High-Affinity Antibodies Designing of SARS-CoV-2 Based on Molecular Dynamics Simulations
Abstract
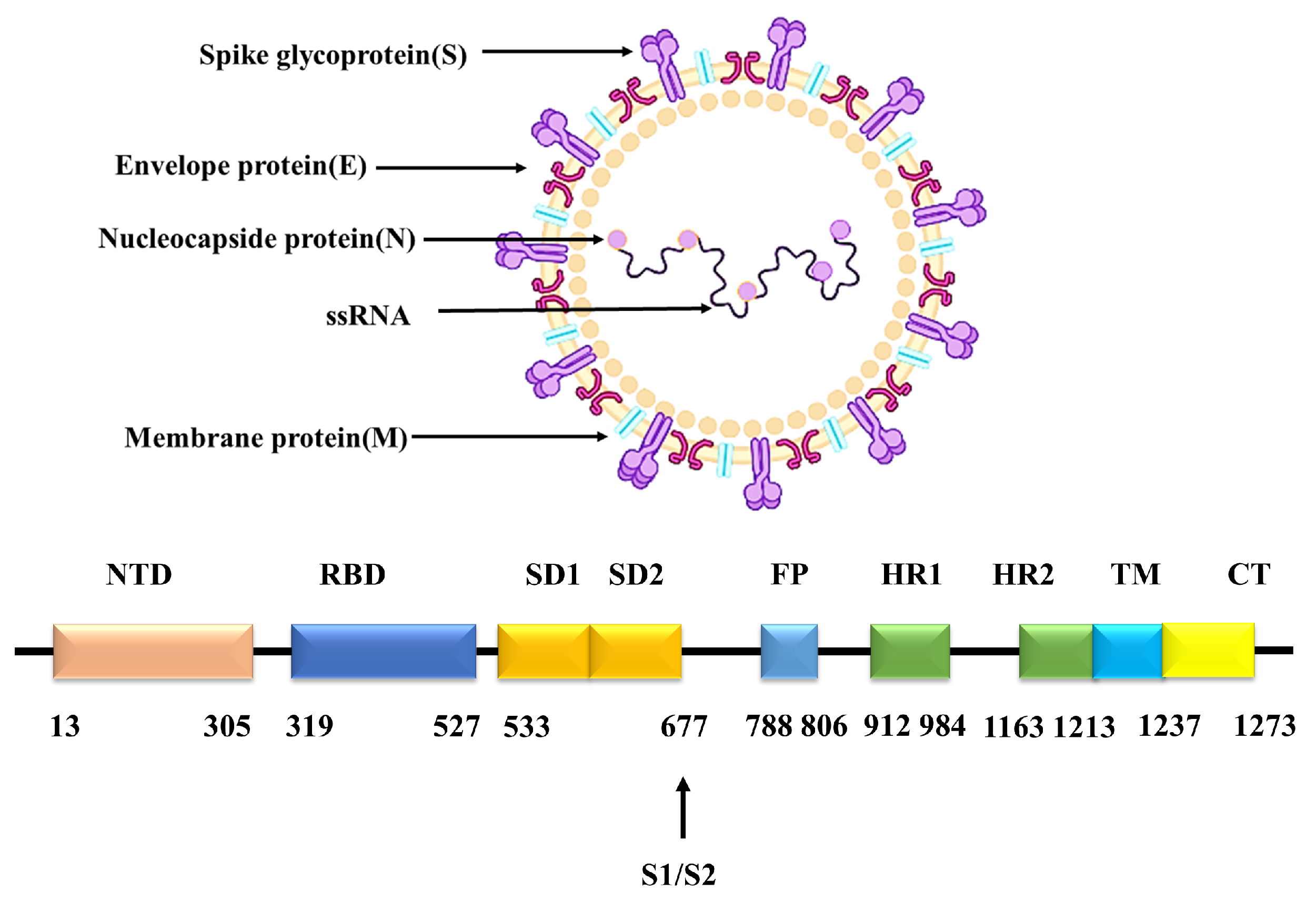
:1. Introduction
2. Results
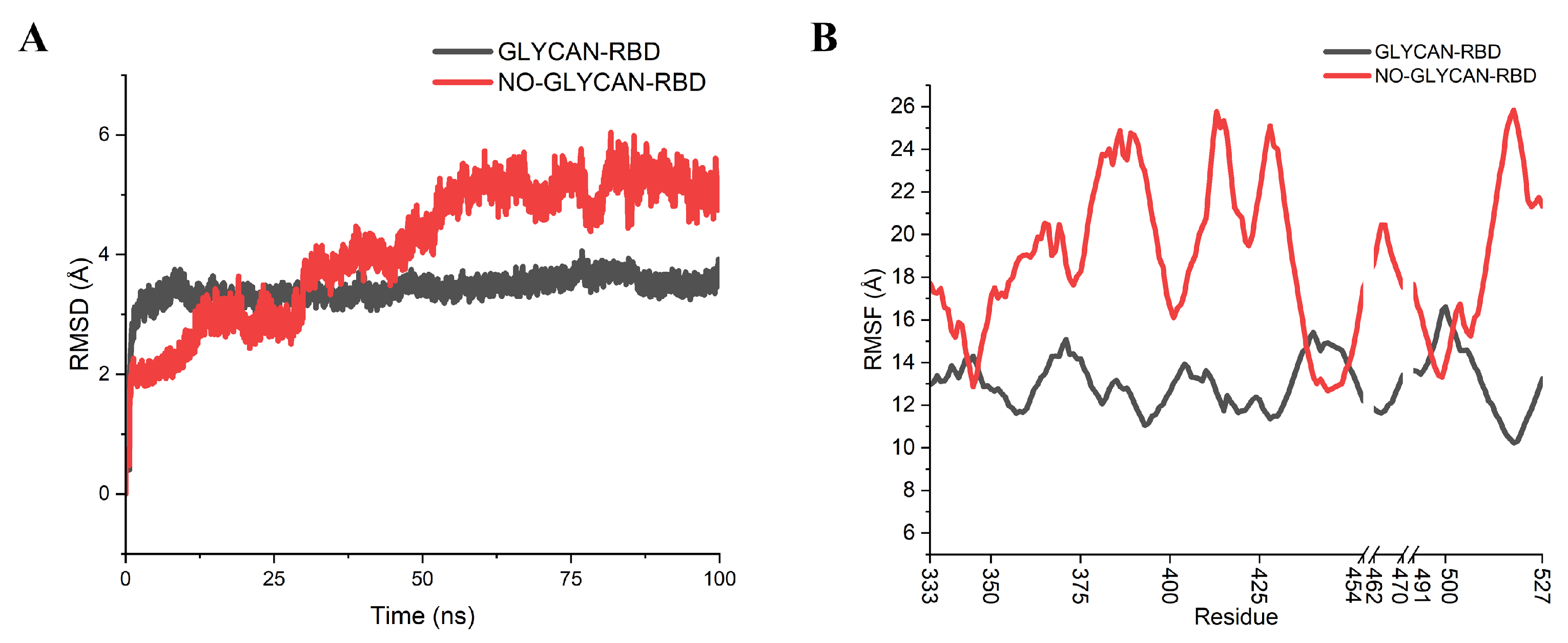
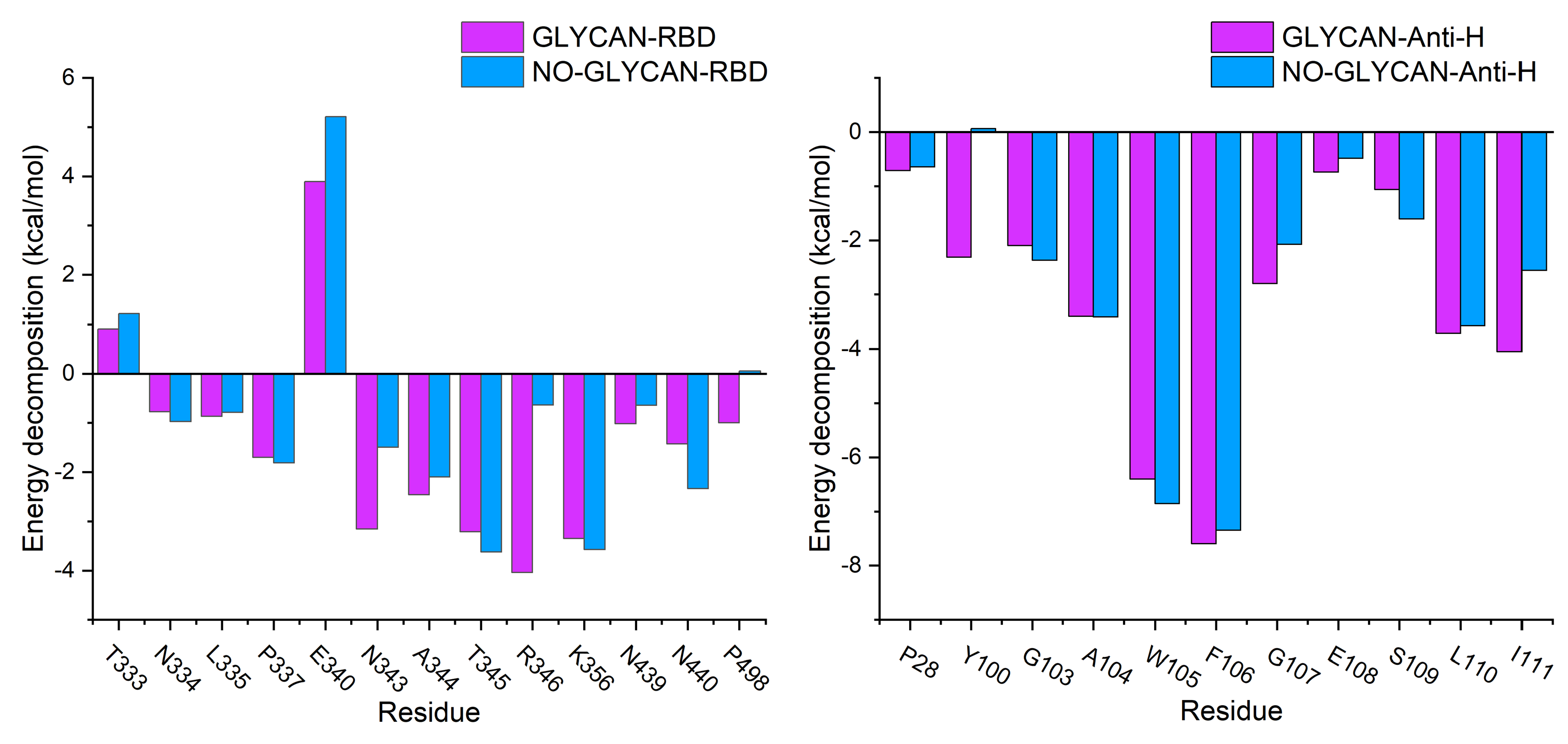
2.1. Polysaccharide Stabilizes the Interaction of Antigen and Antibody
2.2. Antibody Mutation Design
2.2.1. Single Mutant Screening
2.2.2. Single Mutant Affinity Simulation Validation
2.2.3. Representative Conformational Superposition Analysis
3. Discussion
4. Materials and Methods
4.1. Molecular Dynamics Simulation
4.2. MM-GBSA Calculation
4.3. Residue Mutation Prediction
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dong, E.; Du, H.; Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020, 20, 533–534. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chen, P.; Wang, J.; Feng, J.; Zhou, H.; Li, X.; Zhong, W.; Hao, P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020, 63, 457–460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Sun, H.; Yu, H.; Li, S.; Zheng, Q.; Xia, N. Neutralizing antibodies against SARS-CoV-2: Current understanding, challenge and perspective. Antib. Ther. 2020, 3, 285–299. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Li, Y.; Leung, E.L.H.; Liu, X.; Liu, K.; Wang, Q.; Lan, Y.; Li, X.; Yu, H.; Cui, L.; et al. A review of therapeutic agents and Chinese herbal medicines against SARS-COV-2 (COVID-19). Pharmacol. Res. 2020, 158, 104929. [Google Scholar] [CrossRef]
- Zumla, A.; Chan, J.F.; Azhar, E.I.; Hui, D.S.; Yuen, K.Y. Coronaviruses—drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016, 15, 327–347. [Google Scholar] [CrossRef] [Green Version]
- Barnes, C.O.; Jette, C.A.; Abernathy, M.E.; Dam, K.M.A.; Esswein, S.R.; Gristick, H.B.; Malyutin, A.G.; Sharaf, N.G.; Huey-Tubman, K.E.; Lee, Y.E.; et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 2020, 588, 682–687. [Google Scholar] [CrossRef]
- Bagheri, M.; Niavarani, A. Molecular dynamics analysis predicts ritonavir and naloxegol strongly block the SARS-CoV-2 spike protein-hACE2 binding. J. Biomol. Struct. Dyn. 2022, 40, 1597–1606. [Google Scholar] [CrossRef]
- Pinto, D.; Park, Y.J.; Beltramello, M.; Walls, A.C.; Tortorici, M.A.; Bianchi, S.; Jaconi, S.; Culap, K.; Zatta, F.; De Marco, A.; et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 2020, 583, 290–295. [Google Scholar] [CrossRef]
- Treewattanawong, W.; Sitthiyotha, T.; Chunsrivirot, S. Computational redesign of Fab CC12. 3 with substantially better predicted binding affinity to SARS-CoV-2 than human ACE2 receptor. Sci. Rep. 2021, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Cheatham, T.E., III; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M., Jr.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Liu, M.; Gao, J. Enhanced receptor binding of SARS-CoV-2 through networks of hydrogen-bonding and hydrophobic interactions. Proc. Natl. Acad. Sci. USA 2020, 117, 13967–13974. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wu, N.C.; Yuan, M.; Bangaru, S.; Torres, J.L.; Caniels, T.G.; Van Schooten, J.; Zhu, X.; Lee, C.C.D.; Brouwer, P.J.; et al. Cross-neutralization of a SARS-CoV-2 antibody to a functionally conserved site is mediated by avidity. Immunity 2020, 53, 1272–1280. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Shan, C.; Duan, X.; Chen, Z.; Liu, P.; Song, J.; Song, T.; Bi, X.; Han, C.; Wu, L.; et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 2020, 584, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Strokach, A.; Corbi-Verge, C.; Kim, P.M. Predicting changes in protein stability caused by mutation using sequence-and structure-based methods in a CAGI5 blind challenge. Hum. Mutat. 2019, 40, 1414–1423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, K.E.; Kim, J.M.; Rhee, J.; Park, A.K.; Kim, E.J.; Kang, N.S. Molecular Dynamics Studies on the structural characteristics for the stability prediction of SARS-CoV-2. Int. J. Mol. Sci. 2021, 22, 8714. [Google Scholar] [CrossRef]
- Chen, J.; Wu, F.; Lin, D.; Kong, W.; Cai, X.; Yang, J.; Sun, X.; Cao, P. Rational optimization of a human neutralizing antibody of SARS-CoV-2. Comput. Biol. Med. 2021, 135, 104550. [Google Scholar] [CrossRef]
- Shariatifar, H.; Farasat, A. Affinity enhancement of CR3022 binding to RBD; in silico site directed mutagenesis using molecular dynamics simulation approaches. J. Biomol. Struct. Dyn. 2021, 1–10. [Google Scholar] [CrossRef]
- Huerta-Miranda, G.; García-García, W.; Vidal-Limon, A.; Miranda-Hernández, M. Use of simplified models for theoretical prediction of the interactions between available antibodies and the receptor-binding domain of SARS-CoV-2 spike protein. J. Biomol. Struct. Dyn. 2021, 1–10. [Google Scholar] [CrossRef]
- Rolta, R.; Salaria, D.; Sharma, B.; Awofisayo, O.; Fadare, O.A.; Sharma, S.; Patel, C.N.; Kumar, V.; Sourirajan, A.; Baumler, D.J.; et al. Methylxanthines as Potential Inhibitor of SARS-CoV-2: An In Silico Approach. Curr. Pharmacol. Rep. 2022, 8, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Mannar, D.; Leopold, K.; Subramaniam, S. Glycan reactive anti-HIV-1 antibodies bind the SARS-CoV-2 spike protein but do not block viral entry. Sci. Rep. 2021, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hauser, B.M.; Sangesland, M.; Denis, K.J.S.; Lam, E.C.; Case, J.B.; Windsor, I.W.; Feldman, J.; Caradonna, T.M.; Kannegieter, T.; Diamond, M.S.; et al. Rationally designed immunogens enable immune focusing following SARS-CoV-2 spike imprinting. Cell Rep. 2022, 38, 110561. [Google Scholar] [CrossRef] [PubMed]
- Case, J.B.; Mackin, S.; Errico, J.M.; Chong, Z.; Madden, E.A.; Whitener, B.; Guarino, B.; Schmid, M.A.; Rosenthal, K.; Ren, K.; et al. Resilience of S309 and AZD7442 monoclonal antibody treatments against infection by SARS-CoV-2 Omicron lineage strains. Nat. Commun. 2022, 13, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.T.; Badeti, S.; Chen, C.H.; Kim, J.; Choudhary, A.; Honnen, B.; Reichman, C.; Calianese, D.; Pinter, A.; Jiang, Q.; et al. CAR-NK cells effectively target SARS-CoV-2-spike-expressing cell lines in vitro. Front. Immunol. 2021, 12, 652223. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; De Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef] [Green Version]
- Liang, D.; Song, M.; Niu, Z.; Zhang, P.; Rafailovich, M.; Deng, Y. Supervised machine learning approach to molecular dynamics forecast of SARS-CoV-2 spike glycoproteins at varying temperatures. MRS Adv. 2021, 6, 362–367. [Google Scholar] [CrossRef]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef]
- Miller, B.R., III; McGee, T.D., Jr.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA. py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Wang, E.; Weng, G.; Sun, H.; Du, H.; Zhu, F.; Chen, F.; Wang, Z.; Hou, T. Assessing the performance of the MM/PBSA and MM/GBSA methods. 10. Impacts of enhanced sampling and variable dielectric model on protein–protein interactions. Phys. Chem. Chem. Phys. 2019, 21, 18958–18969. [Google Scholar] [CrossRef]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.; Wang, J.; Li, Y.; Wang, W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011, 51, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Gohlke, H.; Case, D.A. Converging free energy estimates: MM-PB (GB) SA studies on the protein–protein complex Ras–Raf. J. Comput. Chem. 2004, 25, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Frenz, B.; Lewis, S.M.; King, I.; DiMaio, F.; Park, H.; Song, Y. Prediction of protein mutational free energy: Benchmark and sampling improvements increase classification accuracy. Front. Bioeng. Biotechnol. 2020, 8, 1175. [Google Scholar] [CrossRef]
- Al-Khafaji, K.; Al-Duhaidahawi, D.; Taskin Tok, T. Using integrated computational approaches to identify safe and rapid treatment for SARS-CoV-2. J. Biomol. Struct. Dyn. 2021, 39, 3387–3395. [Google Scholar] [CrossRef]
- Rolta, R.; Salaria, D.; Sharma, P.; Sharma, B.; Kumar, V.; Rathi, B.; Verma, M.; Sourirajan, A.; Baumler, D.J.; Dev, K. Phytocompounds of Rheum emodi, Thymus serpyllum, and Artemisia annua inhibit spike protein of SARS-CoV-2 binding to ACE2 receptor: In silico approach. Curr. Pharmacol. Rep. 2021, 7, 135–149. [Google Scholar] [CrossRef]



| GLYCAN (kcal/mol) | NO-GLYCAN (kcal/mol) | |
|---|---|---|
| E | −73.279 | −74.057 |
| E | −239.955 | −107.104 |
| G | 270.4689 | 150.9991 |
| E | −12.0327 | −10.9644 |
| E | −313.234 | −181.161 |
| G | 258.4362 | 140.0348 |
| G | −54.7977 | −41.1265 |
| Donor | Acceptor | Occupancy | |
|---|---|---|---|
| GLYCAN | NO-GLYCAN | ||
| ILE111-Main | ASN343-Main | 48.44% | 35.66% |
| ALA104-Main | GLU340-Side | 78.33% | 87.01% |
| TRP105-Main | GLU340-Side | 78.89% | 63.64% |
| PHE106-Main | GLU340-Side | 38.00% | 51.75% |
| LYS356-Side | GLU108-Side | 26.22% | 42.36% |
| THR345-Main | SER109-Main | 37.56% | 42.96% |
| GLY107-Main | GLU340-Side | 39.33% | 17.88% |
| ASN343-Side | TYR100-Side | 41.33% | 2.90% |
| Initial Residues | Predicted Residues | Energy Changes (kcal/mol) |
|---|---|---|
| P28 | W | −14.5437 |
| D99 | T | −40.779 |
| Y100 | W | −12.019 |
| G103 | A | −4.81267 |
| A104 | T | −2.526 |
| W105 | H | −0.075 |
| G107 | H | −2.59663 |
| L111 | T | −2.15933 |
| Systems | Gbind (kcal/mol) |
|---|---|
| WT | −54.7977 |
| Y100W | −47.0290 |
| D99T | −61.5002 |
| G103A | −64.7284 |
| P28W | −60.6685 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, Z.; Liu, H.; Zhou, S.; Xie, Z.; Yuan, S. High-Affinity Antibodies Designing of SARS-CoV-2 Based on Molecular Dynamics Simulations. Int. J. Mol. Sci. 2023, 24, 481. https://doi.org/10.3390/ijms24010481
Tian Z, Liu H, Zhou S, Xie Z, Yuan S. High-Affinity Antibodies Designing of SARS-CoV-2 Based on Molecular Dynamics Simulations. International Journal of Molecular Sciences. 2023; 24(1):481. https://doi.org/10.3390/ijms24010481
Chicago/Turabian StyleTian, Zihui, Hongtao Liu, Shuangyan Zhou, Zengyan Xie, and Shuai Yuan. 2023. "High-Affinity Antibodies Designing of SARS-CoV-2 Based on Molecular Dynamics Simulations" International Journal of Molecular Sciences 24, no. 1: 481. https://doi.org/10.3390/ijms24010481
APA StyleTian, Z., Liu, H., Zhou, S., Xie, Z., & Yuan, S. (2023). High-Affinity Antibodies Designing of SARS-CoV-2 Based on Molecular Dynamics Simulations. International Journal of Molecular Sciences, 24(1), 481. https://doi.org/10.3390/ijms24010481






