Comprehensive Discovery of the Accessible Primary Amino Group-Containing Segments from Cell Surface Proteins by Fine-Tuning a High-Throughput Biotinylation Method
Abstract
1. Introduction
2. Results
2.1. Labeling Condition Optimization and the Effect of Alkylation for the Biotinylation
2.2. Optimization of Labeled Cell Lysis, Membrane Preparation Solubilization and Enrichment of Extracellular Protein Segments
2.3. Cell Surface Peptide Elution Optimization and Their Solid-Phase Extraction Purification
2.4. HPLC Gradient Optimization and Precursor Charge Preference Setting under the HPLC-MS/MS Analysis
2.5. Assessment of Specificity and Validation of the Developed Workflow
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Cell Isolation
4.3. Biotinylation of Accessible Free Amino Groups on the Surface of HL60 Cells
4.4. Mechanical Cell Lysis and Membrane Preparation
4.5. Solubilization of Membrane Preparations and Digestion of Membrane-Associated Proteins
4.6. Enrichment of the Biotinylated Protein Segments on Neutravidin Agarose Resin
4.7. Peptide Purification by Solid-Phase Extraction
4.8. Isolated Peptide Separation by Nanoflow Liquid Chromatography
4.9. Peptide Identification by Tandem Mass Spectrometry
4.10. Assessment of Identified Proteins and Peptides
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Székely, V.; Patik, I.; Ungvári, O.; Telbisz, Á.; Szakács, G.; Bakos, É.; Özvegy-Laczka, C. Fluorescent Probes for the Dual Investigation of MRP2 and OATP1B1 Function and Drug Interactions. Eur. J. Pharm. Sci. 2020, 151, 105395. [Google Scholar] [CrossRef] [PubMed]
- Palm, W.; Thompson, C.B. Nutrient Acquisition Strategies of Mammalian Cells. Nature 2017, 546, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Xu, Z.; Li, S.; Liang, H.; Zhang, C.; Wang, Z.; Li, J.; Li, J.; Yang, H. Systematic Interrogation of Cellular Signaling in Live Cells Using a Membrane-Anchored DNA Multitasking Processor. Angew. Chem. Int. Ed. Engl. 2022, 61, e202113795. [Google Scholar] [CrossRef] [PubMed]
- Barneh, F.; Jafari, M.; Mirzaie, M. Updates on Drug-Target Network; Facilitating Polypharmacology and Data Integration by Growth of DrugBank Database. Brief. Bioinform. 2016, 17, 1070–1080. [Google Scholar] [CrossRef]
- Bausch-Fluck, D.; Goldmann, U.; Müller, S.; van Oostrum, M.; Müller, M.; Schubert, O.T.; Wollscheid, B. The in Silico Human Surfaceome. Proc. Natl. Acad. Sci. USA 2018, 115, E10988–E10997. [Google Scholar] [CrossRef]
- Sau, S.; Alsaab, H.O.; Kashaw, S.K.; Tatiparti, K.; Iyer, A.K. Advances in Antibody-Drug Conjugates: A New Era of Targeted Cancer Therapy. Drug Discov. Today 2017, 22, 1547–1556. [Google Scholar] [CrossRef]
- Martinez-Martin, N. Technologies for Proteome-Wide Discovery of Extracellular Host-Pathogen Interactions. J. Immunol. Res. 2017, 2017, 2197615. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Mao, J.; Yao, Y.; Wang, K.; Qiao, Q.; Fang, Z.; Ye, M. Sensitive Profiling of Cell Surface Proteome by Using an Optimized Biotinylation Method. J. Proteom. 2019, 196, 33–41. [Google Scholar] [CrossRef]
- Kuhlmann, L.; Cummins, E.; Samudio, I.; Kislinger, T. Cell-Surface Proteomics for the Identification of Novel Therapeutic Targets in Cancer. Expert Rev. Proteom. 2018, 15, 259–275. [Google Scholar] [CrossRef]
- Langó, T.; Pataki, Z.G.; Turiák, L.; Ács, A.; Varga, J.K.; Várady, G.; Kucsma, N.; Drahos, L.; Tusnády, G.E. Partial Proteolysis Improves the Identification of the Extracellular Segments of Transmembrane Proteins by Surface Biotinylation. Sci. Rep. 2020, 10, 8880. [Google Scholar] [CrossRef]
- Bausch-Fluck, D.; Hofmann, A.; Wollscheid, B. Cell Surface Capturing Technologies for the Surfaceome Discovery of Hepatocytes. Methods Mol. Biol. 2012, 909, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Qin, H.; Ye, M. An Overview on Enrichment Methods for Cell Surface Proteome Profiling. J. Sep. Sci. 2020, 43, 292–312. [Google Scholar] [CrossRef] [PubMed]
- Geladaki, A.; Kočevar Britovšek, N.; Breckels, L.M.; Smith, T.S.; Vennard, O.L.; Mulvey, C.M.; Crook, O.M.; Gatto, L.; Lilley, K.S. Combining LOPIT with Differential Ultracentrifugation for High-Resolution Spatial Proteomics. Nat. Commun. 2019, 10, 331. [Google Scholar] [CrossRef]
- Wu, C.C.; MacCoss, M.J.; Howell, K.E.; Yates, J.R. A Method for the Comprehensive Proteomic Analysis of Membrane Proteins. Nat. Biotechnol. 2003, 21, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Elschenbroich, S.; Sharma, P.; Sepiashvili, L.; Gramolini, A.O.; Kislinger, T. Use of Colloidal Silica-Beads for the Isolation of Cell-Surface Proteins for Mass Spectrometry-Based Proteomics. Methods Mol. Biol. 2011, 748, 227–241. [Google Scholar] [CrossRef]
- Wollscheid, B.; Bausch-Fluck, D.; Henderson, C.; O’Brien, R.; Bibel, M.; Schiess, R.; Aebersold, R.; Watts, J.D. Mass-Spectrometric Identification and Relative Quantification of N-Linked Cell Surface Glycoproteins. Nat. Biotechnol. 2009, 27, 378–386. [Google Scholar] [CrossRef]
- Zeng, Y.; Ramya, T.N.C.; Dirksen, A.; Dawson, P.E.; Paulson, J.C. High-Efficiency Labeling of Sialylated Glycoproteins on Living Cells. Nat. Methods 2009, 6, 207–209. [Google Scholar] [CrossRef]
- Hörmann, K.; Stukalov, A.; Müller, A.C.; Heinz, L.X.; Superti-Furga, G.; Colinge, J.; Bennett, K.L. A Surface Biotinylation Strategy for Reproducible Plasma Membrane Protein Purification and Tracking of Genetic and Drug-Induced Alterations. J. Proteome Res. 2016, 15, 647–658. [Google Scholar] [CrossRef]
- Langó, T.; Róna, G.; Hunyadi-Gulyás, É.; Turiák, L.; Varga, J.; Dobson, L.; Várady, G.; Drahos, L.; Vértessy, B.G.; Medzihradszky, K.F.; et al. Identification of Extracellular Segments by Mass Spectrometry Improves Topology Prediction of Transmembrane Proteins. Sci. Rep. 2017, 7, 42610. [Google Scholar] [CrossRef]
- Bausch-Fluck, D.; Hofmann, A.; Bock, T.; Frei, A.P.; Cerciello, F.; Jacobs, A.; Moest, H.; Omasits, U.; Gundry, R.L.; Yoon, C.; et al. A Mass Spectrometric-Derived Cell Surface Protein Atlas. PLoS ONE 2015, 10, e0121314. [Google Scholar] [CrossRef]
- Cogger, K.F.; Sinha, A.; Sarangi, F.; McGaugh, E.C.; Saunders, D.; Dorrell, C.; Mejia-Guerrero, S.; Aghazadeh, Y.; Rourke, J.L.; Screaton, R.A.; et al. Glycoprotein 2 Is a Specific Cell Surface Marker of Human Pancreatic Progenitors. Nat. Commun. 2017, 8, 331. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Langó, T.; Turiák, L.; Ács, A.; Várady, G.; Kucsma, N.; Drahos, L.; Tusnády, G.E. Covalently Modified Carboxyl Side Chains on Cell Surface Leads to a Novel Method toward Topology Analysis of Transmembrane Proteins. Sci. Rep. 2019, 9, 15729. [Google Scholar] [CrossRef] [PubMed]
- Nagano, K.; Shinkawa, T.; Kato, K.; Inomata, N.; Yabuki, N.; Haramura, M. Distinct Cell Surface Proteome Profiling by Biotin Labeling and Glycoprotein Capturing. J. Proteom. 2011, 74, 1985–1993. [Google Scholar] [CrossRef] [PubMed]
- Boheler, K.R.; Gundry, R.L. Concise Review: Cell Surface N-Linked Glycoproteins as Potential Stem Cell Markers and Drug Targets. Stem Cells Transl. Med. 2017, 6, 131–138. [Google Scholar] [CrossRef]
- Goshe, M.B.; Blonder, J.; Smith, R.D. Affinity Labeling of Highly Hydrophobic Integral Membrane Proteins for Proteome-Wide Analysis. J. Proteome Res. 2003, 2, 153–161. [Google Scholar] [CrossRef]
- Roesli, C.; Mumprecht, V.; Neri, D.; Detmar, M. Identification of the Surface-Accessible, Lineage-Specific Vascular Proteome by Two-Dimensional Peptide Mapping. FASEB J. 2008, 22, 1933–1944. [Google Scholar] [CrossRef]
- Özkan Küçük, N.E.; Şanal, E.; Tan, E.; Mitchison, T.; Özlü, N. Labeling Carboxyl Groups of Surface-Exposed Proteins Provides an Orthogonal Approach for Cell Surface Isolation. J. Proteome Res. 2018, 17, 1784–1793. [Google Scholar] [CrossRef]
- DeChancie, J.; Houk, K.N. The Origins of Femtomolar Protein-Ligand Binding: Hydrogen-Bond Cooperativity and Desolvation Energetics in the Biotin-(Strept)Avidin Binding Site. J. Am. Chem. Soc. 2007, 129, 5419–5429. [Google Scholar] [CrossRef]
- Rösli, C.; Rybak, J.-N.; Neri, D.; Elia, G. Quantitative Recovery of Biotinylated Proteins from Streptavidin-Based Affinity Chromatography Resins. Methods Mol. Biol. 2008, 418, 89–100. [Google Scholar] [CrossRef]
- Rafiee, M.-R.; Sigismondo, G.; Kalxdorf, M.; Förster, L.; Brügger, B.; Béthune, J.; Krijgsveld, J. Protease-Resistant Streptavidin for Interaction Proteomics. Mol. Syst. Biol. 2020, 16, e9370. [Google Scholar] [CrossRef]
- Karhemo, P.-R.; Ravela, S.; Laakso, M.; Ritamo, I.; Tatti, O.; Mäkinen, S.; Goodison, S.; Stenman, U.-H.; Hölttä, E.; Hautaniemi, S.; et al. An Optimized Isolation of Biotinylated Cell Surface Proteins Reveals Novel Players in Cancer Metastasis. J. Proteom. 2012, 77, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Almahariq, M.; Chao, C.; Mei, F.C.; Hellmich, M.R.; Patrikeev, I.; Motamedi, M.; Cheng, X. Pharmacological Inhibition and Genetic Knockdown of Exchange Protein Directly Activated by CAMP 1 Reduce Pancreatic Cancer Metastasis in Vivo. Mol. Pharmacol. 2015, 87, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Ravenhill, B.J.; Kanjee, U.; Ahouidi, A.; Nobre, L.; Williamson, J.; Goldberg, J.M.; Antrobus, R.; Dieye, T.; Duraisingh, M.T.; Weekes, M.P. Quantitative Comparative Analysis of Human Erythrocyte Surface Proteins between Individuals from Two Genetically Distinct Populations. Commun. Biol. 2019, 2, 350. [Google Scholar] [CrossRef] [PubMed]
- Walton, A.; Stes, E.; Cybulski, N.; Van Bel, M.; Iñigo, S.; Durand, A.N.; Timmerman, E.; Heyman, J.; Pauwels, L.; De Veylder, L.; et al. It’s Time for Some “Site”-Seeing: Novel Tools to Monitor the Ubiquitin Landscape in Arabidopsis Thaliana. Plant Cell 2016, 28, 6–16. [Google Scholar] [CrossRef]
- Niehage, C.; Karbanová, J.; Steenblock, C.; Corbeil, D.; Hoflack, B. Cell Surface Proteome of Dental Pulp Stem Cells Identified by Label-Free Mass Spectrometry. PLoS ONE 2016, 11, e0159824. [Google Scholar] [CrossRef]
- Smolders, K.; Lombaert, N.; Valkenborg, D.; Baggerman, G.; Arckens, L. An Effective Plasma Membrane Proteomics Approach for Small Tissue Samples. Sci. Rep. 2015, 5, 10917. [Google Scholar] [CrossRef]
- Liu, G.; Choi, M.H.; Ma, H.; Guo, X.; Lo, P.-C.; Kim, J.; Zhang, L. Bioorthogonal Conjugation-Assisted Purification Method for Profiling Cell Surface Proteome. Anal. Chem. 2022, 94, 1901–1909. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Dobson, L.; Reményi, I.; Tusnády, G.E. CCTOP: A Consensus Constrained TOPology Prediction Web Server. Nucleic Acids Res. 2015, 43, W408–W412. [Google Scholar] [CrossRef]
- Schäfer, K.; Engstler, C.; Dischinger, K.; Carrie, C. Assessment of Mitochondrial Protein Topology and Membrane Insertion. Methods Mol. Biol. 2022, 2363, 165–181. [Google Scholar] [CrossRef]
- Dobson, L.; Langó, T.; Reményi, I.; Tusnády, G.E. Expediting Topology Data Gathering for the TOPDB Database. Nucleic Acids Res. 2015, 43, D283–D289. [Google Scholar] [CrossRef] [PubMed]
- Dobson, L.; Szekeres, L.I.; Gerdán, C.; Langó, T.; Zeke, A.; Tusnády, G.E. TmAlphaFold Database: Membrane Localization and Evaluation of AlphaFold2 Predicted Alpha-Helical Transmembrane Protein Structures. Nucleic Acids Res. 2022. [Google Scholar] [CrossRef] [PubMed]
- Tey, S.-R.; Mueller, M.; Reilly, M.; Switalski, C.; Robertson, S.; Sakanaka-Yokoyama, M.; Suzuki, M. Cell Surface Proteins for Enrichment and In Vitro Characterization of Human Pluripotent Stem Cell-Derived Myogenic Progenitors. Stem Cells Int. 2022, 2022, 2735414. [Google Scholar] [CrossRef] [PubMed]
- Esbelin, J.; Santos, T.; Ribière, C.; Desvaux, M.; Viala, D.; Chambon, C.; Hébraud, M. Comparison of Three Methods for Cell Surface Proteome Extraction of Listeria Monocytogenes Biofilms. OMICS 2018, 22, 779–787. [Google Scholar] [CrossRef]
- Monteiro, R.; Hébraud, M.; Chafsey, I.; Chambon, C.; Viala, D.; Torres, C.; Poeta, P.; Igrejas, G. Surfaceome and Exoproteome of a Clinical Sequence Type 398 Methicillin Resistant Staphylococcus Aureus Strain. Biochem. Biophys. Rep. 2015, 3, 7–13. [Google Scholar] [CrossRef]
- Nojima, Y.; Iguchi, K.; Suzuki, Y.; Sato, A. The PH-Dependent Formation of PEGylated Bovine Lactoferrin by Branched Polyethylene Glycol (PEG)-N-Hydroxysuccinimide (NHS) Active Esters. Biol. Pharm. Bull. 2009, 32, 523–526. [Google Scholar] [CrossRef]
- Grumbach, I.M.; Veh, R.W. Sulpho-N-Hydroxysuccinimide Activated Long Chain Biotin. A New Microtitre Plate Assay for the Determination of Its Stability at Different PH Values and Its Reaction Rate with Protein Bound Amino Groups. J. Immunol. Methods 1991, 140, 205–210. [Google Scholar] [CrossRef]
- Denoncin, K.; Vertommen, D.; Paek, E.; Collet, J.-F. The Protein-Disulfide Isomerase DsbC Cooperates with SurA and DsbA in the Assembly of the Essential β-Barrel Protein LptD. J. Biol. Chem. 2010, 285, 29425–29433. [Google Scholar] [CrossRef]
- Suttapitugsakul, S.; Xiao, H.; Smeekens, J.; Wu, R. Evaluation and Optimization of Reduction and Alkylation Methods to Maximize Peptide Identification with MS-Based Proteomics. Mol. Biosyst. 2017, 13, 2574–2582. [Google Scholar] [CrossRef]
- Miller, R.M.; Millikin, R.J.; Hoffmann, C.V.; Solntsev, S.K.; Sheynkman, G.M.; Shortreed, M.R.; Smith, L.M. Improved Protein Inference from Multiple Protease Bottom-Up Mass Spectrometry Data. J. Proteome Res. 2019, 18, 3429–3438. [Google Scholar] [CrossRef]
- Dwivedi-Agnihotri, H.; Srivastava, A.; Shukla, A.K. Reversible Biotinylation of Purified Proteins for Measuring Protein-Protein Interactions. Methods Enzymol. 2020, 633, 281–294. [Google Scholar] [CrossRef] [PubMed]
- Nierves, L.; Lange, P.F. Detectability of Biotin Tags by LC-MS/MS. J. Proteome Res. 2021, 20, 3002–3008. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Verma, S.; Modak, S.B.; Chaturvedi, M.M.; Purohit, J.S. Extra-Nuclear Histones: Origin, Significance and Perspectives. Mol. Cell. Biochem. 2022, 477, 507–524. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Gao, F.; Wang, Q.; Wang, K.; Pan, S.; Pan, Z.; Xu, S.; Li, L.; Zhao, D. Differentiation of HL-60 Cells in Serum-Free Hematopoietic Cell Media Enhances the Production of Neutrophil Extracellular Traps. Exp. Ther. Med. 2021, 21, 353. [Google Scholar] [CrossRef] [PubMed]
- Semeraro, F.; Ammollo, C.T.; Morrissey, J.H.; Dale, G.L.; Friese, P.; Esmon, N.L.; Esmon, C.T. Extracellular Histones Promote Thrombin Generation through Platelet-Dependent Mechanisms: Involvement of Platelet TLR2 and TLR4. Blood 2011, 118, 1952–1961. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein Measurement with the Folin Phenol Reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]

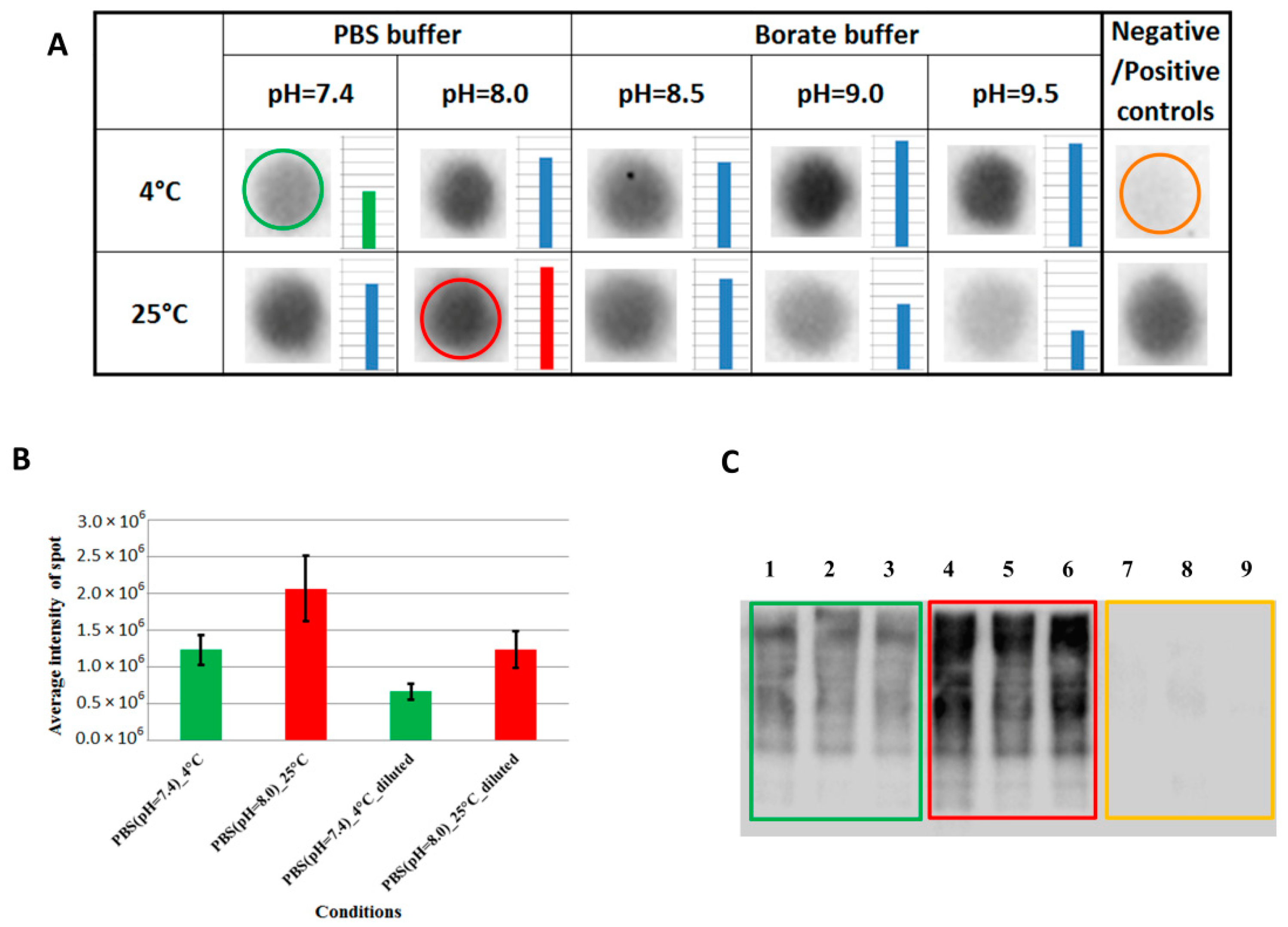
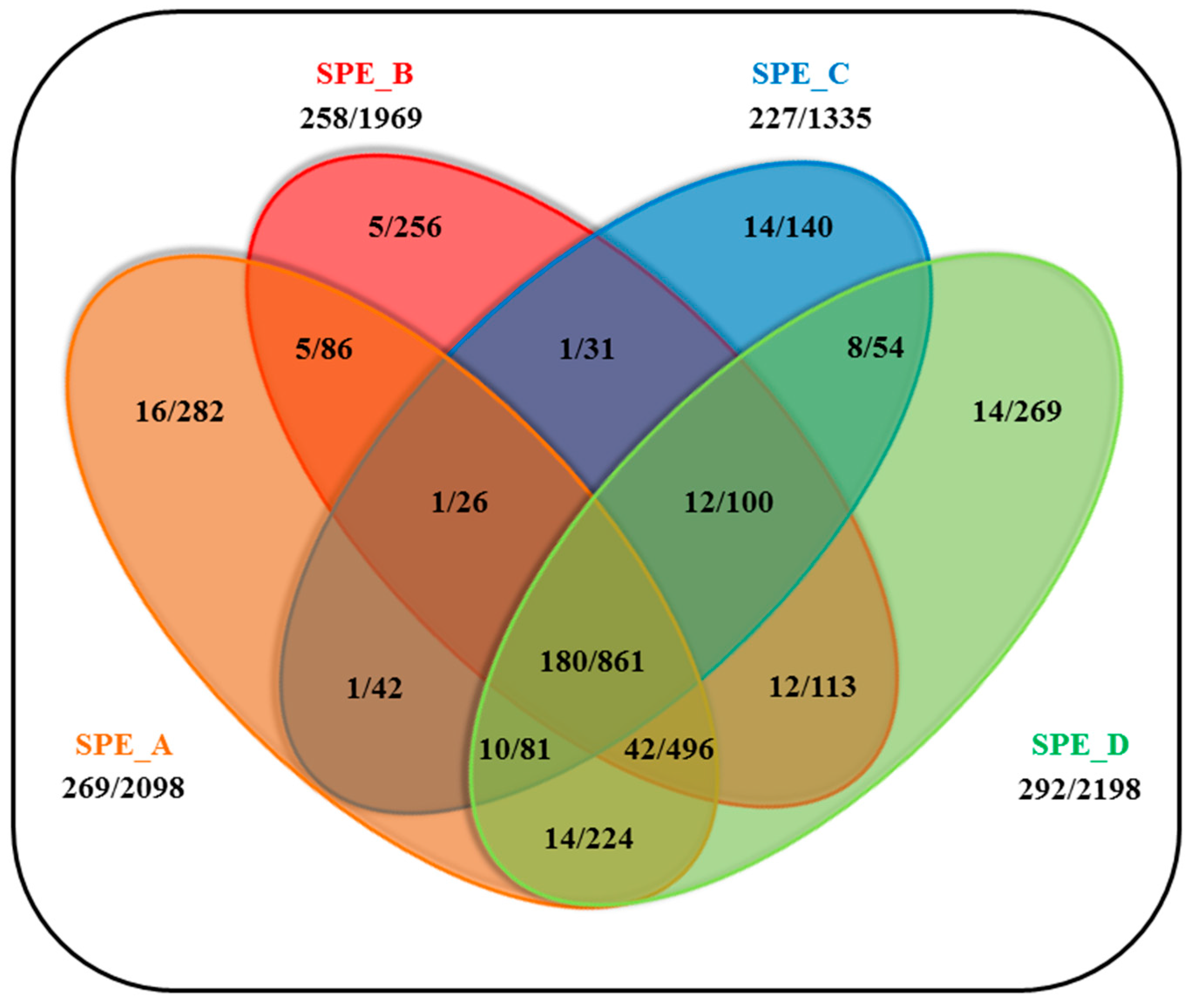
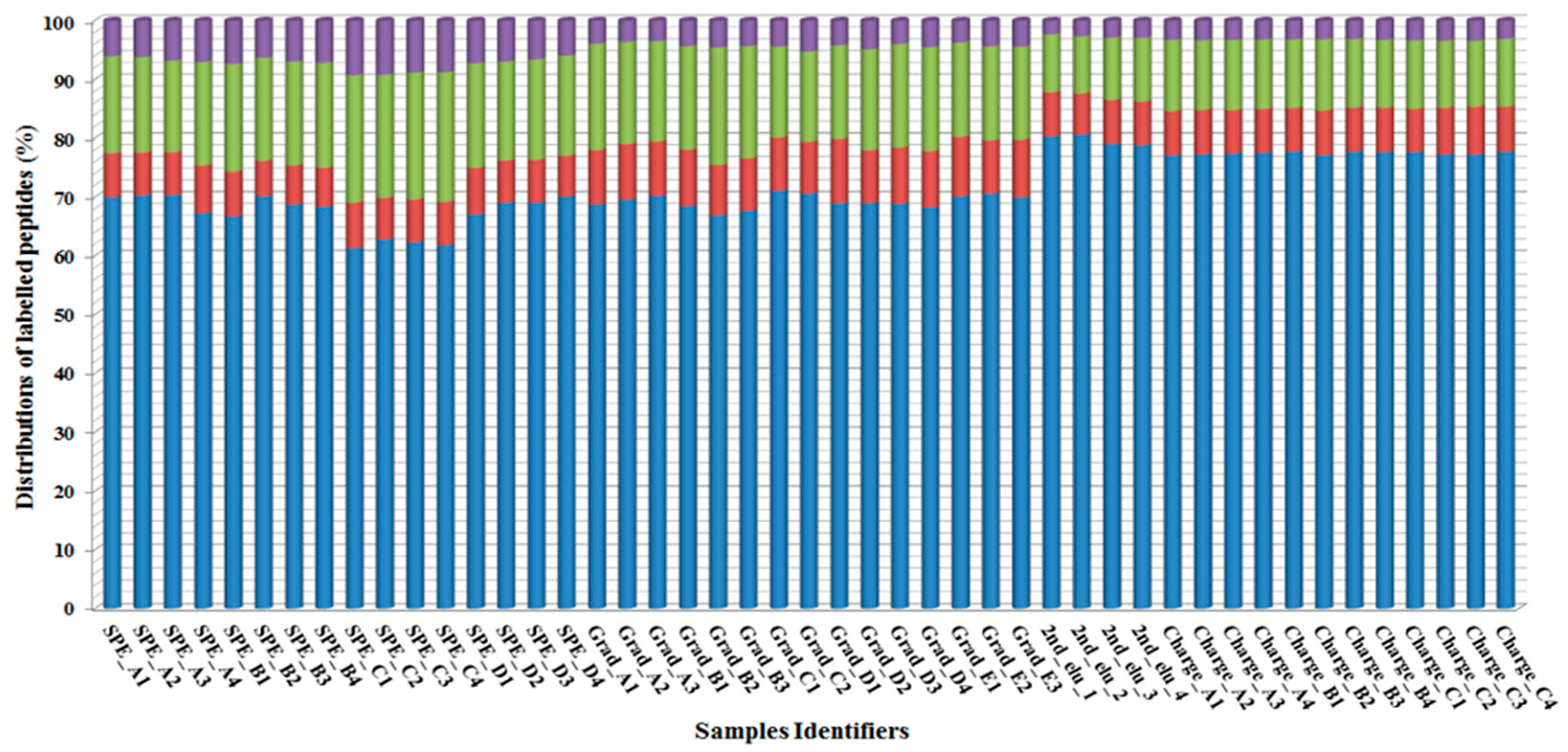
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Langó, T.; Kuffa, K.; Tóth, G.; Turiák, L.; Drahos, L.; Tusnády, G.E. Comprehensive Discovery of the Accessible Primary Amino Group-Containing Segments from Cell Surface Proteins by Fine-Tuning a High-Throughput Biotinylation Method. Int. J. Mol. Sci. 2023, 24, 273. https://doi.org/10.3390/ijms24010273
Langó T, Kuffa K, Tóth G, Turiák L, Drahos L, Tusnády GE. Comprehensive Discovery of the Accessible Primary Amino Group-Containing Segments from Cell Surface Proteins by Fine-Tuning a High-Throughput Biotinylation Method. International Journal of Molecular Sciences. 2023; 24(1):273. https://doi.org/10.3390/ijms24010273
Chicago/Turabian StyleLangó, Tamás, Katalin Kuffa, Gábor Tóth, Lilla Turiák, László Drahos, and Gábor E. Tusnády. 2023. "Comprehensive Discovery of the Accessible Primary Amino Group-Containing Segments from Cell Surface Proteins by Fine-Tuning a High-Throughput Biotinylation Method" International Journal of Molecular Sciences 24, no. 1: 273. https://doi.org/10.3390/ijms24010273
APA StyleLangó, T., Kuffa, K., Tóth, G., Turiák, L., Drahos, L., & Tusnády, G. E. (2023). Comprehensive Discovery of the Accessible Primary Amino Group-Containing Segments from Cell Surface Proteins by Fine-Tuning a High-Throughput Biotinylation Method. International Journal of Molecular Sciences, 24(1), 273. https://doi.org/10.3390/ijms24010273






