with a hydrophobic Alzheimer homology: A structural clue to amyloid assembly. Proc. Natl. Acad. Sci. USA 2000, 97, 9907–9912. [Google Scholar] [CrossRef]
- Schwartz, R.; Istrail, S.; King, J. Frequencies of amino acid strings in globular protein sequences indicate suppression of blocks of consecutive hydrophobic residues. Protein Sci. 2001, 10, 1023–1031. [Google Scholar] [CrossRef]
- West, M.W.; Wang, W.; Patterson, J.; Mancias, J.D.; Beasley, J.R.; Hecht, M.H. De novo amyloid proteins from designed combinatorial libraries. Proc. Natl. Acad. Sci. USA 1999, 96, 11211–11216. [Google Scholar] [CrossRef]
- Pawar, A.P.; DuBay, K.F.; Zurdo, J.; Chiti, F.; Vendruscolo, M.; Dobson, C.M. Prediction of “Aggregation-prone” and “Aggregation-susceptible” Regions in Proteins Associated with Neurodegenerative Diseases. J. Mol. Biol. 2005, 350, 379–392. [Google Scholar] [CrossRef]
- Emily, M.; Talvas, A.; Delamarche, C. MetAmyl: A METa-Predictor for AMYLoid Proteins. PLoS ONE 2013, 8, e79722. [Google Scholar] [CrossRef]
- Hammarström, P.; Wiseman, R.L.; Powers, E.T.; Kelly, J.W. Prevention of Transthyretin Amyloid Disease by Changing Protein Misfolding Energetics. Science 2003, 299, 713–716. [Google Scholar] [CrossRef]
- Laidman, J.; Forse, G.J.; Yeates, T.O. Conformational Change and Assembly through Edge β Strands in Transthyretin and Other Amyloid Proteins. Acc. Chem. Res. 2006, 39, 576–583. [Google Scholar] [CrossRef]
- Olofsson, A.; Ippel, J.H.; Wijmenga, S.S.; Lundgren, E.; Öhman, A. Probing Solvent Accessibility of Transthyretin Amyloid by Solution NMR Spectroscopy. J. Biol. Chem. 2004, 279, 5699–5707. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.D.; Portelius, E.; Kheterpal, I.; Guo, J.-T.; Cook, K.D.; Xu, Y.; Wetzel, R. Mapping Aβ Amyloid Fibril Secondary Structure Using Scanning Proline Mutagenesis. J. Mol. Biol. 2004, 335, 833–842. [Google Scholar] [CrossRef] [PubMed]
- Petkova, A.T.; Ishii, Y.; Balbach, J.J.; Antzutkin, O.N.; Leapman, R.D.; Delaglio, F.; Tycko, R. A structural model for Alzheimer’s β-amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl. Acad. Sci. USA 2002, 99, 16742–16747. [Google Scholar] [CrossRef] [PubMed]
- Török, M.; Milton, S.; Kayed, R.; Wu, P.; McIntire, T.; Glabe, C.G.; Langen, R. Structural and Dynamic Features of Alzheimer’s Aβ Peptide in Amyloid Fibrils Studied by Site-directed Spin Labeling. J. Biol. Chem. 2002, 277, 40810–40815. [Google Scholar] [CrossRef]
- Lansbury, P.T.; Costa, P.R.; Griffiths, J.M.; Simon, E.J.; Auger, M.; Halverson, K.J.; Kocisko, D.A.; Hendsch, Z.S.; Ashburn, T.T.; Spencer, R.G.; et al. Structural model for the β-amyloid fibril based on interstrand alignment of an antiparallel-sheet comprising a C-terminal peptide. Nat. Struct. Mol. Biol. 1995, 2, 990–998. [Google Scholar] [CrossRef]
- Heise, H.; Hoyer, W.; Becker, S.; Andronesi, O.C.; Riedel, D.; Baldus, M. Molecular-level secondary structure, polymorphism, and dynamics of full-length α-synuclein fibrils studied by solid-state NMR. Proc. Natl. Acad. Sci. USA 2005, 102, 15871–15876. [Google Scholar] [CrossRef]
- Miake, H.; Mizusawa, H.; Iwatsubo, T.; Hasegawa, M. Biochemical Characterization of the Core Structure of α-Synuclein Filaments. J. Biol. Chem. 2002, 277, 19213–19219. [Google Scholar] [CrossRef]
- Der-Sarkissian, A.; Jao, C.C.; Chen, J.; Langen, R. Structural Organization of α-Synuclein Fibrils Studied by Site-directed Spin Labeling. J. Biol. Chem. 2003, 278, 37530–37535. [Google Scholar] [CrossRef]
- Illes-Toth, E.; Rempel, D.L.; Gross, M.L. Pulsed Hydrogen–Deuterium Exchange Illuminates the Aggregation Kinetics of α-Synuclein, the Causative Agent for Parkinson’s Disease. ACS Chem. Neurosci. 2018, 9, 1469–1476. [Google Scholar] [CrossRef] [PubMed]
- Fontana, A.; de Laureto, P.P.; De Filippis, V.; Scaramella, E.; Zambonin, M. Probing the partly folded states of proteins by limited proteolysis. Fold. Des. 1997, 2, R17–R26. [Google Scholar] [CrossRef] [PubMed]
- Slamova, I.; Adib, R.; Ellmerich, S.; Golos, M.R.; Gilbertson, J.A.; Botcher, N.; Canetti, D.; Taylor, G.W.; Rendell, N.; Tennent, G.A.; et al. Plasmin activity promotes amyloid deposition in a transgenic model of human transthyretin amyloidosis. Nat. Commun. 2021, 12, 7112. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, C.-C.; Rombouts, F.; Gijsen, H.J. New evolutions in the BACE1 inhibitor field from 2014 to 2018. Bioorg. Med. Chem. Lett. 2019, 29, 761–777. [Google Scholar] [CrossRef] [PubMed]
- Vassar, R.; Kuhn, P.-H.; Haass, C.; Kennedy, M.E.; Rajendran, L.; Wong, P.C.; Lichtenthaler, S.F. Function, therapeutic potential and cell biology of BACE proteases: Current status and future prospects. J. Neurochem. 2014, 130, 4–28. [Google Scholar] [CrossRef]
- Keam, S.J. Inotersen: First Global Approval. Drugs 2018, 78, 1371–1376. [Google Scholar] [CrossRef]
- Adams, D.; Gonzalez-Duarte, A.; O’Riordan, W.D.; Yang, C.-C.; Ueda, M.; Kristen, A.V.; Tournev, I.; Schmidt, H.H.; Coelho, T.; Berk, J.L.; et al. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2018, 379, 11–21. [Google Scholar] [CrossRef]
- Hoy, S.M. Patisiran: First Global Approval. Drugs 2018, 78, 1625–1631. [Google Scholar] [CrossRef]
- Boutajangout, A.; Lindberg, H.; Awwad, A.; Paul, A.; Baitalmal, R.; Almokyad, I.; Höidén-Guthenberg, I.; Gunneriusson, E.; Frejd, F.Y.; Härd, T.; et al. Affibody-Mediated Sequestration of Amyloid β Demonstrates Preventive Efficacy in a Transgenic Alzheimer’s Disease Mouse Model. Front. Aging Neurosci. 2019, 11, 64. [Google Scholar] [CrossRef]


| Amyloidoses | Precursor Fibril Protein | Type of Amyloidosis | ||
|---|---|---|---|---|
| Systemic and/or Localized | Acquired and/or Hereditary | Organ or Tissue Affected | ||
| AL | Immunoglobulin light chain | S, L | A, H | all, no CNS a |
| AH | Immunoglobulin heavy chain | S, L | A | all, no CNS a |
| AA | Apo Serum Amyloid A | S | A | all, no CNS a |
| ATTR | Transthyretin, wild type Transthyretin, mutants | S S | A H | heart, lung, ligaments, PNS b, ANS c, eye |
| Aβ2M | β2-microglobulin, wild type β2-microglobulin, mutants | S S | A H | musculoskeletal system, ANS c |
| AApoAI | Apolipoprotein A I, mutants | S | H | heart, liver, kidney, PNS b, testis, larynx, skin |
| AApoAII | Apolipoprotein A II, mutants | S | H | kidney |
| AApoAIV | Apolipoprotein A IV, wild type | S | A | kidney |
| AApoCII | Apolipoprotein C II, mutants | S | H | kidney |
| AApoCIII | Apolipoprotein C III, mutants | S | H | kidney |
| AGel | Gelsolin, mutants | S | H | kidney, PNS b, cornea |
| ALys | Lysozyme, mutants | S | H | kidney |
| ALECT2 | Leukocyte chemotactic factor-2 | S | A | kidney |
| AFib | Fibrinogen α, mutants | S | H | kidney |
| ACys | Cystatin C, mutants | S | H | CNS a, PNS b, skin |
| ABri | ABriPP d, mutants | S | H | CNS a |
| Adan | ADanPP e, mutants | L | H | CNS a |
| Aβ | Aβ protein precursor, wild type Aβ protein precursor, variant | L L | A H | CNS a |
| AαSyn | α-Synuclein | L | A | CNS a |
| ATau | Tau protein | L | A | CNS a |
| APrP | Prion protein, wild type Prion protein, mutants | L L, S | A H | CNS a, PNS b |
| ACal | (Pro)calcitonin | L, S | A | kidney, thyroid |
| AIAPP | Islet amyloid polypeptide | L | A | Langerhans’ islets, insulinomas |
| AANF | Atrial natriuretic factor | L | A | heart |
| APro | Prolactin | L | A | pituitary gland |
| AIns | Insulin | L | A | skin, muscle |
| ASPC | Lung surfactant protein | L | A | lung |
| ACor | Corneodesmosin | L | A | cornified epithelia, hair follicles |
| AMed | Lactadherin | L | A | senile aorta |
| Aker | Kerato-epithelin | L | A | cornea |
| ALac | Lactoferrin | L | A | cornea |
| AOAAP | Odontogenic ameloblast-associated protein | L | A | tooth forming tissues |
| ASem1 | Semenogelin 1 | L | A | vesicula seminalis |
| AEnf | Enfurvitide | L | A | skin |
| ACatK | Cathepsin K | L | A | kidney, angiomyolipoma |
| AEFEMP1 | EGF-containing fibulin-like extracellular matrix protein 1 (EFEMP1) | L | A | portal veins |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Acquasaliente, L.; De Filippis, V. The Role of Proteolysis in Amyloidosis. Int. J. Mol. Sci. 2023, 24, 699. https://doi.org/10.3390/ijms24010699
Acquasaliente L, De Filippis V. The Role of Proteolysis in Amyloidosis. International Journal of Molecular Sciences. 2023; 24(1):699. https://doi.org/10.3390/ijms24010699
Chicago/Turabian StyleAcquasaliente, Laura, and Vincenzo De Filippis. 2023. "The Role of Proteolysis in Amyloidosis" International Journal of Molecular Sciences 24, no. 1: 699. https://doi.org/10.3390/ijms24010699
APA StyleAcquasaliente, L., & De Filippis, V. (2023). The Role of Proteolysis in Amyloidosis. International Journal of Molecular Sciences, 24(1), 699. https://doi.org/10.3390/ijms24010699








