Differential Plasma Metabolites between High- and Low-Grade Meningioma Cases
Abstract
1. Introduction
2. Results
2.1. Patients
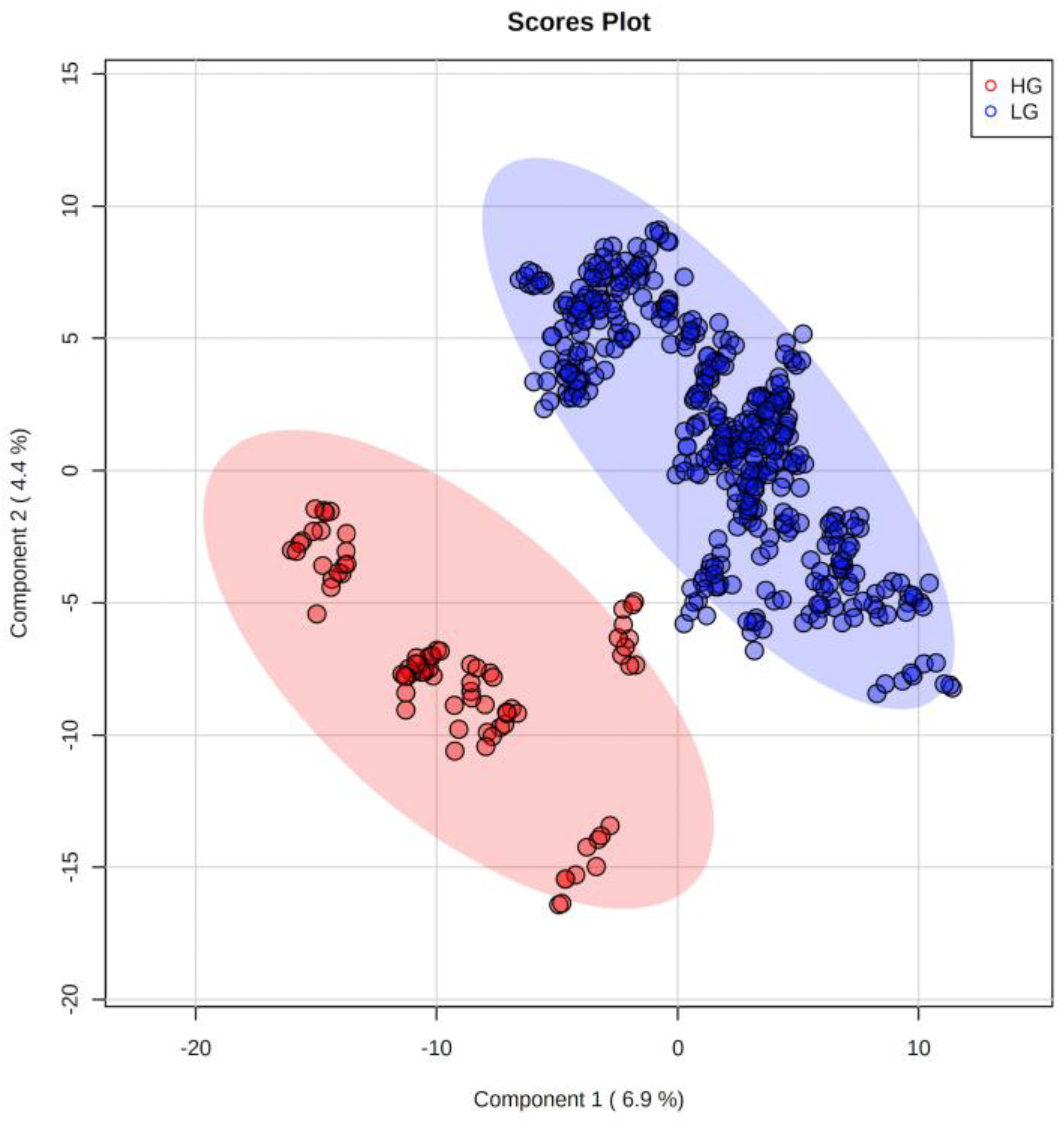
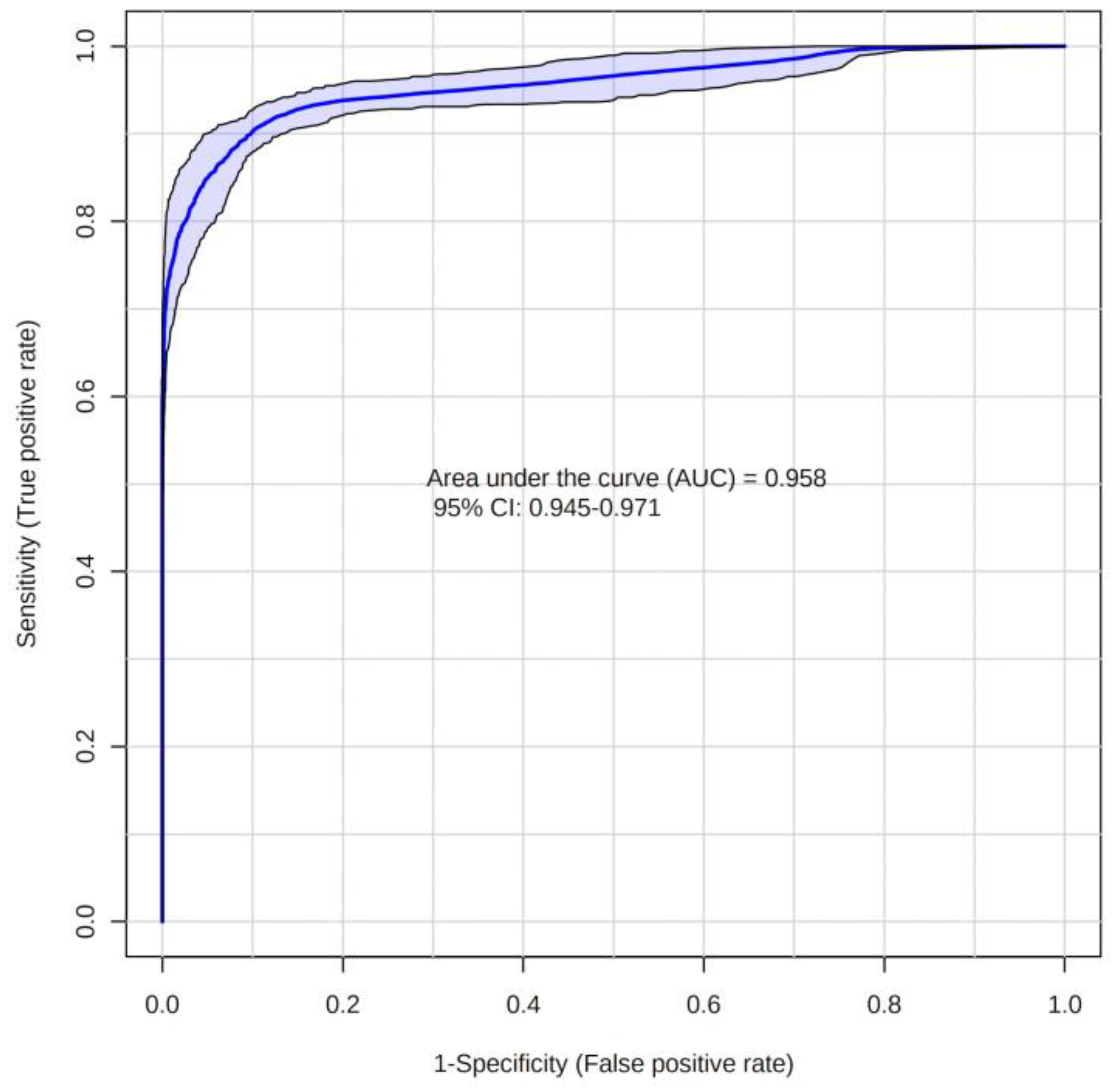
2.2. Metabolites
3. Discussion
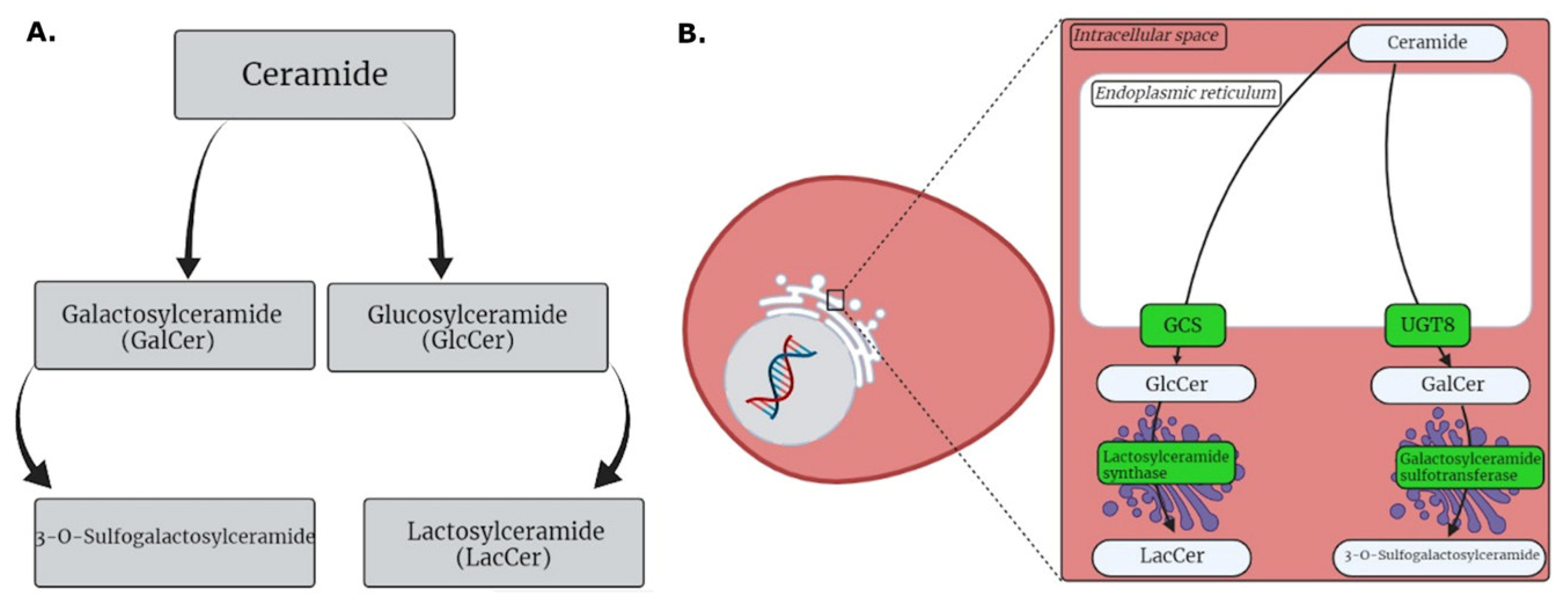
3.1. HG MGM Metabolites
3.2. LG MGM Metabolites
4. Materials and Methods
4.1. Patients and Samples
4.2. Sample Preparation
4.3. MS Analysis
4.4. Statistical Analysis and Molecular Identification
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Benson, V.S.; Pirie, K.; Green, J.; Casabonne, D.; Beral, V. Lifestyle Factors and Primary Glioma and Meningioma Tumours in the Million Women Study Cohort. Br. J. Cancer 2008, 99, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Fathi, A.R.; Roelcke, U. Meningioma. Curr. Neurol. Neurosci. Rep. 2013, 13, 337. [Google Scholar] [CrossRef]
- Wang, N.; Osswald, M. Meningiomas: Overview and New Directions in Therapy. Semin. Neurol. 2018, 38, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Lamszus, K. Meningioma Pathology, Genetics, and Biology. J. Neuropathol. Exp. Neurol. 2004, 63, 275–286. [Google Scholar] [CrossRef]
- Marosi, C.; Hassler, M.; Roessler, K.; Reni, M.; Sant, M.; Mazza, E.; Vecht, C. Meningioma. Crit. Rev. Oncol. Hematol. 2008, 67, 153–171. [Google Scholar] [CrossRef]
- Wiemels, J.; Wrensch, M.; Claus, E.B. Epidemiology and Etiology of Meningioma. J. Neurooncol. 2010, 99, 307–314. [Google Scholar] [CrossRef]
- Mashayekhi, F.; Saberi, A.; Mashayekhi, S. Serum TIMP1 and TIMP2 Concentration in Patients with Different Grades of Meningioma. Clin. Neurol. Neurosurg. 2018, 170, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Buerki, R.A.; Horbinski, C.M.; Kruser, T.; Horowitz, P.M.; James, C.D.; Lukas, R.V. An Overview of Meningiomas. Future Oncol. 2018, 14, 2161–2177. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Ogasawara, C.; Philbrick, B.D.; Cory Adamson, D. Meningioma: A Review of Epidemiology, Pathology, Diagnosis, Treatment, and Future Directions. Biomedicines 2021, 9, 319. [Google Scholar] [CrossRef]
- Goldbrunner, R.; Minniti, G.; Preusser, M.; Jenkinson, M.D.; Sallabanda, K.; Houdart, E.; von Deimling, A.; Stavrinou, P.; Lefranc, F.; Lund-Johansen, M.; et al. EANO Guidelines for the Diagnosis and Treatment of Meningiomas. Lancet Oncol. 2016, 17, e383–e391. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Kim, S.H.; Kim, J.H.; Hwang, S.; Yoo, H.J. Understanding Metabolomics in Biomedical Research. Endocrinol. Metab. 2016, 31, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Gowda, G.A.N.; Zhang, S.; Gu, H.; Asiago, V.; Shanaiah, N.; Raftery, D. Metabolomics-Based Methods for Early Disease Diagnostics. Expert Rev. Mol. Diagn. 2008, 8, 617–633. [Google Scholar] [CrossRef] [PubMed]
- Medina, S.; Dominguez-Perles, R.; Gil, J.I.; Ferreres, F.; Gil-Izquierdo, A. Metabolomics and the Diagnosis of Human Diseases -A Guide to the Markers and Pathophysiological Pathways Affected. Curr. Med. Chem. 2014, 21, 823–848. [Google Scholar] [CrossRef]
- Ogretmen, B.; Hannun, Y.A. Biologically Active Sphingolipids in Cancer Pathogenesis and Treatment. Nat. Rev. Cancer 2004, 4, 604–616. [Google Scholar] [CrossRef]
- Saddoughi, S.A.; Ogretmen, B. Diverse Functions of Ceramide in Cancer Cell Death and Proliferation, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2013; Volume 117. [Google Scholar] [CrossRef]
- Grösch, S.; Schiffmann, S.; Geisslinger, G. Chain Length-Specific Properties of Ceramides. Prog. Lipid. Res. 2012, 51, 50–62. [Google Scholar] [CrossRef]
- Morad, S.A.F.; Cabot, M.C. Ceramide-Orchestrated Signalling in Cancer Cells. Nat. Rev. Cancer 2013, 13, 51–65. [Google Scholar] [CrossRef]
- Spinedi, A.; Di Bartolomeo, S.; Piacentini, M. Apoptosis Induced by N-Hexanoylsphingosine in CHP-100 Cells Associates with Accumulation of Endogenous Ceramide and Is Potentiated by Inhibition of Glucocerebroside Synthesis. Cell Death Differ. 1998, 5, 785–791. [Google Scholar] [CrossRef][Green Version]
- Ogretmen, B.; Hannun, Y.A. Updates on Functions of Ceramide in Chemotherapy-Induced Cell Death and in Multidrug Resistance. Drug Resist. Updat. 2001, 4, 368–377. [Google Scholar] [CrossRef]
- Ryland, L.K.; Fox, T.E.; Liu, X.; Loughran, T.P.; Kester, M. Dysregulation of Sphingolipid Metabolism in Cancer. Cancer Biol. 2011, 11, 138–149. [Google Scholar] [CrossRef]
- Bleicher, R.J.; Cabot, M.C. Glucosylceramide Synthase and Apoptosis. Biochim. Biophys. Acta 2002, 1585, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Lavie, Y.; Cao, H.T.; Bursten, S.L.; Giuliano, A.E.; Cabot, M.C. Accumulation of Glucosylceramides in Multidrug-Resistant Cancer Cells. J. Biol. Chem. 1996, 271, 19530–19536. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Han, T.Y.; Giuliano, A.E.; Ichikawa, S.; Hirabayashi, Y.; Cabot, M.C. Glycosylation of Ceramide Potentiates Cellular Resistance to Tumor Necrosis Factor-α-Induced Apoptosis. Exp. Cell Res. 1999, 252, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Lavie, Y.; Cao, H.T.; Volner, A.; Lucci, A.; Han, T.Y.; Geffen, V.; Giuliano, A.E.; Cabot, M.C. Agents That Reverse Multidrug Resistance, Tamoxifen, Verapamil, and Cyclosporin A, Block Glycosphingolipid Metabolism by Inhibiting Ceramide Glycosylation in Human Cancer Cells. J. Biol. Chem. 1997, 272, 1682–1687. [Google Scholar] [CrossRef] [PubMed]
- Gouazé, V.; Yu, J.Y.; Bleicher, R.J.; Han, T.-Y.; Liu, Y.-Y.; Wang, H.; Gottesman, M.M.; Bitterman, A.; Giuliano, A.E.; Cabot, M.C. Overexpression of Glucosylceramide Synthase and P-Glycoprotein in Cancer Cells Selected for Resistance to Natural Product Chemotherapy. Mol. Cancer 2004, 3, 633–639. [Google Scholar] [CrossRef]
- Beier, U.H.; Görögh, T. Implications of Galactocerebrosidase and Galactosylcerebroside Metabolism in Cancer Cells. Int. J. Cancer 2005, 115, 6–10. [Google Scholar] [CrossRef]
- Sprong, H.; Kruithof, B.; Leijendekker, R.; Slot, J.W.; Van Meer, G.; Van Der Sluijs, P. UDP-Galactose:Ceramide Galactosyltransferase is a Class I Integral Membrane Protein of the Endoplasmic Reticulum. J. Biol. Chem. 1998, 273, 25880–25888. [Google Scholar] [CrossRef]
- Dziȩgiel, P.; Owczarek, T.; Plazùk, E.; Gomukiewicz, A.; Majchrzak, M.; Podhorska-Okoów, M.; Driouch, K.; Lidereau, R.; Ugorski, M. Ceramide Galactosyltransferase (UGT8) Is a Molecular Marker of Breast Cancer Malignancy and Lung Metastases. Br. J. Cancer 2010, 103, 524–531. [Google Scholar] [CrossRef] [PubMed]
- Owczarek, T.B.; Suchanski, J.; Pula, B.; Kmiecik, A.M.; Chadalski, M.; Jethon, A.; Dziegiel, P.; Ugorski, M. Galactosylceramide Affects Tumorigenic and Metastatic Properties of Breast Cancer Cells as an Anti-Apoptotic Molecule. PLoS ONE 2013, 8, e84191. [Google Scholar] [CrossRef]
- Kok, J.W.; Veldman, R.J.; Klappe, K.; Koning, H.; Filipeanu, C.M.; Müller, M.M. Differential Expression of Sphingolipids in MRP1 Overexpressing HT29 Cells. Int. J. Cancer 2000, 87, 172–178. [Google Scholar] [CrossRef]
- Wegner, M.S.; Gruber, L.; Mattjus, P.; Geisslinger, G.; Grösch, S. The UDP-Glucose Ceramide Glycosyltransferase (UGCG) and the Link to Multidrug Resistance Protein 1 (MDR1). BMC Cancer 2018, 18, 153. [Google Scholar] [CrossRef]
- Chatterjee, S. Regulation of Synthesis of Lactosylceramide in Normal and Tumor Proximal Tubular Cells. Biochim. Biophys. Acta Lipids Lipid Metab. 1993, 1167, 339–344. [Google Scholar] [CrossRef]
- Hummel, I.; Klappe, K.; Kok, J.W. Up-Regulation of Lactosylceramide Synthase in MDR1 Overexpressing Human Liver Tumour Cells. FEBS Lett. 2005, 579, 3381–3384. [Google Scholar] [CrossRef]
- Peng, W.; Tan, S.; Xu, Y.; Wang, L.; Qiu, D.; Cheng, C.; Lin, Y.; Liu, C.; Li, Z.; Li, Y.; et al. LC-MS/MS Metabolome Analysis Detects the Changes in the Lipid Metabolic Profiles of DMMR and PMMR Cells. Oncol. Rep. 2018, 40, 1026–1034. [Google Scholar] [CrossRef] [PubMed]
- Gasa, S.; Časl, M.-T.; Makita, A.; Sakakibara, N.; KoyanagiI, T.; Atsuta, T. Presence and Characterization of Glycolipid Sulfotransferase in Human Cancer Serum. Eur. J. Biochem. 1990, 189, 301–306. [Google Scholar] [CrossRef]
- Gasa, S.; Casl, M.T.; Jin, T.; Kamio, K.; Uehara, Y.; Miyazaki, T.; Makita, A. Elevated Serum Level of Glycolipid Sulfotransferase in Patients with Hepatocellular Carcinoma. Cancer Lett. 1991, 59, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Honke, K.; Tsuda, M.; Hirahara, Y.; Miyao, N.; Tsukamoto, T.; Satoh, M.; Wada, Y. Cancer-Associated Expression of Glycolipid Sulfotransferase Gene in Human Renal Cell Carcinoma Cells. Cancer Res. 1998, 58, 3800–3805. [Google Scholar]
- Shi, B.-Z.; Hu, P.; Geng, F.; He, P.-J.; Wu, X.-Z. Gal3ST-2 Involved in Tumor Metastasis Process by Regulation of Adhesion Ability to Selectins and Expression of Integrins. Biochem. Biophys. Res. Commun. 2005, 332, 934–940. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Xu, Y.; Fitch, W.L.; Zheng, M.; Merritt, R.E.; Shrager, J.B.; Zhang, W.; Dill, D.L.; Peltz, G.; Hoang, C.D. Liquid Chromatography/Mass Spectrometry Methods for Measuring Dipeptide Abundance in Non-Small-Cell Lung Cancer. Rapid. Commun. Mass Spectrom. 2013, 27, 2091–2098. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, H.; Hirayama, A.; Shoji, F.; Maruyama, M.; Suzuki, K.; Yamanaka-Okumura, H.; Tatano, H.; Morine, Y.; Soga, T.; Shimada, M.; et al. Comprehensive Dipeptide Analysis Revealed Cancer-Specific Profile in the Liver of Patients with Hepatocellular Carcinoma and Hepatitis. Metabolites 2020, 10, 442. [Google Scholar] [CrossRef]
- Kuo, M.T.; Chen, H.H.W.; Feun, L.G.; Savaraj, N. Targeting the Proline–Glutamine–Asparagine–Arginine Metabolic Axis in Amino Acid Starvation Cancer Therapy. Pharmaceuticals 2021, 14, 72. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Luo, L.; Xie, Y.; Zhao, Y.; Yao, J.; Liu, X. PYCR1 Knockdown Inhibits the Proliferation, Migration, and Invasion by Affecting JAK/STAT Signaling Pathway in Lung Adenocarcinoma. Mol. Carcinog. 2020, 59, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Dillon, B.J.; Prieto, V.G.; Curley, S.A.; Ensor, C.M.; Holtsberg, F.W.; Bomalaski, J.S.; Clark, M.A. Incidence and Distribution of Argininosuccinate Synthetase Deficiency in Human Cancers: A Method for Identifying Cancers Sensitive to Arginine Deprivation. Cancer 2004, 100, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Krahling, S.; Callahan, M.K.; Williamson, P.; Schlegel, R.A. Exposure of Phosphatidylserine Is a General Feature in the Phagocytosis of Apoptotic Lymphocytes by Macrophages. Cell Death Differ. 1999, 6, 183–189. [Google Scholar] [CrossRef]
- Birge, R.B.; Boeltz, S.; Kumar, S.; Carlson, J.; Wanderley, J.; Calianese, D.; Barcinski, M.; Brekken, R.A.; Huang, X.; Hutchins, J.T.; et al. Phosphatidylserine Is a Global Immunosuppressive Signal in Efferocytosis, Infectious Disease, and Cancer. Cell Death Differ. 2016, 23, 962–978. [Google Scholar] [CrossRef]
- Chang, W.; Fa, H.; Xiao, D.; Wang, J. Targeting Phosphatidylserine for Cancer Therapy: Prospects and Challenges. Theranostics 2020, 10, 9214–9229. [Google Scholar] [CrossRef]
- Martin, S.J.; Reutelingsperger, C.P.M.; McGahon, A.J.; Rader, J.A.; Van Schie, R.C.A.A.; LaFace, D.M.; Green, D.R. Early Redistribution of Plasma Membrane Phosphatidylserine Is a General Feature of Apoptosis Regardless of the Initiating Stimulus: Inhibition by Overexpression of BCL-2 and Abl. J. Exp. Med. 1995, 182, 1545–1556. [Google Scholar] [CrossRef]
- Fadok, V.A.; Bratton, D.L.; Rose, D.M.; Pearson, A.; Ezekewitz, R.A.B.; Henson, P.M. A Receptor for Phosphatidylserine-Specific Clearance of Apoptotic Cells. Nature 2000, 405, 85–90. [Google Scholar] [CrossRef]
- Huynh, M.-L.N.; Fadok, V.A.; Henson, P.M. Phosphatidylserine-Dependent Ingestion of Apoptotic Cells Promotes TGF-Β1 Secretion and the Resolution of Inflammation. J. Clin. Investig. 2002, 109, 41–50. [Google Scholar] [CrossRef]
- Voll, R.E.; Herrmann, M.; Roth, E.A.; Stach, C.; Kalden, J.R.; Girkontaite, I. Immunosuppressive Effects of Apoptotic Cells. Nature 1997, 390, 350–351. [Google Scholar] [CrossRef]
- Utsugi, T.; Schroit, A.J.; Connor, J.; Bucana, C.D.; Fidler, I.J. Elevated Expression of Phosphatidylserine in the Outer Membrane Leaflet of Human Tumor Cells and Recognition by Activated Human Blood Monocytes. Cancer Res. 1991, 51, 3062–3066. [Google Scholar] [PubMed]
- Vallabhapurapu, S.D.; Blanco, V.M.; Sulaiman, M.K.; Vallabhapurapu, S.L.; Chu, Z.; Franco, R.S.; Qi, X. Variation in Human Cancer Cell External Phosphatidylserine Is Regulated by Flippase Activity and Intracellular Calcium. Oncotarget 2015, 6, 34375–34388. [Google Scholar] [CrossRef] [PubMed]
- Chalasani, P.; Marron, M.; Roe, D.; Clarke, K.; Iannone, M.; Livingston, R.B.; Shan, J.S.; Stopeck, A.T. A Phase I Clinical Trial of Bavituximab and Paclitaxel in Patients with HER2 Negative Metastatic Breast Cancer. Cancer Med. 2015, 4, 1051–1059. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Li, L.; Thorpe, P.E.; Yopp, A.C.; Brekken, R.A.; Huang, X. Antibody-Mediated Blockade of Phosphatidylserine Enhances the Antitumor Effect of Sorafenib in Hepatocellular Carcinomas Xenografts. Ann. Surg. Oncol. 2016, 23 (Suppl. S5), 583–591. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.; Arriaga, Y.; Anandam, J.; Karri, S.; Syed, S.; Verma, U.; Abdelnaby, A.; Raja, G.; Dong, Y.; Beg, M.S.; et al. A Phase I Clinical Trial of the Phosphatidylserine-Targeting Antibody Bavituximab in Combination with Radiation Therapy and Capecitabine in the Preoperative Treatment of Rectal Adenocarcinoma. Am. J. Clin. Oncol. Cancer Clin. Trials 2018, 41, 972–976. [Google Scholar] [CrossRef] [PubMed]
- Elvas, F.; Stroobants, S.; Wyffels, L. Phosphatidylethanolamine Targeting for Cell Death Imaging in Early Treatment Response Evaluation and Disease Diagnosis. Apoptosis 2017, 22, 971–987. [Google Scholar] [CrossRef]
- Broughton, L.J.; Crow, C.; Maraveyas, A.; Madden, L.A. Duramycin-Induced Calcium Release in Cancer Cells. Anticancer Drugs 2016, 27, 173–182. [Google Scholar] [CrossRef]
- Cheng, F.; Wen, Z.; Feng, X.; Wang, X.; Chen, Y. A Serum Lipidomic Strategy Revealed Potential Lipid Biomarkers for Early-Stage Cervical Cancer. Life Sci. 2020, 260, 118489. [Google Scholar] [CrossRef]
- Lu, Y.; Chen, J.; Huang, C.; Li, N.; Zou, L.; Chia, S.E.; Chen, S.; Yu, K.; Ling, Q.; Cheng, Q.; et al. Comparison of Hepatic and Serum Lipid Signatures in Hepatocellular Carcinoma Patients Leads to the Discovery of Diagnostic and Prognostic Biomarkers. Oncotarget 2018, 9, 5032–5043. [Google Scholar] [CrossRef]
- Goto, K.; Hozumi, Y.; Nakano, T.; Saino-Saito, S.; Martelli, A.M. Lipid Messenger, Diacylglycerol, and Its Regulator, Diacylglycerol Kinase, in Cells, Organs, and Animals: History and Perspective. Tohoku J. Exp. Med. 2008, 214, 199–212. [Google Scholar] [CrossRef]
- Wakelam, M.J.O. Diacylglycerol—When Is It an Intracellular Messenger? Biochim. Biophys. Acta 1998, 1436, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Fazio, A.; Obeng, E.O.; Rusciano, I.; Marvi, M.V.; Zoli, M.; Mongiorgi, S.; Ramazzotti, G.; Follo, M.Y.; McCubrey, J.A.; Cocco, L.; et al. Subcellular Localization Relevance and Cancer-Associated Mechanisms of Diacylglycerol Kinases. Int. J. Mol. Sci. 2020, 21, 5297. [Google Scholar] [CrossRef] [PubMed]
- Filigheddu, N.; Cutrupi, S.; Porporato, P.E.; Riboni, F.; Baldanzi, G.; Chianale, F.; Fortina, E.; Piantanida, P.; De Bortoli, M.; Vacca, G.; et al. Diacylglycerol Kinase Is Required for HGF-Induced Invasiveness and Anchorage-Independent Growth of MDA-MB-231 Breast Cancer Cells. Anticancer Res. 2007, 27, 1489–1492. [Google Scholar] [PubMed]
- Filigheddu, N.; Sampietro, S.; Chianale, F.; Porporato, P.E.; Gaggianesi, M.; Gregnanin, I.; Rainero, E.; Ferrara, M.; Perego, B.; Riboni, F.; et al. Diacylglycerol Kinase α Mediatses 17-β-Estradiol-Induced Proliferation, Motility, and Anchorage-Independent Growth of Hec-1A Endometrial Cancer Cell Line through the G Protein-Coupled Estrogen Receptor GPR30. Cell Signal 2011, 23, 1988–1996. [Google Scholar] [CrossRef]
- Yanagisawa, K.; Yasuda, S.; Kai, M.; Imai, S.; Yamada, K.; Yamashita, T.; Jimbow, K.; Kanoh, H.; Sakane, F. Diacylglycerol Kinase α Suppresses Tumor Necrosis Factor-α-Induced Apoptosis of Human Melanoma Cells through NF-ΚB Activation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2007, 1771, 462–474. [Google Scholar] [CrossRef]
- Prinz, P.U.; Mendler, A.N.; Masouris, I.; Durner, L.; Oberneder, R.; Noessner, E. High DGK-α and Disabled MAPK Pathways Cause Dysfunction of Human Tumor-Infiltrating CD8 + T Cells That Is Reversible by Pharmacologic Intervention. J. Immunol. 2012, 188, 5990–6000. [Google Scholar] [CrossRef]
- Prinz, P.U.; Mendler, A.N.; Brech, D.; Masouris, I.; Oberneder, R.; Noessner, E. NK-Cell Dysfunction in Human Renal Carcinoma Reveals Diacylglycerol Kinase as Key Regulator and Target for Therapeutic Intervention. Int. J. Cancer 2014, 135, 1832–1841. [Google Scholar] [CrossRef]
- Jung, I.-Y.; Kim, Y.-Y.; Yu, H.-S.; Lee, M.; Kim, S.; Lee, J. CRISPR/Cas9-Mediated Knockout of DGK Improves Antitumor Activities of Human T Cells. Cancer Res. 2018, 78, 4692–4703. [Google Scholar] [CrossRef]
- Dominguez, C.L.; Floyd, D.H.; Xiao, A.; Mullins, G.R.; Kefas, B.A.; Xin, W.; Yacur, M.N.; Abounader, R.; Lee, J.K.; Wilson, G.M.; et al. Diacylglycerol Kinase α Is a Critical Signaling Node and Novel Therapeutic Target in Glioblastoma and Other Cancers. Cancer Discov. 2013, 3, 782–797. [Google Scholar] [CrossRef]
- Takeishi, K.; Taketomi, A.; Shirabe, K.; Toshima, T.; Motomura, T.; Ikegami, T.; Yoshizumi, T.; Sakane, F.; Maehara, Y. Diacylglycerol Kinase Alpha Enhances Hepatocellular Carcinoma Progression by Activation of Ras-Raf-MEK-ERK Pathway. J. Hepatol. 2012, 57, 77–83. [Google Scholar] [CrossRef]
- Torres-Ayuso, P.; Daza-Martín, M.; Martín-Pérez, J.; Ávila-Flores, A.; Mérida, I. Diacylglycerol Kinase a Promotes 3D Cancer Cell Growth and Limits Drug Sensitivity through Functional Interaction with Src. Oncotarget 2014, 5, 9710–9726. [Google Scholar] [CrossRef] [PubMed]
- Yew, A.; Trang, A.; Nagasawa, D.T.; Spasic, M.; Choy, W.; Garcia, H.M.; Yang, I. Chromosomal Alterations, Prognostic Factors, and Targeted Molecular Therapies for Malignant Meningiomas. J. Clin. Neurosci. 2013, 20, 17–22. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2014-2018. Neuro Oncol. 2021, 23 (Suppl. S2), III1–III105. [Google Scholar] [CrossRef] [PubMed]
- Melo, C.F.O.R.; Delafiori, J.; de Oliveira, D.N.; Guerreiro, T.M.; Esteves, C.Z.; Lima, E.O.; Pando-Robles, V.; Catharino, R.R.; Milanez, G.P.; do Nascimento, G.M.; et al. Serum Metabolic Alterations upon ZIKA Infection. Front. Microbiol. 2017, 8, 1954, Correction in Front. Microbiol. 2017, 8, 2373. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the Gap between Raw Spectra and Functional Insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
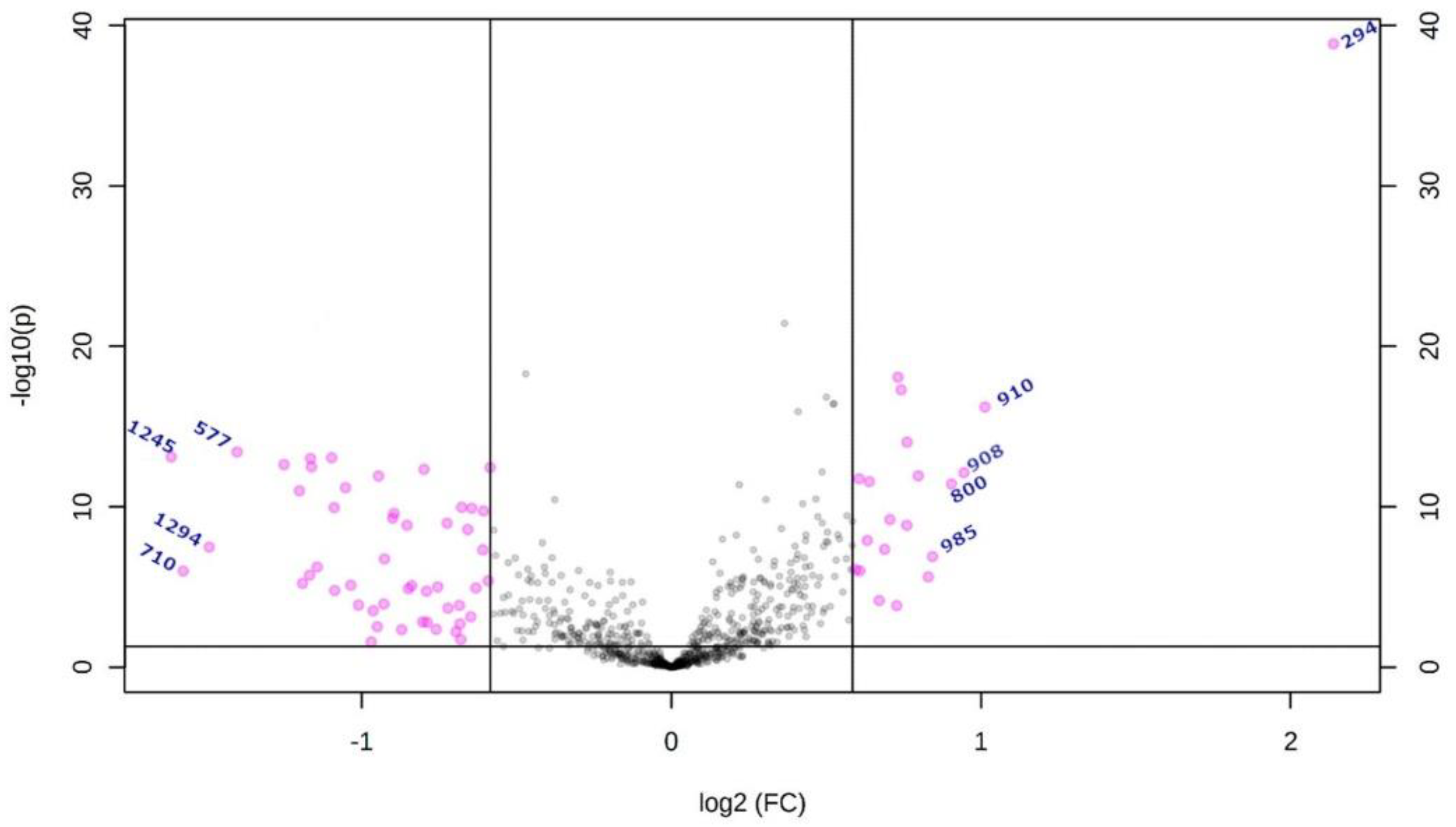
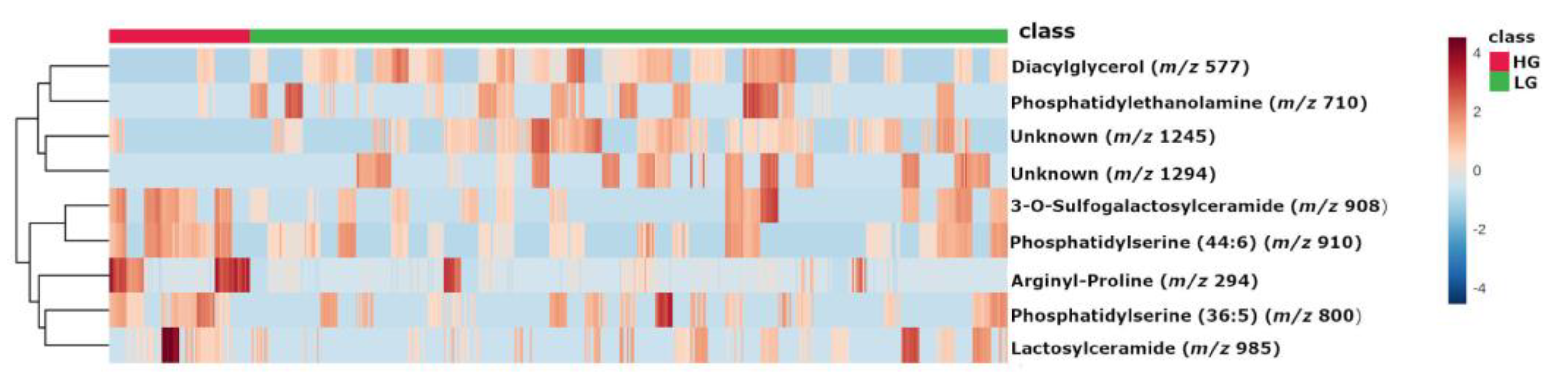
| m/z | Database ID | Metabolite | Formula | Adducts | MS/MS | Log2(FC) | |
|---|---|---|---|---|---|---|---|
| High-grade | 294 | MID85632 | Arginyl-Proline | C11H21N5O3 | [M + Na]+ | 268-254-250-236-266 | 2.1383 |
| 910 | HMDB0116777 | PS (44:6) # | C50H86NO10P | [M + NH4]+ | 184-104-125-720-495 | 1.0127 | |
| 908 | HMDB0012318 | 3-O-Sulfogalactosylceramide (42:2) # | C48H91NO11S | [M + NH4]+ | 494-95-81-604-109-184 | 0.94786 | |
| 800 | LMGP03010137 | PS (36:5) # | C42H72NO10P | [M + NH4]+ | 184-119-368-135-86-437 | 0.90497 | |
| 985 | HMDB0011594 | LacCer (40:1) # | C52H99NO13 | [M + K]+ | 360-338-369-648-437 | 0.84326 | |
| Low-grade | 577 | MID58656 MID58684 | DG (34:1) # | C37H70O5 | [M + H-H2O]+ | 95-81-57-69-109-339-121-419-135 | −1.4045 |
| 1294 | - | Unknown | - | - | 1275-1262-1243-798 | −1.4927 | |
| 710 | MID40644 | PE (32:0) # | C37H74NO8P | [M + NH4]+ | 661-655-678-692-674-642-341-454 | −1.5768 | |
| 1245 | - | Unknown | - | - | 112-907-184-720-338-625-303 | −1.6156 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurokawa, G.A.; Hamamoto Filho, P.T.; Delafiori, J.; Galvani, A.F.; de Oliveira, A.N.; Dias-Audibert, F.L.; Catharino, R.R.; Pardini, M.I.M.C.; Zanini, M.A.; Lima, E.d.O.; et al. Differential Plasma Metabolites between High- and Low-Grade Meningioma Cases. Int. J. Mol. Sci. 2023, 24, 394. https://doi.org/10.3390/ijms24010394
Kurokawa GA, Hamamoto Filho PT, Delafiori J, Galvani AF, de Oliveira AN, Dias-Audibert FL, Catharino RR, Pardini MIMC, Zanini MA, Lima EdO, et al. Differential Plasma Metabolites between High- and Low-Grade Meningioma Cases. International Journal of Molecular Sciences. 2023; 24(1):394. https://doi.org/10.3390/ijms24010394
Chicago/Turabian StyleKurokawa, Gabriel A., Pedro T. Hamamoto Filho, Jeany Delafiori, Aline F. Galvani, Arthur N. de Oliveira, Flávia L. Dias-Audibert, Rodrigo R. Catharino, Maria Inês M. C. Pardini, Marco A. Zanini, Estela de O. Lima, and et al. 2023. "Differential Plasma Metabolites between High- and Low-Grade Meningioma Cases" International Journal of Molecular Sciences 24, no. 1: 394. https://doi.org/10.3390/ijms24010394
APA StyleKurokawa, G. A., Hamamoto Filho, P. T., Delafiori, J., Galvani, A. F., de Oliveira, A. N., Dias-Audibert, F. L., Catharino, R. R., Pardini, M. I. M. C., Zanini, M. A., Lima, E. d. O., & Ferrasi, A. C. (2023). Differential Plasma Metabolites between High- and Low-Grade Meningioma Cases. International Journal of Molecular Sciences, 24(1), 394. https://doi.org/10.3390/ijms24010394








