LaDAL1 Coordinates Age and Environmental Signals in the Life Cycle of Larix kaempferi
Abstract
1. Introduction
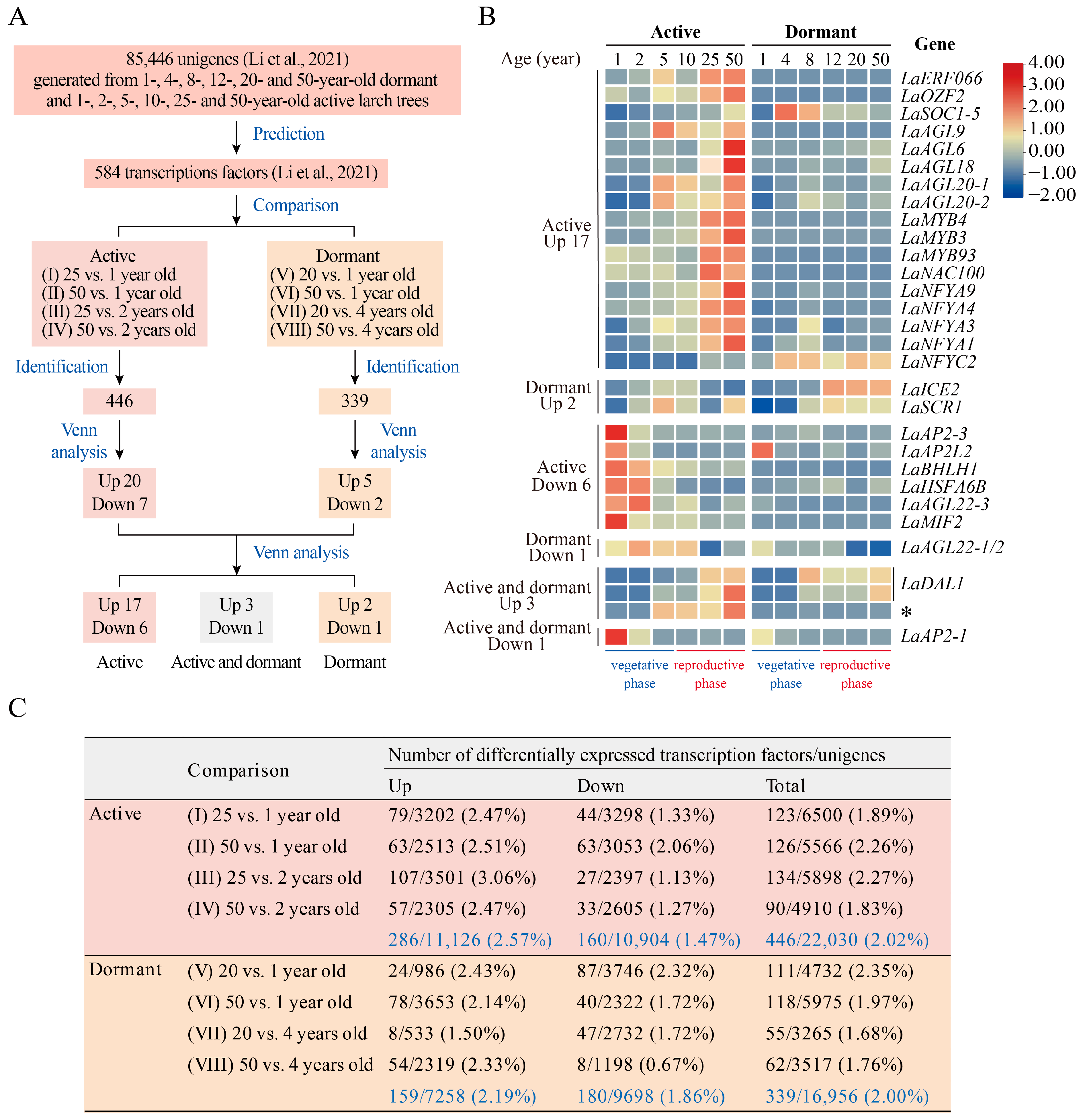
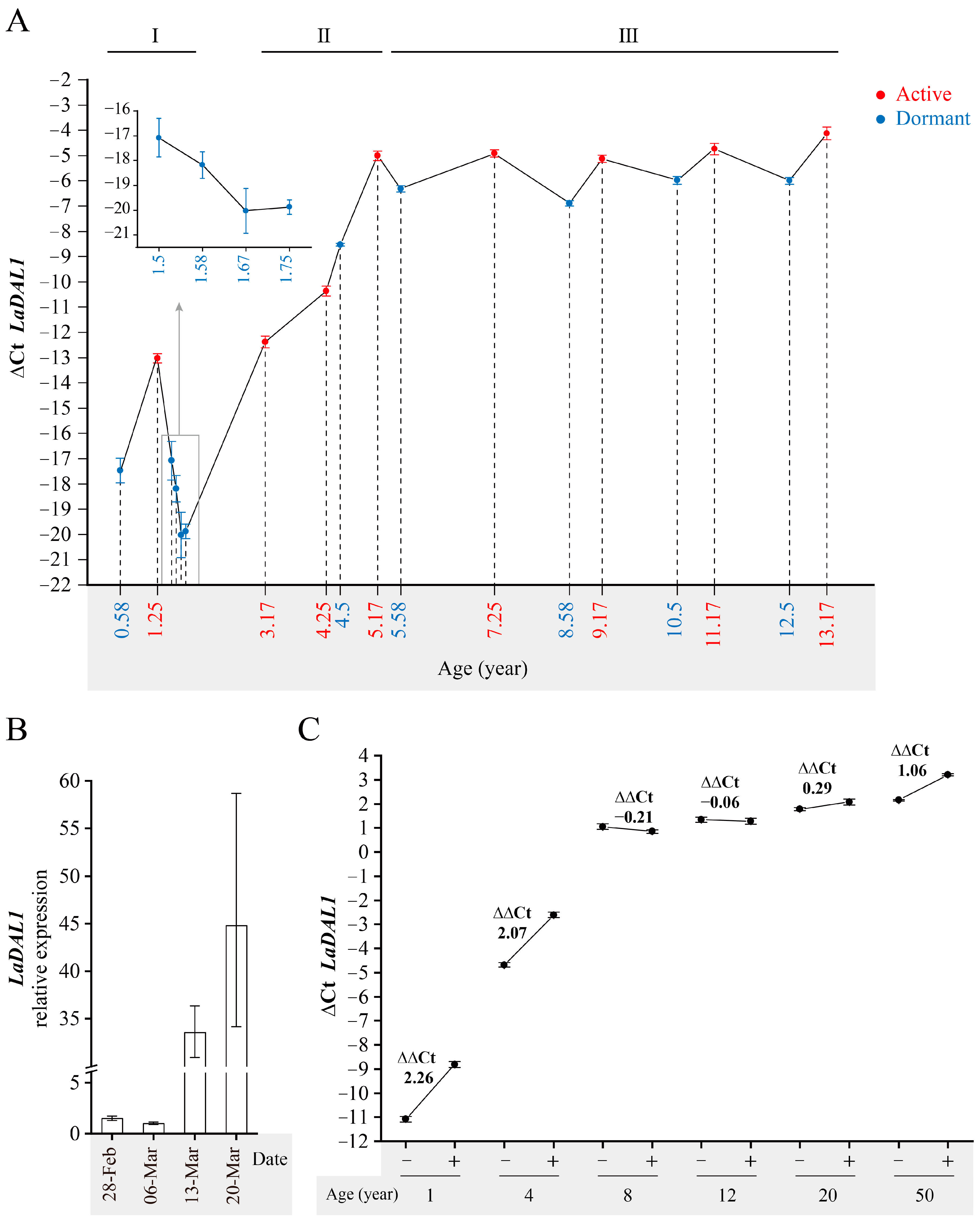
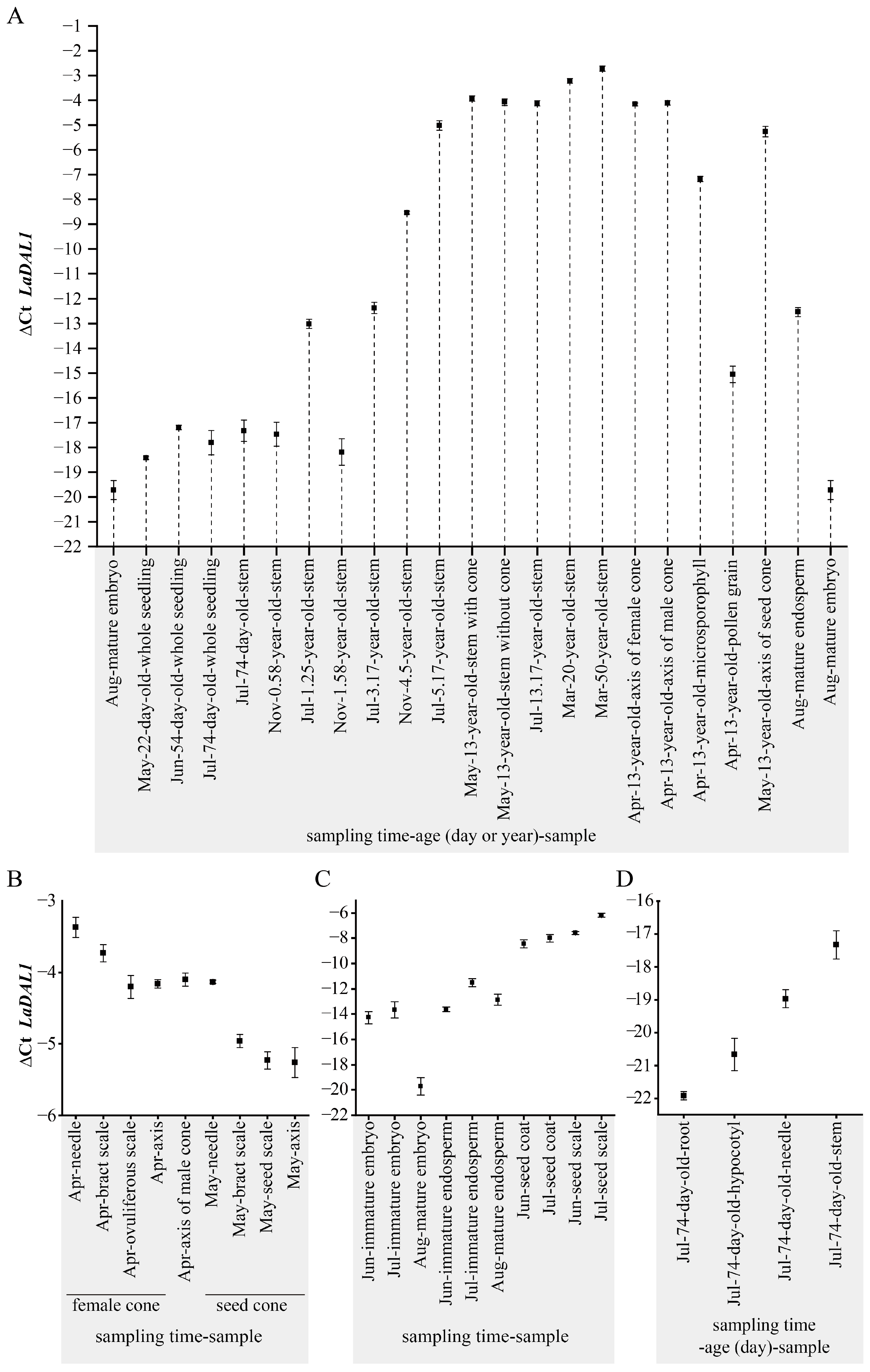
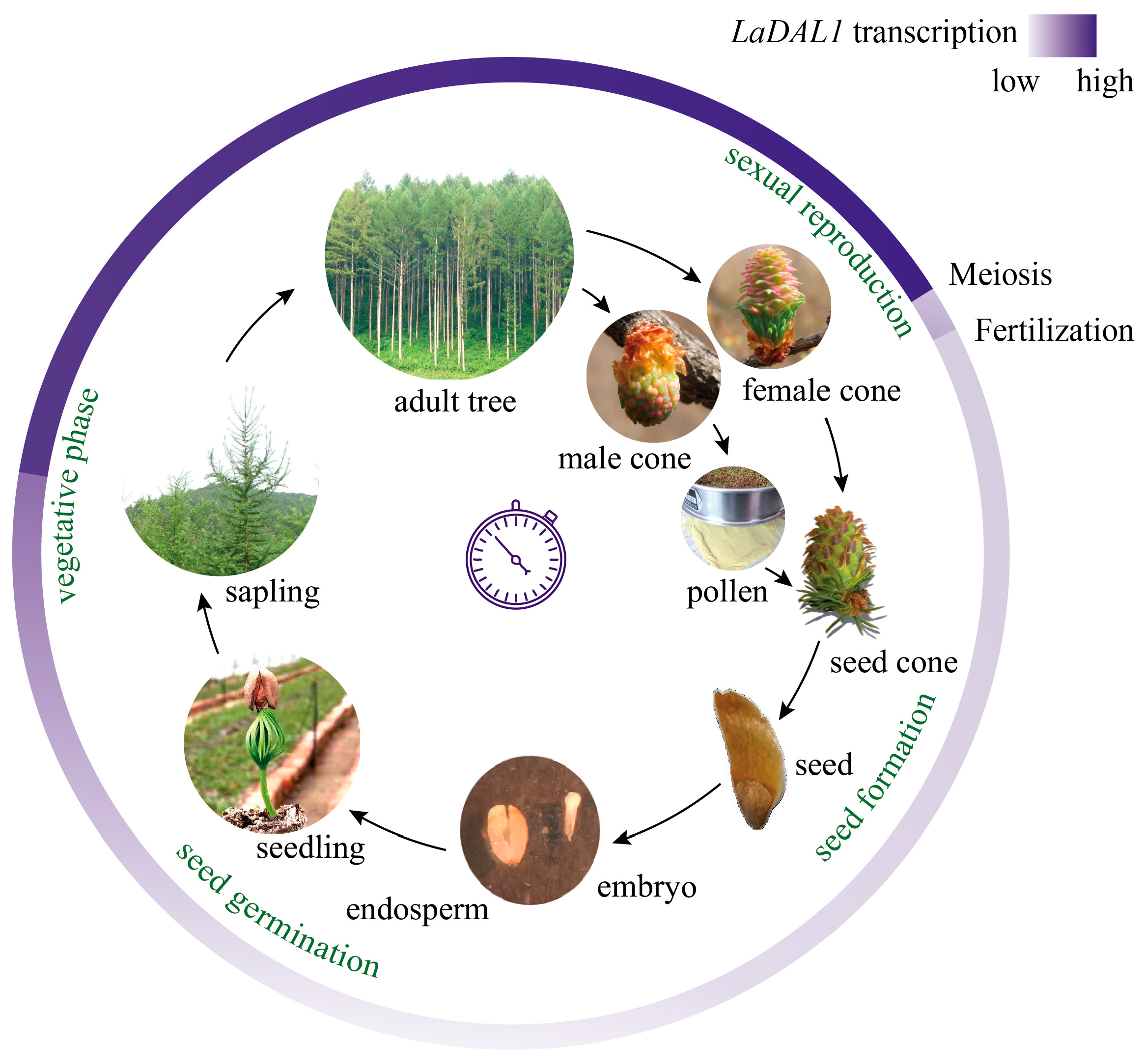
2. Results and Discussion
3. Materials and Methods
3.1. Transcriptome Data
3.2. Identification of DEUs
3.3. Plant Materials
3.3.1. The Materials Used for Determining the Dynamic Temporal Expression Pattern of LaDAL1 in the Annual Growth Cycle of Larix kaempferi Trees of Different Ages
3.3.2. The Materials Used for Determining the Dynamic Spatiotemporal Expression Pattern of LaDAL1 during the L. kaempferi Life Cycle
Seeds and Seedlings
Saplings and Mature Trees
Cones
3.4. Water Culture Experiments
3.5. Sequence Analysis, Full-Length cDNA Cloning, and Annotation
3.6. RNA Extraction and cDNA Synthesis
3.7. QRT-PCR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xiang, W.B.; Li, W.F.; Zhang, S.G.; Qi, L.W. Transcriptome-wide analysis to dissect the transcription factors orchestrating the phase change from vegetative to reproductive development in Larix kaempferi. Tree Genet. Genomes 2019, 15, 68. [Google Scholar] [CrossRef]
- Zhang, Y.; Zang, Q.L.; Qi, L.W.; Han, S.Y.; Li, W.F. Effects of cutting, pruning, and grafting on the expression of age-related genes in Larix kaempferi. Forests 2020, 11, 218. [Google Scholar] [CrossRef]
- Ye, Z.L.; Zang, Q.L.; Cheng, D.X.; Li, X.Y.; Qi, L.W.; Li, W.F. Over-expression of larch DAL1 accelerates life-cycle progression in Arabidopsis. Forests 2022, 13, 953. [Google Scholar] [CrossRef]
- Carlsbecker, A.; Tandre, K.; Johanson, U.; Englund, M.; Engström, P. The MADS-box gene DAL1 is a potential mediator of the juvenile-to-adult transition in Norway spruce (Picea abies). Plant J. 2004, 40, 546–557. [Google Scholar] [CrossRef]
- Shi, S.L.; Yan, S.Y.; Zhao, C.; Zhang, P.; Yang, L.; Wang, C.; Shen, H.L. Deep sequencing and analysis of transcriptomes of Pinus koraiensis Sieb. & Zucc. Forests 2020, 11, 350. [Google Scholar] [CrossRef]
- Ma, J.J.; Chen, X.; Song, Y.T.; Zhang, G.F.; Zhou, X.Q.; Que, S.P.; Mao, F.; Pervaiz, T.; Lin, J.X.; Li, Y.; et al. MADS-box transcription factors MADS11 and DAL1 interact to mediate the vegetative-to-reproductive transition in pine. Plant Physiol. 2021, 187, 247–262. [Google Scholar] [CrossRef] [PubMed]
- Li, W.F.; Kang, Y.; Zhang, Y.; Zang, Q.L.; Qi, L.W. Concerted control of the LaRAV1-LaCDKB1;3 module by temperature during dormancy release and reactivation of larch. Tree Physiol. 2021, 41, 1918–1937. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Han, S.; Qi, L.; Zhang, S. Transcriptome resources and genome-wide marker development for Japanese larch (Larix kaempferi). Front. Agric. Sci. Eng. 2014, 1, 77–84. [Google Scholar] [CrossRef]
- Li, W.F.; Yang, W.H.; Zhang, S.G.; Han, S.Y.; Qi, L.W. Transcriptome analysis provides insights into wood formation during larch tree aging. Tree Genet. Genomes 2017, 13, 19. [Google Scholar] [CrossRef]
- Li, A.; Yu, X.; Cao, B.B.; Peng, L.X.; Gao, Y.; Feng, T.; Li, H.; Ren, Z.Y. LkAP2L2, an AP2/ERF transcription factor gene of Larix kaempferi, with pleiotropic roles in plant branch and seed development. Russ. J. Genet. 2017, 53, 1335–1342. [Google Scholar] [CrossRef]
- Zhang, L.F.; Li, W.F.; Xu, H.; Qi, L.W.; Han, S.Y. Cloning and characterization of four differentially expressed cDNAs encoding NFYA homologs involved in responses to ABA during somatic embryogenesis in Japanese larch (Larix leptolepis). Plant Cell Tissue Organ Cult. 2014, 117, 293–304. [Google Scholar] [CrossRef]
- Alejano, R.; Domínguez-Delmás, M.; García-González, I.; Wazny, T.; Vázquez-Piqué, J.; Fernández-Martínez, M. The age of black pine (Pinus nigra Arn. ssp. salzmannii (Dunal) Franco) mother trees has no effect on seed germination and on offspring seedling performance. Ann. For. Sci. 2019, 76, 15. [Google Scholar] [CrossRef]
- Katahata, S.; Futamura, N.; Igasaki, T.; Shinohara, K. Functional analysis of SOC1-like and AGL6-like MADS-box genes of the gymnosperm Cryptomeria japonica. Tree Genet. Genomes 2014, 10, 317–327. [Google Scholar] [CrossRef]
- Gao, J.; Zhang, K.; Cheng, Y.J.; Yu, S.; Shang, G.D.; Wang, F.X.; Wu, L.Y.; Xu, Z.G.; Mai, Y.X.; Zhao, X.Y.; et al. A robust mechanism for resetting juvenility during each generation in Arabidopsis. Nat. Plants 2022, 8, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Nikolayeva, O.; Robinson, M.D. edgeR for differential RNA-seq and ChIP-seq analysis: An application to stem cell biology. Methods Mol. Biol. 2014, 1150, 45–79. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Meyer, C.A.; Wang, Q.; Jun, S.; Liu, X.; Liu, S.; Zhang, Y. GFOLD: A generalized fold change for ranking differentially expressed genes from RNA-seq data. Bioinformatics 2012, 28, 272–2788. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Li, W.F.; Zhang, S.G.; Han, S.Y.; Wu, T.; Zhang, J.H.; Qi, L.W. The post-transcriptional regulation of LaSCL6 by miR171 during maintenance of embryogenic potential in Larix kaempferi (Lamb.) Carr. Tree Genet. Genomes 2014, 10, 223–229. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.-Y.; Ye, Z.-L.; Cheng, D.-X.; Zang, Q.-L.; Qi, L.-W.; Li, W.-F. LaDAL1 Coordinates Age and Environmental Signals in the Life Cycle of Larix kaempferi. Int. J. Mol. Sci. 2023, 24, 426. https://doi.org/10.3390/ijms24010426
Li X-Y, Ye Z-L, Cheng D-X, Zang Q-L, Qi L-W, Li W-F. LaDAL1 Coordinates Age and Environmental Signals in the Life Cycle of Larix kaempferi. International Journal of Molecular Sciences. 2023; 24(1):426. https://doi.org/10.3390/ijms24010426
Chicago/Turabian StyleLi, Xiang-Yi, Zha-Long Ye, Dong-Xia Cheng, Qiao-Lu Zang, Li-Wang Qi, and Wan-Feng Li. 2023. "LaDAL1 Coordinates Age and Environmental Signals in the Life Cycle of Larix kaempferi" International Journal of Molecular Sciences 24, no. 1: 426. https://doi.org/10.3390/ijms24010426
APA StyleLi, X.-Y., Ye, Z.-L., Cheng, D.-X., Zang, Q.-L., Qi, L.-W., & Li, W.-F. (2023). LaDAL1 Coordinates Age and Environmental Signals in the Life Cycle of Larix kaempferi. International Journal of Molecular Sciences, 24(1), 426. https://doi.org/10.3390/ijms24010426





