Abstract
The stemness and metastasis of cancer cells are crucial features in determining cancer progression. Argonaute-2 (AGO2) overexpression was reported to be associated with microRNA (miRNA) biogenesis, supporting the self-renewal and differentiation characteristics of cancer stem cells (CSCs). Ursolic acid (UA), a triterpene compound, has multiple biological functions, including anticancer activity. In this study, we find that UA inhibits the proliferation of MDA-MB-231 and MCF-7 breast cancer cell lines using the CCK-8 assay. UA induced a significant decrease in the fraction of CSC in which it was examined by changes in the expression of stemness biomarkers, including the Nanog and Oct4 genes. UA altered invasion and migration capacities by significant decreases in the levels of epithelial-to-mesenchymal transition (EMT) proteins of slug and vimentin. Furthermore, the co-reduction in oncogenic miRNA levels (miR-9 and miR-221) was a result of the down-modulation in AGO2 in breast cancer cells in vitro. Mechanically, UA increases PTEN expression to inactivate the FAK/PI3K/Akt/mTOR signaling pathway and the decreased level of c-Myc in quantitative RT-PCR and Western blot imaging analyses. Our current understanding of the anticancer potential of UA in interrupting between EMT programming and the state of CSC suggests that UA can contribute to improvements in the clinical practice of breast cancer.
1. Introduction
The dissemination and colonization of distant lymph node organs in cancer cells that acquire genetic and/or epigenetic alterations in primary tumor cells are the primary causes of most cancer deaths [1,2]. Cancer stem cell (CSC) scenarios are supported by small side population (SP) cells, located within the tumor niche, and are responsible for maintaining the propagation of new tumor cells [3]. SP cells enable self-renewal, initiation, and differentiation in the cancer colony setting that has the advantage of highly invasive, metastatic, and drug-resistant potential of new tumor ones. The multidrug-resistant transporter protein, ATP-binding cassette superfamily G member 2 (ABCG2), has been implicated as enriched expression in CSC chemoresistance [4]. In our previous studies, in an attempt to isolate breast CSC cells (BCSCs), we separated tumor SP cells based on this transport gene with the high enzymatic activity phenotype responsible for the efflux of the fluorescent dye, Hoechst 33342, and then validated by examination of overexpression of stem cell transcription factors, including Nanog, Oct4, and CD133, which are reported as specific markers in representative CSCs [5,6]. Furthermore, CSCs with high levels of pluripotency are associated with oncogenic microRNA (miRNA) molecules, their aberrant expressions provided proliferative and repair capacities of DNA to increase resistance to apoptosis and allow them to survive in the face of anticancer treatment [7].
During recent decades, scientific research has made great efforts to identify deregulated RNA-binding proteins (RBPs) in cancer cell development. Argonaute-2 (AGO2), one of the members of the Argonaute family, is the crucial component of the RNA-induced silencing complex (RISC) with catalytic activity during small-RNA-guided gene silencing processes [8]. As a key regulator of miRNA biogenesis, AGO2 plays multiple roles in aspects of tumor cell tumorigenesis and progression, as well as has been reported to mediate tumor-promoting transcriptomic changes during carcinogenesis, including prostate, breast, gastrointestinal, ovarian, and endometrial cancers of human organs [9,10]. Mechanically, the AGO2-mediated RISC complex promotes cancer cell outgrowth, survival, and motility by suppressing the tumor suppressor PTEN (phosphatase and tensin homologue on chromosome 10) and activates phosphatidylinositol-3-kinase (PI3K)/AKT signaling [11]. The loss of PTEN is correlated with a human breast and prostate stem/progenitor subpopulation accompanied by mesenchymal characteristics with epithelial-mesenchymal transition (EMT) and macrometastasis [12,13].
Pentacyclic triterpenes (PT) are derived from five carbon isoprene units of medicinal herbs, fruits, and vegetables that are widely recognized to have a wide range of biological activities in the treatment of diseases, including diabetic complications [14,15], cardiovascular diseases [16], as well as potential immunomodulators to express anti-inflammatory effects [17,18]. One of the PT molecules, ursolic acid (UA, 3β-hydroxy-urs-12-en-28-oic acid), has been devoted to the antitumor characterization of in vitro and in vivo studies and clinical applications, supporting multifunctional bioactivities in cancer [19,20]. Among its numerous biological properties, in the present study, the antitumor effect of UA is of particular interest due to the diversity that interferes with BCSC biogenesis, shedding light on the pilot of UA for the restriction of AGO2-mediated miRNA expression in triple-negative breast cancer (TNBC). TNBC encompasses a subtype of breast cancers that lack the expression of the estrogen receptor (ER), the progesterone receptor (PR), and the human epidermal growth factor receptor 2 (HER2). Compared with other human breast cancer cell lines, TNBC MDA-MB-231 cells are known to have a higher proportion of CSC and have more invasive phenotypes [21]. Deregulation of stem cell-related signaling pathways, such as NF-kB, Wnt/β-catenin, Notch, and PI3K/p-Akt, lead to chemoresistance, recurrence, and metastasis in TNBC [22]. In this study, we explored the down-modulation of AGO2 in breast cancer treated with UA, which led us to understand the increase in the level of PTEN inversely associated with AGO2 expression in inhibiting breast cancer invasiveness. These findings are essential for the establishment of UA-permissive cancer treatment relevant to the novel role of the anticancer action of this PT compound in the down-modulation of AGO2 to improve the invasiveness of CSC-imposed tumor cells and the development of therapeutic approaches to improve future clinical cancer therapy.
2. Results
2.1. UA Inhibited Cell Proliferation in Breast Cancer Cell Lines
The effect of UA on the cell viability of different breast cell lines was evaluated using the CCK-8 assay. We used an achievably soluble dosing and tried several concentrations of UA for 24 and 48 h, respectively. UA caused a time (24 and 48 h)- and dosage (0–50 μM)-dependent inhibition of cell proliferation in MDA-MB-231 (Figure 1A) and MCF-7 (Figure 1B) cell lines (p < 0.001). At 24 h, the IC50 value of UA was greater than 30 μM for these two breast cancer cell lines. When incubation with UA was extended for 48 h, the IC50 values of UA on MDA-MB-231 and MCF-7 were calculated as 24.0 ± 1.8 μM and 29.2 ± 2.1 μM, respectively. On the contrary, as shown in Figure 1C, for HBL-100 human breast epithelial cells, treatment with more than 40 μM UA for 48 h decreased cell viability by less than 30% (74.7 ± 8.7% and 70.0 ± 6.8 with respect to 40 and 50 μM UA). These findings suggested that the MCF-7 and MDA-MB-231 carcinoma cell lines were more sensitive to UA treatment compared with the non-carcinoma cell line HBL-100. Furthermore, according to the status of the three most commonly used conventionally, namely ER, PR, and HER2, luminal A subtype MCF-7 and TNBC subtype MDA-MB-231 were selected for further study to investigate the inhibitory effects of UA on CSC, EMT and metastasis and to elucidate its role in the decrease in the signaling pathway with respect to the properties of CSC.
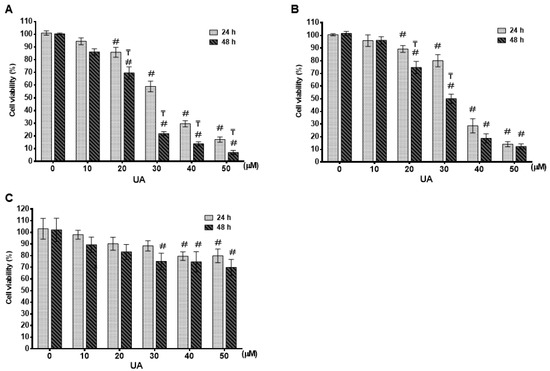
Figure 1.
Effect of UA on cell survival in different breast cancer cells: Cell survival was evaluated by MTT assay after 24 h and 48 h of UA treatment in breast cancer cell lines of MDA-MB-231 in (A) and MCF-7 in (B) and normal mammary epithelial HBL-100 cell line in (C) with different concentrations as indicated. The control group (0 µM) did not receive UA treatment (100% cell viability). Data are shown as mean ± S.D for three independent experiments. #, indicates significant differences from the control; ₸, indicates significant differences compared to the respective concentrations; # and ₸ indicate significant differences at p < 0.05.
2.2. UA Decreases the Fraction of Breast Cancer Stem Cells
In adult tissues or organs, the proportion of normal somatic stem cells is extremely rare. As previously stated, less than 0.01% (only an estimated 1 in 10,000 to 15,000) of hematopoietic cells are hematopoietic stem cells (HSC) in the HSC niche [23]. However, these induced pluripotent stem cells can be directed to CSC where an appropriate tumor microenvironment is present in the cancer niche and then lead to the fate of self-renewal and differentiation to perpetuate tumor regeneration of clonal evolution in cancer progression, and eventually developing drug resistance of the newly re-initiated cancer cell clone [24]. Therefore, we examine whether UA could influence the decreased proportion of CSC genesis of the MCF-7 and MDA-MB-231 breast cancer cell lines. Using Hoechst 33342 dye and fluorescence-activated cell sorting (FACS) of cancer cells treated with 20 µM reserpine, we concluded that after 48 h of incubation, UA significantly decreased the percentages of ABCG2-positive BCSCs, representing approximately 86% (from 2.3% to 0.4%) in MDA-MB-231 cells and 78% (from 2.6% to 0.6%) in the flow cytometry assay (Figure 2A,B). Furthermore, the significantly decreased level of AGO2 due to UA treatment in both breast cancer cell lines was estimated from Western blot (Figure 2C) and qRT-PCR analyzes (Figure 2D).
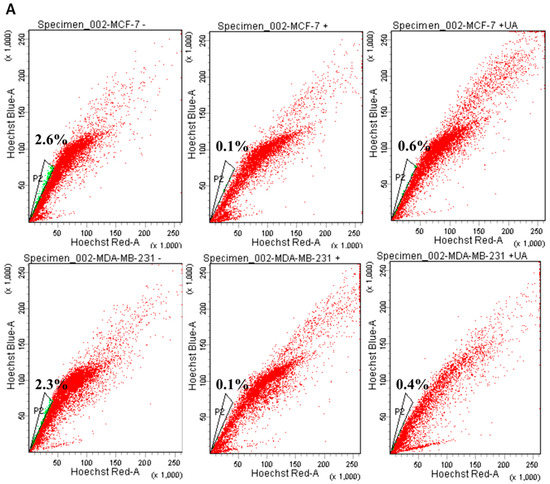
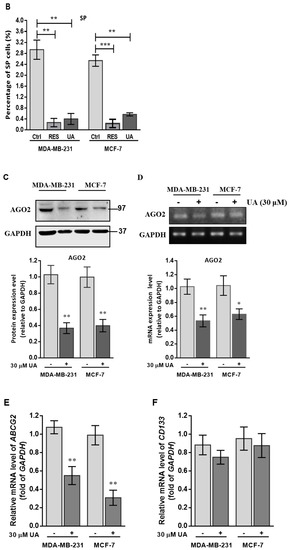
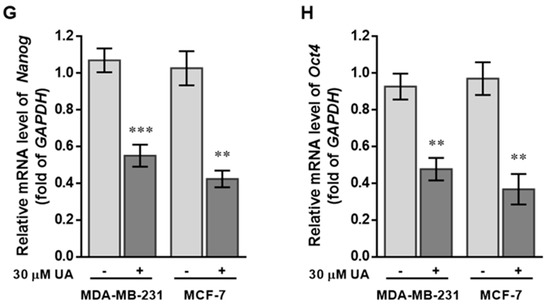
Figure 2.
UA reduces the fraction of SP in breast cancer cell lines: (A) Representative examples of cancer stem cells (SP cells) were exploited by the flow-activated cell sorting (FACS) technique. Cells were stained with Hoechst 33342 dye in the absence (left) or presence (middle) of 50 µM reserpine and 30 μM UA (right) was evaluated by FACS. The uptake of Hoechst 33342 dye occurs uniformly in all cells through passive diffusion (shown in red), SP was detected by the loss of the fluorescent population (shown in green) after treatment with the ABCG2 inhibitor (RES) or UA. (B) The relative percentage of SP was estimated in human MDA-MB-231 and MCF-7 cell lines stained with Hoechst 33342 after incubation with UA for 48 h. (C) Western blot image and (D) reverse transcription-PCR (RT-PCR) data indicate that AGO2 levels in MDA-MB-231 and MCF-7 cell lines were significantly reduced after treatment with UA relative to control cells. (E) Reduced ABCG2 associated with lower levels of CSC-related genes encoding (F) CD133, (G) Nanog, and (H) Oct4 in breast cancer cell line after UA treatment compared with the same cell line without UA treatment. GAPDH was used as an internal control for the detection of mRNA transcripts. Data were presented as mean ± SD from three independent experiments. * p < 0.05; ** p < 0.01; *** p < 0.001 vs. the untreated control.
Increasing evidence has shown that CSCs are represented by SP cells associated with elevated levels of AGO2 and embryonic stem cell-specific factors, including the ABCG2, Nanog, Oct4, and CD133 genes [25], we determined the relative transcripts of the genes in both cell lines, using real-time RT-PCR analysis and β-actin gene as internal standard for quantitative comparison (Figure 2E–H). Significantly decreased levels of mRNA transcripts were about 0.5-fold lower with respect to the ABCG2 gene detected in both BC cells treated with UA compared to the control group (Figure 2E), as well as a significant reduction in CSC markers, including the Nanog and Oct4 genes (by >60%) in cell lines treated with 30 μM UA compared to the control (Figure 2G,H). Together, these findings indicated that UA down-modulates AGO2 to restrict the renewal properties of CSC.
2.3. Sustained Exposure to UA Suppresses Migration and Invasion of Breast Cancer
The CSC theory of cancer progression indicated that this subpopulation of cells plays a key role in driving tumor aggressiveness within the new tumor subclone, and therefore we examined the suppressive function of UA by reducing CSC to inhibit tumor cell metastasis of both breast cancer cell lines. As shown in Figure 3A, invasion and migration capabilities decreased significantly in both MDA-MB-231 and MCF-7 cells treated with UA for 48 h using the Transwell chambers assay. Compared with the control group, MDA-MB-231 cells treated with UA have significantly lower percentages of invasion and migration capacity by >40%. Similar results were obtained in MCF-7 cells treated with UA compared to those detected in the control group, showing a decrease in invasion and migration by >60% (Figure 3B,C).
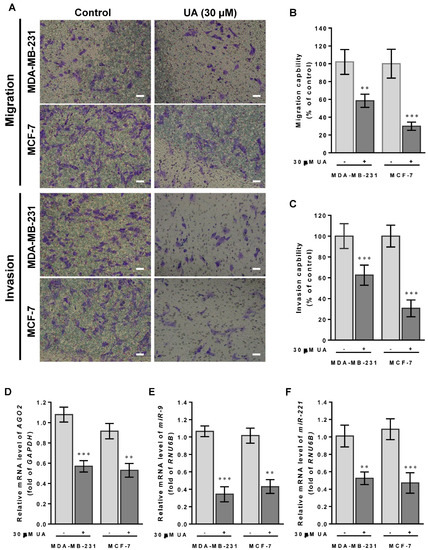
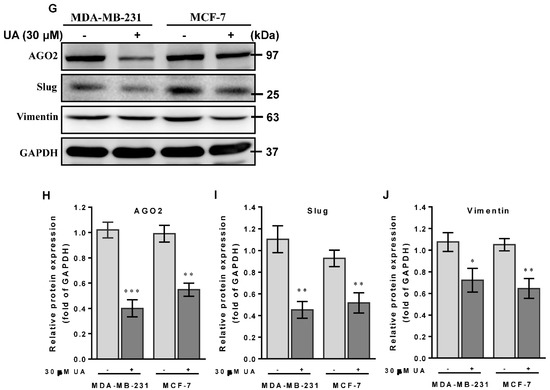
Figure 3.
UA suppresses breast cancer cell migration and invasion: (A) Effects of UA on inhibition of cancer cell migration (top panel) and invasion (lower panel) for MDA-MB-231 and MCF-7 cells that were analyzed by the Transwell assay; scale bar = 50 μm. Quantification of migrated cells in (B) and invaded cells in (C) was presented as mean ± SD of three independent experiments. ** p < 0.01; *** p < 0.001 vs. untreated control. (D) Reduction in the AGO2 gene and EMT-related miRNA in (E) miR-9 and (F) miR-221 in breast cancer cell lines treated with UA compared with the control. Relative expression of miRNAs is quantified using TaqMan-based real-time PCR along with RNU6B. (G) Representative images of Western blot of mesenchymal proteins and densitometric analyzes of AGO2 in (H), slug in (I), and vimentin in (J) were performed using Digital Protein DNA Imagineware. GAPDH was used as an internal control. All experiments were carried out in triplicate and all data are expressed as mean ± SD. * p < 0.05; ** p < 0.01; *** p < 0.001 vs. the untreated control group.
2.4. UA Impedes Tumor Cell Migration and Invasion by Inhibiting EMT-Related miRNAs and Proteins
Furthermore, we have previously shown that miR-9 and miR-221 play an important role in contributing to the invasive phenotype due to the promotion of EMT in breast cancer [5]. Our hypothesis of the mechanism underlying the effects of UA on the decreased ability to invade and migrate casually to link miR-9 and miR-221 was due to the inhibition of AGO2 in breast cancer cells. In addition, because the MDA-MB-231 cell line is intrinsically deficient in E-cadherin expression [26] and the slug protein acts as a promoting transcription factor capable of increasing vimentin in invasive and metastatic cancers [27], we examined whether UA reduces slug and vimentin levels to prohibit migration and invasion of both breast cancer cell lines. As expected, we found a significant reduction in miR-9 (Figure 3E) and miR-221 transcripts (Figure 3F) when AGO2 expression is repressed in the presence of UA treatment compared to the absence of UA in both breast cancer cell lines (Figure 3D). Given that decreased expression of these two miRNAs leads to EMT interruption and inhibits tumor metastasis, along with a reduction in AGO2 (Figure 3G), representative expression levels relevant to mesenchymal proteins, including slug and vimentin, were significantly lower in both breast cancer cell lines treated with UA than in negative control cells in Western blot image analysis (Figure 3H–J). Together, these data determined that UA treatment allows inhibition of breast CSC characteristics and the EMT process to eventually prevent breast cancer aggression.
2.5. UA Enhances PTEN to Downmodulate the FAK/PI3K/Akt/mTOR Signaling Pathway
Recent studies have shown that PTEN gain of function could prevent ABCG2 activity with respect to tumor stem cell characteristics [28]. On the contrary, FAK acts as a key mediator in cell-ECM interactions that contributed to tumorigenesis and progression by stimulating AGO2 to activate the FAK/PI3K/Akt signaling pathway in head and neck squamous cell carcinomas [29]. Mechanically, we examined whether the reduced level of AGO2 due to UA could inactivate FAK/PI3K/Akt/mTOR signaling that retarded cell migration and invasion of breast cancer cells. As shown in Figure 4A, protein data from Western blot analysis showed that, in accordance with the reduction in AGO2 (Figure 4B), an increase in the level of PTEN protein (Figure 4C) was inversely associated with FAK expression levels (Figure 4D) and the phosphorylated status of PI3K (p-P85αTyr467/Tyr199), Akt (p-AktSer473) and mTOR (p-mTOR2481) in the experimental group compared to those of MCF-7 and MA-MB-231 cells without UA treatment (Figure 4F–H). Furthermore, since AGO2 has been reported to stabilize c-Myc to facilitate stem cell renewal and differentiation, leading to tumorigenesis and progression of hepatocellular carcinomas [30]. Consistently, we also determined c-Myc expression levels that decreased along with AGO2 reduction after UA treatment (Figure 4I). These data support the underlying mechanism of UA that improves PTEN to decrease AGO2-mediated FAK/PI3K/Akt/mTOR signaling in conjunction with a reduction in c-Myc and pluripotent stem cell biomarkers, which is dedicated to the potential of UA as an effective natural phytochemical suppressor for breast cancer progression.
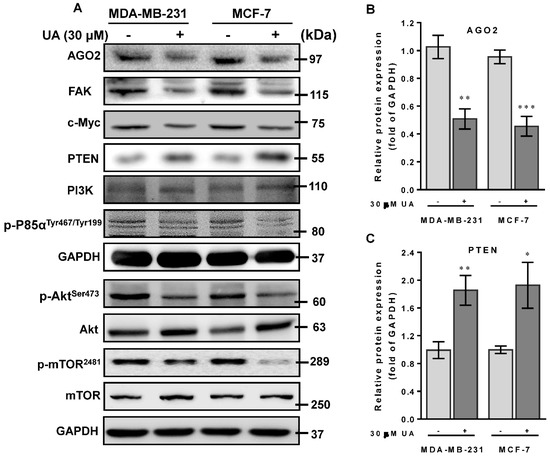
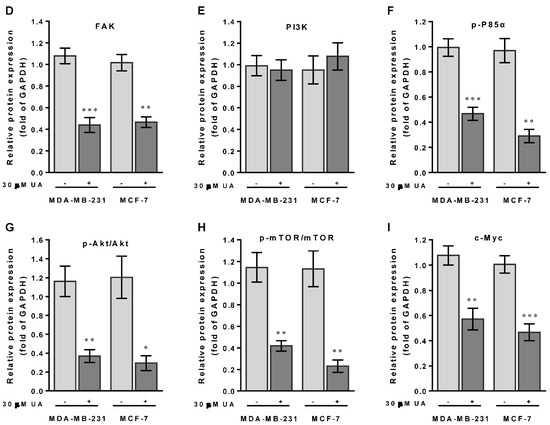
Figure 4.
UA activates PTEN to modulate the FAK/PI3K/Akt signaling pathway: (A) Breast cancer cell lines were treated with UA for 48 h. Western blot analysis with antibodies to tumor progression-relevant proteins as indicated. GAPDH was used to normalize the amount of protein in each lane. (B) Incubation of breast cancer cells with UA (30 μM) significantly decreases AGO2. (C) Increased PTEN negatively regulates the expression levels of (D) FAK and (E) PI3-kinase p110 (PI3K), and inactivates the phosphorylated (activated) forms of (F) PI3K (p-P85αTyr467/Tyr199), (G) Akt (p-AktSer473), and (H) mTOR (p-mTOR2481), and reduced level of (I) c-Myc in both cells treated with UA. Densitometric analyzes for Western blot were performed as described in Materials and Methods. Data are shown as mean ± SD for three independent experiments. * p < 0.05; ** p < 0.01; *** p < 0.001 vs. control group (Ctrl, 0 µM).
3. Discussion
In the present study, UA suppresses CSC and EMT to alleviate metastatic phenotypes by inhibiting AGO2 in breast cancer cell lines. As previously reported, AGO2 overexpression is crucial to turn on the transcription of numerous oncogenic proteins and RNA transcripts to promote pluripotent cell development and reprogram the tumorigenic potential of several types of cancer [31,32]. In addition, AGO2 overexpression contributes to the biogenesis of miRNAs that endorse biological activities in terms of self-renewal and differentiation of CSC [33,34]. In this paper, we explore that UA interferes with CSC characteristics through inhibition of AGO2; Downregulation of AGO2-mediated biogenesis of CSC-related miRNAs, such as miR-9 and miR-221, by which the change in miRNA transcripts correlates with the reduction in slug and vimentin proteins during MET and prevents metastasis in MDA-MB-231 and MCF-7 breast cancer cell lines treated with UA.
UA has been shown to induce apoptosis and antimetastatic activity by destructing the PI3K/Akt/mTOR signaling pathways in human chronic myelogenous leukemia and breast cancer [35,36]. In this study, the metastatic potential of MDA-MB-231 and MCF-7 cells was significantly suppressed when both cell lines were treated with UA at a concentration of 30 μM. Mechanically, the effect of UA on the inhibition of breast cancer cell progression was determined by an elevated level of PTEN, as a consequence, the reduction in FAK associated with concomitant suppression of PI3K, Akt, and mTOR phosphorylation, leading to decreased capacity for invasion and migration of tumor cells. Interestingly, decreased levels of AGO2 and the phosphorylated form of m-TOR protein were detected in MDA-MB-231 cells treated with UA in which endocrine receptors, including ER, PR, and HER2, are deficient. This finding is consistent with the previous study showing that UA treatment inhibited phosphorylation of Akt and mTOR proteins in human LNCaP and PC-3 prostate cancer cells regardless of their status as androgen receptor or sensitivity to androgen [37]. Taken together, these data provide evidence to support the a priori theory that UA not only confers antitumor activities, but also prohibits an antimetastatic phenotype through suppression of AGO2 and inactivation of the FAK/PI3K/Akt pathway regardless of the status of the hormone receptor.
Recent studies have revealed that AGO2 acts as an essential mediator for the function of oncogenic miRNAs, such as miR-19, miR-203, and miR-221/-222 which acquire the invasion and dissemination capabilities of mesenchymal stem cells [38,39,40]. Furthermore, the actions of miR-9 and -221 have been validated to correlate tumor clonal evolution and metastasis, manifesting an increase in the percentage of cells with CSC characteristics [5]. A distinguishing characteristic of neural stem cell (NSC) also showed increased levels of AGO2 associated with miR-9 in the nucleus, leading to cell survival through the Notch signaling pathway of NSC cells [41]. In this study, cells treated with 30 μM UA demonstrated a significant reduction in miR-9 and -221 in SP cells of MDA-MB-231 and MCF-7. The molecular mechanism underlying the protection made by UA in antimetastatic properties is ascribed to a lesser extent to miR-9, shedding light on our understanding of miRNA-associated CSC genesis that correlates the expression level of the AGO2 protein. To our knowledge, the present study is the first of its kind investigation to demonstrate that UA impedes cancer cell stemming and tumor cell fates by inhibiting the AGO2-miRNA mechanism in human breast cancer.
FAK acts as an important protein in cell–ECM interactions that affect cell proliferation, migration, and metastasis. Our results explored that inhibition of EMT-driven breast cancer cell metastasis by UA is endorsed by an increased level of PTEN in subsequent inactivation of the FAK/PI3K signaling pathway (Figure 5). Furthermore, AGO2 has been shown to decrease PTEN expression in HCC angiogenesis [42], whereas PTEN caused apoptotic death by repression of the c-Myc transcript in human breast cancer cells [43]. Consistently, we found that an increase in the level of PTEN is inversely associated with a decrease in the c-Myc transcript in MDA-MB-231 and MCF-7 cells treated with UA. The data confirmed that PTEN modulates the c-myc gene in which the significance of UA interferes with the malignant transformation of breast cancer. However, it should be noted that the broad spectrum of UA in anticancer activity is to interfere with proteins related to tumor cell characteristics of proliferation, adhesion, migration, and survival through numerous signaling pathways, including hypoxia-, PI3K/AKT-, Wnt/β-catenin signaling [44,45,46], as well as JAK2/STAT3 transcription in changing pro-tumor microenvironments [47]. In particular, when the effect of UA on breast cancer was examined, one major concern in evaluating this phytochemical in anticancer activity was that genetic contributions in relation to CSC characteristics and the current understanding of tumor cell pluripotency in breast cancer progression provides solid justification for our results.
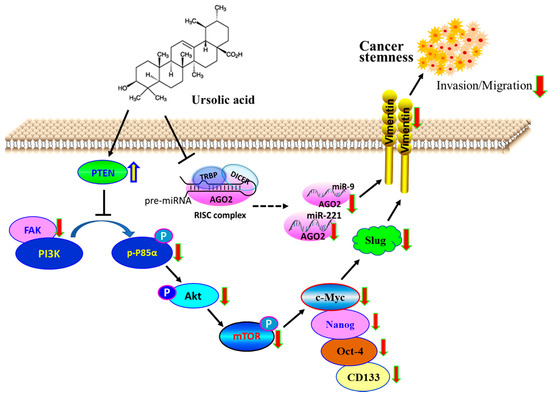
Figure 5.
Schematic representation of anticancer activity through decreased BCSC. Tumor suppressive role of PTEN, whose gain in responding to UA decreases the fraction of CSC by downregulating the expression of AGO2 to inactivate the FAK/PI3K/Akt/mTOR and c-Myc signaling pathway. The model also suggests that UA decreased the AGO2-mediated biogenesis of miR-9 and -221 that contribute to the reduction in slug and vimentin, leading to the inhibition of breast cancer invasion and migration during EMT.
4. Materials and Methods
4.1. Cell Culture
Two breast cancer cell lines, including invasive TNBC MDA-MB-231 (HTB-26) and early-stage MCF-7 [HTB-22, estrogen receptor positive, ER(+)] cell lines, and a non-tumorigenic human breast epithelial cell lines, HBL-100 (HTB-124) were obtained from the American Type Culture Collection (Manassas, VA, USA) and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Life Technologies, Inc., Grand Island, NY, USA) containing 0.1 mM sodium pyruvate, 10% FBS, 2 mM L-glutamine, 100 IU/mL penicillin, 100 µg/mL streptomycin (Biosource, Rockvile, MD, USA) and supplemented with 10% fetal bovine serum. Cells were cultured at a constant temperature of 37 °C in a humidified atmosphere of 5% CO2.
4.2. Characterization and Isolation of CSC
Details for breast CSC preparation have been previously described [4]. Briefly, dissociated breast cancer cells (2 × 106 cell/mL) were suspended in SP medium (calcium- and magnesium-free Hanks balanced salt solution with 2% FBS, 1% penicillin/streptomycin and 10 mM HEPES). Hoechst 33342 (Sigma-Aldrich, St. Louis, MO, USA) was optimized (5 μg/mL) on the respective breast tumor cells. The specific ABCG2 transporter inhibitor, reserpine (RES, Sigma-Aldrich) with a concentration of 50 μM was added before adding the Hoechst 33342 fluorescent dye. Cells were kept on ice in the dark for FACS analysis. The SP from NSP was then counted on a Becton Dickinson FACS Digital Vantage (Diva) cell sorter (BD Biosciences, San Jose, CA, USA). Hoechst was excited at a wavelength of 355 nm and a fluorescent profile was measured at dual-wavelength analysis (450/50 nm and 675/20 nm). Cells cotreated with Hoechst and RES were used as the control group for the determination of SP.
4.3. Cell Viability
The MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay was performed exactly to estimate cell viability as previously described [48]. After UA treatment at the indicated times (24 and 48 h), cells (5 × 104 cells/mL/well) were suspended and rinsed with phosphate-buffered saline (PBS) twice before incubation with MTT for an additional 4 h. The blue formazan generated, proportional to the number of viable cells in the MTT assay, was dissolved in isopropyl alcohol and measured spectrophotometrically at 563 nm. A percentage of cell viability with untreated cells was defined as 100%.
4.4. Matrigel Invasion and Migration Assays
The transwell cell migration assay that estimates the capacity for cell motility and invasiveness towards a chemo-attractant was performed exactly as previously stated [5]. Briefly, cells (5 × 104) were seeded into 48-well modified Boyden chambers (Neuro Probe, Cabin John, MD, USA) with 8 μm pore size polyarbonate membrane filters to detect cell migration (without Matrigel) and invasion (containing Matrigel) for 24 h. Penetrated cells that were attached to the lower surface of the membrane were fixed with methanol and stained with Giemsa solution (Sigma-Aldrich, St. Louis, MO, USA). The invaded cells were stained with crystal violet dye and quantified by counting five random high-power visions using an Olympus Ckx41 light microscope (Tokyo, Japan).
4.5. RNA Extraction, cDNA Transcription and Real-Time PCR Analysis
Total RNA was extracted from breast cancer cells using TRIzol reagent (, Invitrogen Co., Carlsbad, CA, USA). First-strand cDNA synthesis was performed in the presence of random primers using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster city, CA, USA) according to the manufacturer’s instructions. The sequences of the primer sets used for the real-time PCR analysis to detect each mRNA transcript are listed in Table 1. Real-time PCR was performed using the KAPATM SYBR® FAST qPCR kit (KAPA Biosystems, Woburn, MA, USA) and the results were normalized to those of the glyceraldehyde-3-phophate dehydrogenase (GAPDH) gene. The sequences of probes and primer sets used to detect miR-9 (ID 000583) and miR-221 (ID 000524) in the microRNA assay were purchased from Applied Biosystems Inc. (Foster, CA, USA). The level of U6 small nuclear RNA (encoded by RNU6B) was used to normalize the relative expression level of the miRNAs. The comparative CT method (-ddCt) was used to estimate the relative expression (fold change) of mRNA and miRNA transcripts in tumor and non-tumor cells in each case (2−ddCt, where ddCt = dCttarget − dCtinternal control). All qPCR experiments were performed in triplicate in the samples to determine the levels of target transcripts.

Table 1.
Primer sets used for qRT-PCR analysis.
4.6. Western Blot Analysis
The preparation of whole cell extract and Western blot is performed as stated [5]. Protein concentrations were determined by BCA reagent following the manufacturer’s instructions (Bio-Rad, Richmond, CA, USA). All proteins were resolved on sodium dodecyl sulfate-polyacrylamide gels and transferred to PVDF membranes. Primary antibodies for AGO2 (no. 2897), phosphor-Akt (Ser473) (no. 9271S), phospho-mTOR (Ser2481) (no. 2974), slug (no. 9585) and vimentin (no. 5741) were purchased from Cell Signaling Technology (Danvers, MA, USA); ABCG2 (no. GTX12131), FAK (no. GTX100764), GAPDH (no. GTX100118) and PTEN (no. GTX101025) were purchased from GeneTex International Co., Ltd. (Hsinchu City 300 Taiwan.); phospho-PI3 kinase p85 (Tyr467/Tyr199)(# bs-3332R, Bioss, Inc., China); mTOR (# 66888-1, Proteintech) and PI3K (P110, #SC-8010) and c-Myc (#SC-40) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Horseradish-peroxidase-conjugated goat anti-mouse IgG antibody (Jackson Immunoresearch, West Grove, PA, USA) was used as the second antibody, and the signals were detected using the enhanced chemiluminescence (ECL) system (Millipore, Temecula, CA, USA). Protein band intensities quantified by densitometry (Digital Protein Imagineware, Huntington Station, NY, USA).
4.7. Statistical Analyses
All statistical analyzes were performed with GraphPad Pro Prism 8.01 (GraphPad, CA, USA). Continuous and categorical variables were displayed as mean ± standard deviation (SD) and percentages, respectively. The expression levels of the gene transcripts and the intensity of the Western blots of interest from the UA treatment were compared with the results from the control group using an unpaired two-tailed t test with Welch’s correction (Welch’s t test). The p value < 0.05 was considered statistically significant.
5. Conclusions
In conclusion, our findings provide evidence to support the tumor-suppressive role of PTEN, whose gain in responding to UA could decrease the CSC portion by downregulating the expression of AGO2. PTEN caused decreases in the levels of the FAK/PI3K/Akt/mTOR signaling pathway in association with EMT conversion in UA-treated breast cancer cells. Through this, UA has the ability to kill breast cancer cells, and animal studies are needed to prove this in vivo. Collectively, this may not only improve a signature of decreased cancer cell stemness that alters the communicative nature of EMT with respect to down-modulated AGO2 expression of UA, but also provide clinicians with information on new applications in alternative effective therapeutic routes for breast cancer.
Author Contributions
Conceptualization, W.-L.L., Y.-F.L., C.-Y.S. and C.-W.C.; Data curation, Y.-F.L., T.-H.Y., J.-C.S., Y.-T.H. and H.-J.L.; Formal analysis, W.-L.L., Y.-F.L., T.-H.Y. and J.-C.S.; Funding acquisition, C.-W.C.; Methodology, J.-C.S., Y.-T.H. and H.-J.L.; Project administration, C.-Y.S. and C.-W.C.; Supervision, C.-Y.S. and C.-W.C.; Writing—original draft, W.-L.L., Y.-F.L. and C.-W.C.; Writing—review and editing, C.-Y.S. and C.-W.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Ministry of Science and Technology of Taiwan (grant number MOST 107-2320-B-040-012, MOST 110-2314-B-040-025, and NSTC 111-2314-B-001-003). The authors declare that no funds, grants, or other support was received during the preparation of this manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| UA | ursolic acid |
| AGO2 | Argonaute-2 |
| CSC | cancer stem cell |
| SP | side population |
| ABCG2 | ATP-binding cassette superfamily G member 2 |
| EMT | epithelial-to-mesenchymal transition |
| miRNA | microRNA |
| PTEN | phosphatase and tensin homologue on chromosome 10 |
| qRT-PCR | quantitative reverse transcription PCR |
References
- Chiang, A.C.; Massague, J. Molecular basis of metastasis. N. Engl. J. Med. 2008, 359, 2814–2823. [Google Scholar] [CrossRef] [PubMed]
- Hunter, K.W.; Crawford, N.P.; Alsarraj, J. Mechanisms of metastasis. Breast Cancer Res. 2008, 10 (Suppl. 1), S2. [Google Scholar] [CrossRef] [PubMed]
- Yokota, J. Tumor progression and metastasis. Carcinogenesis 2000, 21, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Turnquist, H.; Jackson, J.; Sgagias, M.; Yan, Y.; Gong, M.; Dean, M.; Sharp, J.G.; Cowan, K. The multidrug resistance transporter ABCG2 (breast cancer resistance protein 1) effluxes Hoechst 33342 and is overexpressed in hematopoietic stem cells. Clin. Cancer Res. 2002, 8, 22–28. [Google Scholar]
- Cheng, C.W.; Yu, J.C.; Hsieh, Y.H.; Liao, W.L.; Shieh, J.C.; Yao, C.C.; Lee, H.J.; Chen, P.M.; Wu, P.E.; Shen, C.Y. Increased Cellular Levels of MicroRNA-9 and MicroRNA-221 Correlate with Cancer Stemness and Predict Poor Outcome in Human Breast Cancer. Cell. Physiol. Biochem. 2018, 48, 2205–2218. [Google Scholar] [CrossRef]
- Cheng, C.W.; Liao, W.L.; Chen, P.M.; Yu, J.C.; Shiau, H.P.; Hsieh, Y.H.; Lee, H.J.; Cheng, Y.C.; Wu, P.E.; Shen, C.Y. MiR-139 Modulates Cancer Stem Cell Function of Human Breast Cancer through Targeting CXCR4. Cancers 2021, 13, 2582. [Google Scholar] [CrossRef]
- Theunissen, T.W.; Jaenisch, R. Molecular control of induced pluripotency. Cell. Stem Cell. 2014, 14, 720–734. [Google Scholar] [CrossRef]
- Romero-Cordoba, S.L.; Salido-Guadarrama, I.; Rodriguez-Dorantes, M.; Hidalgo-Miranda, A. miRNA biogenesis: Biological impact in the development of cancer. Cancer Biol. Ther. 2014, 15, 1444–1455. [Google Scholar] [CrossRef]
- Lee, J.H.; Schutte, D.; Wulf, G.; Fuzesi, L.; Radzun, H.J.; Schweyer, S.; Engel, W.; Nayernia, K. Stem-cell protein Piwil2 is widely expressed in tumors and inhibits apoptosis through activation of Stat3/Bcl-XL pathway. Hum. Mol. Genet. 2006, 15, 201–211. [Google Scholar] [CrossRef]
- Chen, Z.; Che, Q.; He, X.; Wang, F.; Wang, H.; Zhu, M.; Sun, J.; Wan, X. Stem cell protein Piwil1 endowed endometrial cancer cells with stem-like properties via inducing epithelial-mesenchymal transition. BMC Cancer. 2015, 15, 811. [Google Scholar] [CrossRef]
- Ciuffreda, L.; Falcone, I.; Incani, U.C.; Del Curatolo, A.; Conciatori, F.; Matteoni, S.; Vari, S.; Vaccaro, V.; Cognetti, F.; Milella, M. PTEN expression and function in adult cancer stem cells and prospects for therapeutic targeting. Adv. Biol. Regul. 2014, 56, 66–80. [Google Scholar] [CrossRef]
- Montales, M.T.; Rahal, O.M.; Kang, J.; Rogers, T.J.; Prior, R.L.; Wu, X.; Simmen, R.C. Repression of mammosphere formation of human breast cancer cells by soy isoflavone genistein and blueberry polyphenolic acids suggests diet-mediated targeting of cancer stem-like/progenitor cells. Carcinogenesis 2012, 33, 652–660. [Google Scholar] [CrossRef]
- Mulholland, D.J.; Kobayashi, N.; Ruscetti, M.; Zhi, A.; Tran, L.M.; Huang, J.; Gleave, M.; Wu, H. Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer Res. 2012, 72, 1878–1889. [Google Scholar] [CrossRef]
- Wu, P.P.; Zhang, K.; Lu, Y.J.; He, P.; Zhao, S.Q. In vitro and in vivo evaluation of the antidiabetic activity of ursolic acid derivatives. Eur. J. Med. Chem. 2014, 80, 502–508. [Google Scholar] [CrossRef]
- Kim, K.D.; Jung, H.Y.; Ryu, H.G.; Kim, B.; Jeon, J.; Yoo, H.Y.; Park, C.H.; Choi, B.H.; Hyun, C.K.; Kim, K.T.; et al. Betulinic acid inhibits high-fat diet-induced obesity and improves energy balance by activating AMPK. Nutr. Metab. Cardiovasc. Dis. 2019, 29, 409–420. [Google Scholar] [CrossRef]
- Erdmann, J.; Kujacinski, M.; Wicinski, M. Beneficial Effects of Ursolic Acid and Its Derivatives-Focus on Potential Biochemical Mechanisms in Cardiovascular Conditions. Nutrients 2021, 13, 3900. [Google Scholar] [CrossRef]
- Lee, J.Y.; Choi, J.K.; Jeong, N.H.; Yoo, J.; Ha, Y.S.; Lee, B.; Choi, H.; Park, P.H.; Shin, T.Y.; Kwon, T.K.; et al. Anti-inflammatory effects of ursolic acid-3-acetate on human synovial fibroblasts and a murine model of rheumatoid arthritis. Int. Immunopharmacol. 2017, 49, 118–125. [Google Scholar] [CrossRef]
- Dhakal, H.; Kim, M.J.; Lee, S.; Choi, Y.A.; Kim, N.; Kwon, T.K.; Khang, D.; Kim, S.H. Ursolic acid inhibits FcepsilonRI-mediated mast cell activation and allergic inflammation. Int. Immunopharmacol. 2021, 99, 107994. [Google Scholar] [CrossRef]
- Lorincz, A.; Mihaly, J.; Wacha, A.; Nemeth, C.; Besztercei, B.; Gyulavari, P.; Varga, Z.; Petak, I.; Bota, A. Combination of multifunctional ursolic acid with kinase inhibitors for anti-cancer drug carrier vesicles. Mater. Sci. Eng. C Mater. Biol. Appl. 2021, 131, 112481. [Google Scholar] [CrossRef]
- Jin, H.; Pi, J.; Yang, F.; Jiang, J.; Wang, X.; Bai, H.; Shao, M.; Huang, L.; Zhu, H.; Yang, P.; et al. Folate-Chitosan Nanoparticles Loaded with Ursolic Acid Confer Anti-Breast Cancer Activities in vitro and in vivo. Sci. Rep. 2016, 6, 0782. [Google Scholar] [CrossRef]
- Liu, T.J.; Sun, B.C.; Zhao, X.L.; Zhao, X.M.; Sun, T.; Gu, Q.; Yao, Z.; Dong, X.Y.; Zhao, N.; Liu, N. CD133+ cells with cancer stem cell characteristics associates with vasculogenic mimicry in triple-negative breast cancer. Oncogene 2013, 32, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Yousefnia, S.; Seyed Forootan, F.; Seyed Forootan, S.; Nasr Esfahani, M.H.; Gure, A.O.; Ghaedi, K. Mechanistic Pathways of Malignancy in Breast Cancer Stem Cells. Front. Oncol. 2020, 10, 452. [Google Scholar] [CrossRef] [PubMed]
- Weissman, I.L. Translating stem and progenitor cell biology to the clinic: Barriers and opportunities. Science 2000, 287, 1442–1446. [Google Scholar] [CrossRef] [PubMed]
- Lindeman, G.J.; Visvader, J.E. Insights into the cell of origin in breast cancer and breast cancer stem cells. Asia-Pac. J. Clin. Oncol. 2010, 6, 89–97.21. [Google Scholar] [CrossRef] [PubMed]
- Alberti, L.; Losi, L.; Leyvraz, S.; Benhattar, J. Different Effects of BORIS/CTCFL on Stemness Gene Expression, Sphere Formation and Cell Survival in Epithelial Cancer Stem Cells. PLoS ONE 2015, 10, e0132977. [Google Scholar] [CrossRef]
- Hollestelle, A.; Peeters, J.K.; Smid, M.; Timmermans, M.; Verhoog, L.C.; Westenend, P.J.; Heine, A.A.; Chan, A.; Sieuwerts, A.M.; Wiemer, E.A.C.; et al. Loss of E-cadherin is not a necessity for epithelial to mesenchymal transition in human breast cancer. Breast Cancer Res. Treat. 2013, 138, 47–57. [Google Scholar] [CrossRef]
- Vuoriluoto, K.; Haugen, H.; Kiviluoto, S.; Mpindi, J.P.; Nevo, J.; Gjerdrum, C.; Tiron, C.; Lorens, J.B.; Ivaska, J. Vimentin regulates EMT induction by Slug and oncogenic H-Ras and migration by governing Axl expression in breast cancer. Oncogene 2011, 30, 1436–1448. [Google Scholar] [CrossRef]
- Bleau, A.M.; Hambardzumyan, D.; Ozawa, T.; Fomchenko, E.I.; Huse, J.T.; Brennan, C.W.; Holland, E.C. PTEN/PI3K/Akt pathway regulates the side population phenotype and ABCG2 activity in glioma tumor stem-like cells. Cell. Stem Cell. 2009, 4, 226–235. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, B.; Chen, X.; Li, W.; Dong, P. AGO2 involves the malignant phenotypes and FAK/PI3K/AKT signaling pathway in hypopharyngeal-derived FaDu cells. Oncotarget 2017, 8, 54735–54746. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X. Targeting the Wnt/beta-catenin signaling pathway in cancer. J. Hematol. Oncol. 2020, 13, 165. [Google Scholar] [CrossRef]
- Nowak, I.; Sarshad, A.A. Argonaute Proteins Take Center Stage in Cancers. Cancers 2021, 13, 788. [Google Scholar] [CrossRef]
- Hanina, S.A.; Mifsud, W.; Down, T.A.; Hayashi, K.; O’Carroll, D.; Lao, K.; Miska, E.A.; Surani, M.A. Genome-wide identification of targets and function of individual MicroRNAs in mouse embryonic stem cells. PLoS Genet. 2010, 6, e1001163. [Google Scholar] [CrossRef]
- Takahashi, R.U.; Miyazaki, H.; Ochiya, T. The role of microRNAs in the regulation of cancer stem cells. Front. Genet. 2014, 4, 295. [Google Scholar] [CrossRef]
- Khan, A.Q.; Ahmed, E.I.; Elareer, N.R.; Junejo, K.; Steinhoff, M.; Uddin, S. Role of miRNA-Regulated Cancer Stem Cells in the Pathogenesis of Human Malignancies. Cells 2019, 8, 840. [Google Scholar] [CrossRef]
- Wu, B.; Wang, X.; Chi, Z.F.; Hu, R.; Zhang, R.; Yang, W.; Liu, Z.G. Ursolic acid-induced apoptosis in K562 cells involving upregulation of PTEN gene expression and inactivation of the PI3K/Akt pathway. Arch. Pharm. Res. 2012, 35, 543–548. [Google Scholar] [CrossRef]
- Yeh, C.T.; Wu, C.H.; Yen, G.C. Ursolic acid, a naturally occurring triterpenoid, suppresses migration and invasion of human breast cancer cells by modulating c-Jun N-terminal kinase, Akt and mammalian target of rapamycin signaling. Mol. Nutr. Food. Res. 2010, 54, 1285–1295. [Google Scholar] [CrossRef]
- Meng, Y.; Lin, Z.M.; Ge, N.; Zhang, D.L.; Huang, J.; Kong, F. Ursolic Acid Induces Apoptosis of Prostate Cancer Cells via the PI3K/Akt/mTOR Pathway. Am. J. Chin. Med. 2015, 43, 1471–1486. [Google Scholar] [CrossRef]
- Obayashi, M.; Yoshida, M.; Tsunematsu, T.; Ogawa, I.; Sasahira, T.; Kuniyasu, H.; Imoto, I.; Abiko, Y.; Xu, D.; Fukunaga, S.; et al. microRNA-203 suppresses invasion and epithelial-mesenchymal transition induction via targeting NUAK1 in head and neck cancer. Oncotarget 2016, 7, 8223–8239. [Google Scholar] [CrossRef]
- Sternburg, E.L.; Estep, J.A.; Nguyen, D.K.; Li, Y.; Karginov, F.V. Antagonistic and cooperative AGO2-PUM interactions in regulating mRNAs. Sci. Rep. 2018, 8, 15316. [Google Scholar] [CrossRef]
- Zhang, X.; Yu, H.; Lou, J.R.; Zheng, J.; Zhu, H.; Popescu, N.I.; Lupu, F.; Lind, S.E.; Ding, W.Q. MicroRNA-19 (miR-19) regulates tissue factor expression in breast cancer cells. J. Biol. Chem. 2011, 286, 1429–1435. [Google Scholar] [CrossRef]
- Katz, S.; Cussigh, D.; Urban, N.; Blomfield, I.; Guillemot, F.; Bally-Cuif, L.; Coolen, M. A Nuclear Role for miR-9 and Argonaute Proteins in Balancing Quiescent and Activated Neural Stem Cell States. Cell Rep. 2016, 17, 1383–1398. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.L.; Huang, Y.; Li, L.F.; Zhu, H.L.; Gao, H.X.; Liu, H.; Lv, S.Q.; Xu, Z.H.; Zheng, L.N.; Liu, T.; et al. Argonaute 2 promotes angiogenesis via the PTEN/VEGF signaling pathway in human hepatocellular carcinoma. Acta Pharmacol. Sin. 2015, 36, 1237–1245. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Grigorieva, I.; Steele, R.; Hoover, R.G.; Ray, R.B. PTEN transcriptionally modulates c-myc gene expression in human breast carcinoma cells and is involved in cell growth regulation. Gene 1999, 235, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.L.; Shui, Y.M.; Jiang, W.; Huang, E.Y.; Shou, Q.Y.; Ji, X.; He, B.C.; Lv, G.Y.; He, T.C. Hypoxia pathway and hypoxia-mediated extensive extramedullary hematopoiesis are involved in ursolic acid’s anti-metastatic effect in 4T1 tumor bearing mice. Oncotarget 2016, 7, 71802–71816. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.D. Ursolic Acid Decreases the Proliferation of MCF-7 Cell-Derived Breast Cancer Stem-Like Cells by Modulating the ERK and PI3K/AKT Signaling Pathways. Prev. Nutr. Food Sci. 2021, 26, 434–444. [Google Scholar] [CrossRef]
- Zhang, R.X.; Li, Y.; Tian, D.D.; Liu, Y.; Nian, W.; Zou, X.; Chen, Q.Z.; Zhou, L.Y.; Deng, Z.L.; He, B.C. Ursolic acid inhibits proliferation and induces apoptosis by inactivating Wnt/beta-catenin signaling in human osteosarcoma cells. Int. J. Oncol. 2016, 49, 1973–1982. [Google Scholar] [CrossRef]
- Kang, D.Y.; Sp, N.; Lee, J.M.; Jang, K.J. Antitumor Effects of Ursolic Acid through Mediating the Inhibition of STAT3/PD-L1 Signaling in Non-Small Cell Lung Cancer Cells. Biomedicines 2021, 9, 297. [Google Scholar] [CrossRef]
- Liao, W.L.; Lin, J.Y.; Shieh, J.C.; Yeh, H.F.; Hsieh, Y.H.; Cheng, Y.C.; Lee, H.J.; Shen, C.Y.; Cheng, C.W. Induction of G2/M Phase Arrest by Diosgenin via Activation of Chk1 Kinase and Cdc25C Regulatory Pathways to Promote Apoptosis in Human Breast Cancer Cells. Int. J. Mol. Sci. 2019, 21, 172. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).