RAGE Inhibitors for Targeted Therapy of Cancer: A Comprehensive Review
Abstract
1. Introduction
2. Immunotherapy for Cancer Treatment
3. Carbonyl Stress and RAGE Ligands in Cancer
3.1. Advanced Glycation End Products (AGEs)
3.2. High-Mobility Group Box1
3.3. S100/Calgranulin Family of Proteins
3.3.1. S100A4/Calvasculin
3.3.2. S100A6/Calcyclin
3.3.3. S100A7/Psoriasin
3.3.4. S100A8/Calgranulin-A and S100A9/Calgranulin-B
3.3.5. S100A14/S100 Calcium-Binding Protein A14
3.3.6. S100A16/S100 Calcium-Binding Protein A16
3.3.7. S100B/S100 Calcium-Binding Protein B
3.3.8. S100P/S100 Calcium-Binding Protein P
3.4. Adhesion Molecules
3.5. Complement Component
3.6. Advanced Lipoxidation end Products (ALEs)
3.7. Lipopolysaccharide
4. RAGE-Specific Signaling in Cancer Progression
5. Anti-RAGE Therapeutics in Cancer Management
5.1. siRNA Inhibitor
5.2. shRNA Inhibitor
5.3. RBGO1 and ADC
5.4. Sorafenib
5.5. Duloxetine
5.6. Heparin
5.7. S100P-Derived RAGE Inhibitor
5.8. FPS-ZM1 and LY294002
5.9. Gefitinib
5.10. Metformin
5.11. RAP (RAGE Antagonist Peptide)
5.12. Cromolyn
5.13. Ethyl Pyruvate
5.14. Papaverine
5.15. Hispidin and Ergothioneine
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 6 September 2022).
- Neeper, M.; Schmidt, A.M.; Brett, J.; Yan, S.D.; Wang, F.; Pan, Y.C.; Elliston, K.; Stern, D.; Shaw, A. Cloning and Expression of a Cell Surface Receptor for Advanced Glycosylation End Products of Proteins. J. Biol. Chem. 1992, 267, 14998–15004. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Song, Y.; Zhou, L.; Li, W.; Zhu, X. Downregulation of RAGE Inhibits Cell Proliferation and Induces Apoptosis via Regulation of PI3K/AKT Pathway in Cervical Squamous Cell Carcinoma. OncoTargets Ther. 2020, 13, 2385. [Google Scholar] [CrossRef] [PubMed]
- Buckley, S.T.; Ehrhardt, C. The Receptor for Advanced Glycation End Products (RAGE) and the Lung. J. Biomed. Biotechnol. 2010, 2010, 917108. [Google Scholar] [CrossRef]
- Lizotte, P.-P.; Hanford, L.E.; Enghild, J.J.; Nozik-Grayck, E.; Giles, B.-L.; Oury, T.D. Developmental Expression of the Receptor for Advanced Glycation End-Products (RAGE) and Its Response to Hyperoxia in the Neonatal Rat Lung. BMC Dev. Biol. 2007, 7, 15. [Google Scholar] [CrossRef] [PubMed]
- Schraml, P.; Bendik, I.; Ludwig, C.U. Differential Messenger RNA and Protein Expression of the Receptor for Advanced Glycosylated End Products in Normal Lung and Non-Small Cell Lung Carcinoma. Cancer Res. 1997, 57, 3669–3671. [Google Scholar]
- Bartling, B.; Demling, N.; Silber, R.-E.; Simm, A. Proliferative Stimulus of Lung Fibroblasts on Lung Cancer Cells Is Impaired by the Receptor for Advanced Glycation End-Products. Am. J. Respir. Cell Mol. Biol. 2006, 34, 83–91. [Google Scholar] [CrossRef]
- Bartling, B.; Hofmann, H.-S.; Weigle, B.; Silber, R.-E.; Simm, A. Down-Regulation of the Receptor for Advanced Glycation End-Products (RAGE) Supports Non-Small Cell Lung Carcinoma. Carcinogenesis 2005, 26, 293–301. [Google Scholar] [CrossRef]
- Hofmann, H.-S.; Hansen, G.; Burdach, S.; Bartling, B.; Silber, R.-E.; Simm, A. Discrimination of Human Lung Neoplasm from Normal Lung by Two Target Genes. Am. J. Respir. Crit. Care Med. 2004, 170, 516–519. [Google Scholar] [CrossRef]
- Hsieh, H.-L.; Schäfer, B.W.; Sasaki, N.; Heizmann, C.W. Expression Analysis of S100 Proteins and RAGE in Human Tumors Using Tissue Microarrays. Biochem. Biophys. Res. Commun. 2003, 307, 375–381. [Google Scholar] [CrossRef]
- Taguchi, A.; Blood, D.C.; del Toro, G.; Canet, A.; Lee, D.C.; Qu, W.; Tanji, N.; Lu, Y.; Lalla, E.; Fu, C. Blockade of RAGE–Amphoterin Signalling Suppresses Tumour Growth and Metastases. Nature 2000, 405, 354–360. [Google Scholar] [CrossRef]
- Kwak, T.; Drews-Elger, K.; Ergonul, A.; Miller, P.C.; Braley, A.; Hwang, G.H.; Zhao, D.; Besser, A.; Yamamoto, Y.; Yamamoto, H. Targeting of RAGE-Ligand Signaling Impairs Breast Cancer Cell Invasion and Metastasis. Oncogene 2017, 36, 1559–1572. [Google Scholar] [CrossRef] [PubMed]
- Logsdon, C.D.; Fuentes, M.K.; Huang, E.H.; Arumugam, T. RAGE and RAGE Ligands in Cancer. Curr. Mol. Med. 2007, 7, 777–789. [Google Scholar] [CrossRef] [PubMed]
- Kang, R.; Tang, D.; Schapiro, N.E.; Livesey, K.M.; Farkas, A.; Loughran, P.; Bierhaus, A.; Lotze, M.T.; Zeh, H.J. The Receptor for Advanced Glycation End Products (RAGE) Sustains Autophagy and Limits Apoptosis, Promoting Pancreatic Tumor Cell Survival. Cell Death Differ. 2010, 17, 666–676. [Google Scholar] [CrossRef]
- Chavakis, T.; Bierhaus, A.; Al-Fakhri, N.; Schneider, D.; Witte, S.; Linn, T.; Nagashima, M.; Morser, J.; Arnold, B.; Preissner, K.T. The Pattern Recognition Receptor (RAGE) Is a Counterreceptor for Leukocyte Integrins a Novel Pathway for Inflammatory Cell Recruitment. J. Exp. Med. 2003, 198, 1507–1515. [Google Scholar] [CrossRef] [PubMed]
- Fehrenbach, H.; Kasper, M.; Tschernig, T.; Shearman Ms, S.D.; Müller, M. Receptor Foradvanced Glycation Endproducts (Rage) Exillbits Idghly Differential Cellular and Subcellular Localisation in Rat and Human Lung. Cell. Mol. Biol. 1998, 44, 1147–1157. [Google Scholar] [PubMed]
- Bierhaus, A.; Humpert, P.M.; Morcos, M.; Wendt, T.; Chavakis, T.; Arnold, B.; Stern, D.M.; Nawroth, P.P. Understanding RAGE, the Receptor for Advanced Glycation End Products. J. Mol. Med. 2005, 83, 876–886. [Google Scholar] [CrossRef]
- Schmidt, A.M.; Du Yan, S.; Yan, S.F.; Stern, D.M. The Multiligand Receptor RAGE as a Progression Factor Amplifying Immune and Inflammatory Responses. J. Clin. Investig. 2001, 108, 949–955. [Google Scholar] [CrossRef]
- Ishiguro, H.; Nakaigawa, N.; Miyoshi, Y.; Fujinami, K.; Kubota, Y.; Uemura, H. Receptor for Advanced Glycation End Products (RAGE) and Its Ligand, Amphoterin Are Overexpressed and Associated with Prostate Cancer Development. Prostate 2005, 64, 92–100. [Google Scholar] [CrossRef]
- Kuniyasu, H.; Chihara, Y.; Takahashi, T. Co-Expression of Receptor for Advanced Glycation End Products and the Ligand Amphoterin Associates Closely with Metastasis of Colorectal Cancer. Oncol. Rep. 2003, 10, 445–448. [Google Scholar] [CrossRef]
- Kuniyasu, H.; Oue, N.; Wakikawa, A.; Shigeishi, H.; Matsutani, N.; Kuraoka, K.; Ito, R.; Yokozaki, H.; Yasui, W. Expression of Receptors for Advanced Glycation End-products (RAGE) Is Closely Associated with the Invasive and Metastatic Activity of Gastric Cancer. J. Pathol. 2002, 196, 163–170. [Google Scholar] [CrossRef]
- Lotze, M.T.; Tracey, K.J. High-Mobility Group Box 1 Protein (HMGB1): Nuclear Weapon in the Immune Arsenal. Nat. Rev. Immunol. 2005, 5, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Huttunen, H.J.; Fages, C.; Kuja-Panula, J.; Ridley, A.J.; Rauvala, H. Receptor for Advanced Glycation End Products-Binding COOH-Terminal Motif of Amphoterin Inhibits Invasive Migration and Metastasis. Cancer Res. 2002, 62, 4805–4811. [Google Scholar] [PubMed]
- Sakaguchi, T.; Yan, S.F.; Du Yan, S.; Belov, D.; Rong, L.L.; Sousa, M.; Andrassy, M.; Marso, S.P.; Duda, S.; Arnold, B. Central Role of RAGE-Dependent Neointimal Expansion in Arterial Restenosis. J. Clin. Investig. 2003, 111, 959–972. [Google Scholar] [CrossRef]
- Hofmann, M.A.; Drury, S.; Fu, C.; Qu, W.; Taguchi, A.; Lu, Y.; Avila, C.; Kambham, N.; Bierhaus, A.; Nawroth, P. RAGE Mediates a Novel Proinflammatory Axis: A Central Cell Surface Receptor for S100/Calgranulin Polypeptides. Cell 1999, 97, 889–901. [Google Scholar] [CrossRef]
- Kislinger, T.; Fu, C.; Huber, B.; Qu, W.; Taguchi, A.; Du Yan, S.; Hofmann, M.; Yan, S.F.; Pischetsrieder, M.; Stern, D. N ε-(Carboxymethyl) Lysine Adducts of Proteins Are Ligands for Receptor for Advanced Glycation End Products That Activate Cell Signaling Pathways and Modulate Gene Expression. J. Biol. Chem. 1999, 274, 31740–31749. [Google Scholar] [CrossRef]
- Lander, H.M.; Tauras, J.M.; Ogiste, J.S.; Hori, O.; Moss, R.A.; Schmidt, A.M. Activation of the Receptor for Advanced Glycation End Products Triggers a P21 Ras-Dependent Mitogen-Activated Protein Kinase Pathway Regulated by Oxidant Stress. J. Biol. Chem. 1997, 272, 17810–17814. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Guh, J.; Chen, H.; Hung, W.; Lai, Y.; Chuang, L. Role of Receptor for Advanced Glycation End-product (RAGE) and the JAK/STAT-signaling Pathway in AGE-induced Collagen Production in NRK-49F Cells. J. Cell. Biochem. 2001, 81, 102–113. [Google Scholar] [CrossRef]
- Yeh, C.-H.; Sturgis, L.; Haidacher, J.; Zhang, X.-N.; Sherwood, S.J.; Bjercke, R.J.; Juhasz, O.; Crow, M.T.; Tilton, R.G.; Denner, L. Requirement for P38 and P44/P42 Mitogen-Activated Protein Kinases in RAGE-Mediated Nuclear Factor-ΚB Transcriptional Activation and Cytokine Secretion. Diabetes 2001, 50, 1495–1504. [Google Scholar] [CrossRef]
- Arumugam, T.; Simeone, D.M.; Schmidt, A.M.; Logsdon, C.D. S100P Stimulates Cell Proliferation and Survival via Receptor for Activated Glycation End Products (RAGE). J. Biol. Chem. 2004, 279, 5059–5065. [Google Scholar] [CrossRef]
- Arumugam, T.; Simeone, D.M.; Van Golen, K.; Logsdon, C.D. S100P Promotes Pancreatic Cancer Growth, Survival, and Invasion. Clin. Cancer Res. 2005, 11, 5356–5364. [Google Scholar] [CrossRef]
- Huttunen, H.J.; Fages, C.; Rauvala, H. Receptor for Advanced Glycation End Products (RAGE)-Mediated Neurite Outgrowth and Activation of NF-ΚB Require the Cytoplasmic Domain of the Receptor but Different Downstream Signaling Pathways. J. Biol. Chem. 1999, 274, 19919–19924. [Google Scholar] [CrossRef] [PubMed]
- Bierhaus, A.; Schiekofer, S.; Schwaninger, M.; Andrassy, M.; Humpert, P.M.; Chen, J.; Hong, M.; Luther, T.; Henle, T.; Klöting, I. Diabetes-Associated Sustained Activation of the Transcription Factor Nuclear Factor-ΚB. Diabetes 2001, 50, 2792–2808. [Google Scholar] [CrossRef] [PubMed]
- Moysa, A.; Hammerschmid, D.; Szczepanowski, R.H.; Sobott, F.; Dadlez, M. Enhanced Oligomerization of Full-Length RAGE by Synergy of the Interaction of Its Domains. Sci. Rep. 2019, 9, 20332. [Google Scholar] [CrossRef]
- Fritz, G. RAGE: A Single Receptor Fits Multiple Ligands. Trends Biochem. Sci. 2011, 36, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Dattilo, B.M.; Fritz, G.; Leclerc, E.; Vander Kooi, C.W.; Heizmann, C.W.; Chazin, W.J. The Extracellular Region of the Receptor for Advanced Glycation End Products Is Composed of Two Independent Structural Units. Biochemistry 2007, 46, 6957–6970. [Google Scholar] [CrossRef]
- Xue, J.; Rai, V.; Singer, D.; Chabierski, S.; Xie, J.; Reverdatto, S.; Burz, D.S.; Schmidt, A.M.; Hoffmann, R.; Shekhtman, A. Advanced Glycation End Product Recognition by the Receptor for AGEs. Structure 2011, 19, 722–732. [Google Scholar] [CrossRef]
- Leclerc, E.; Fritz, G.; Vetter, S.W.; Heizmann, C.W. Binding of S100 Proteins to RAGE: An Update. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2009, 1793, 993–1007. [Google Scholar] [CrossRef]
- Zong, H.; Madden, A.; Ward, M.; Mooney, M.H.; Elliott, C.T.; Stitt, A.W. Homodimerization Is Essential for the Receptor for Advanced Glycation End Products (RAGE)-Mediated Signal Transduction. J. Biol. Chem. 2010, 285, 23137–23146. [Google Scholar] [CrossRef]
- Yatime, L.; Andersen, G.R. Structural Insights into the Oligomerization Mode of the Human Receptor for Advanced Glycation End-products. FEBS J. 2013, 280, 6556–6568. [Google Scholar] [CrossRef]
- Kierdorf, K.; Fritz, G. RAGE Regulation and Signaling in Inflammation and Beyond. J. Leukoc. Biol. 2013, 94, 55–68. [Google Scholar] [CrossRef]
- Xie, J.; Burz, D.S.; He, W.; Bronstein, I.B.; Lednev, I.; Shekhtman, A. Hexameric Calgranulin C (S100A12) Binds to the Receptor for Advanced Glycated End Products (RAGE) Using Symmetric Hydrophobic Target-Binding Patches. J. Biol. Chem. 2007, 282, 4218–4231. [Google Scholar] [CrossRef] [PubMed]
- Iwamura, M.; Yamamoto, Y.; Kitayama, Y.; Higuchi, K.; Fujimura, T.; Hase, T.; Yamamoto, H. Epidermal Expression of Receptor for Advanced Glycation End Products (RAGE) Is Related to Inflammation and Apoptosis in Human Skin. Exp. Dermatol. 2016, 25, 235–237. [Google Scholar] [CrossRef] [PubMed]
- Hudson, B.I.; Carter, A.M.; Harja, E.; Kalea, A.Z.; Arriero, M.; Yang, H.; Grant, P.J.; Schmidt, A.M. Identification, Classification, and Expression of RAGE Gene Splice Variants. FASEB J. 2008, 22, 1572–1580. [Google Scholar] [CrossRef] [PubMed]
- Foell, D.; Wittkowski, H.; Vogl, T.; Roth, J. S100 Proteins Expressed in Phagocytes: A Novel Group of Damage-associated Molecular Pattern Molecules. J. Leukoc. Biol. 2007, 81, 28–37. [Google Scholar] [CrossRef]
- Ding, Q.; Keller, J.N. Evaluation of Rage Isoforms, Ligands, and Signaling in the Brain. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2005, 1746, 18–27. [Google Scholar] [CrossRef]
- Win, M.T.T.; Yamamoto, Y.; Munesue, S.; Saito, H.; Han, D.; Motoyoshi, S.; Kamal, T.; Ohara, T.; Watanabe, T.; Yamamoto, H. Regulation of RAGE for Attenuating Progression of Diabetic Vascular Complications. Exp. Diabetes Res. 2011, 2012, 894605. [Google Scholar] [CrossRef]
- Du Yan, S.; Chen, X.; Fu, J.; Chen, M.; Zhu, H.; Roher, A.; Slattery, T.; Zhao, L.; Nagashima, M.; Morser, J. RAGE and Amyloid-β Peptide Neurotoxicity in Alzheimer’s Disease. Nature 1996, 382, 685–691. [Google Scholar] [CrossRef]
- Tsang, D.K.; Crowe, D.L. The Mitogen Activated Protein Kinase Pathway Is Required for Proliferation but Not Invasion of Human Squamous Cell Carcinoma Lines. Int. J. Oncol. 1999, 15, 519–542. [Google Scholar] [CrossRef]
- Flohr, A.M.; Rogalla, P.; Meiboom, M.; Borrmann, L.; Krohn, M.; Thode-Halle, B.; Bullerdiek, J. Variation of HMGB1 Expression in Breast Cancer. Anticancer. Res. 2001, 21, 3881–3885. [Google Scholar]
- Reed, J.; Jurgensmeier, J.; Matsuyama, S. Bcl-2 Family Proteins and Mitochondria. Biochim. Biophys. Acta (BBA)-Bioenerg. 1998, 1366, 127–137. [Google Scholar] [CrossRef]
- Liao, S.; Li, J.; Wei, W.; Wang, L.; Zhang, Y.; Li, J.; Wang, C.; Sun, S. Association between Diabetes Mellitus and Breast Cancer Risk: A Meta-Analysis of the Literature. Asian Pac. J. Cancer Prev. 2011, 12, 1061–1065. [Google Scholar] [PubMed]
- Larsson, S.C.; Mantzoros, C.S.; Wolk, A. Diabetes Mellitus and Risk of Breast Cancer: A Meta-analysis. Int. J. Cancer 2007, 121, 856–862. [Google Scholar] [CrossRef] [PubMed]
- Marenholz, I.; Heizmann, C.W.; Fritz, G. S100 Proteins in Mouse and Man: From Evolution to Function and Pathology (Including an Update of the Nomenclature). Biochem. Biophys. Res. Commun. 2004, 322, 1111–1122. [Google Scholar] [CrossRef] [PubMed]
- Donato, R. S100: A Multigenic Family of Calcium-Modulated Proteins of the EF-Hand Type with Intracellular and Extracellular Functional Roles. Int. J. Biochem. Cell Biol. 2001, 33, 637–668. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, M.; Thornalley, P.J.; Giardino, I.; Beisswenger, P.; Thorpe, S.R.; Onorato, J.; Brownlee, M. Overexpression of Glyoxalase-I in Bovine Endothelial Cells Inhibits Intracellular Advanced Glycation Endproduct Formation and Prevents Hyperglycemia-Induced Increases in Macromolecular Endocytosis. J. Clin. Investig. 1998, 101, 1142–1147. [Google Scholar] [CrossRef] [PubMed]
- Reddy, S.; Bichler, J.; Wells-Knecht, K.J.; Thorpe, S.R.; Baynes, J.W.N. Epsilon.-(Carboxymethyl) Lysine Is a Dominant Advanced Glycation End Product (AGE) Antigen in Tissue Proteins. Biochemistry 1995, 34, 10872–10878. [Google Scholar] [CrossRef]
- Thomas, J.O.; Travers, A.A. HMG1 and 2, and Related ‘Architectural’DNA-Binding Proteins. Trends Biochem. Sci. 2001, 26, 167–174. [Google Scholar] [CrossRef]
- Wang, H.; Bloom, O.; Zhang, M.; Vishnubhakat, J.M.; Ombrellino, M.; Che, J.; Frazier, A.; Yang, H.; Ivanova, S.; Borovikova, L.; et al. HMG-1 as a late mediator of endotoxin lethality in mice. Science 1999, 285, 248–251. [Google Scholar] [CrossRef]
- Scaffidi, P.; Misteli, T.; Bianchi, M.E. Release of Chromatin Protein HMGB1 by Necrotic Cells Triggers Inflammation. Nature 2002, 418, 191–195. [Google Scholar] [CrossRef]
- Klune, J.R.; Dhupar, R.; Cardinal, J.; Billiar, T.R.; Tsung, A. HMGB1: Endogenous Danger Signaling. Mol. Med. 2008, 14, 476–484. [Google Scholar] [CrossRef]
- Ulloa, L.; Messmer, D. High-Mobility Group Box 1 (HMGB1) Protein: Friend and Foe. Cytokine Growth Factor Rev. 2006, 17, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, R.; Galvez, B.G.; Pusterla, T.; De Marchis, F.; Cossu, G.; Marcu, K.B.; Bianchi, M.E. Cells Migrating to Sites of Tissue Damage in Response to the Danger Signal HMGB1 Require NF-ΚB Activation. J. Cell Biol. 2007, 179, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Türeci, Ö. Personalized Vaccines for Cancer Immunotherapy. Science 2018, 359, 1355–1360. [Google Scholar] [CrossRef] [PubMed]
- June, C.H.; O’Connor, R.S.; Kawalekar, O.U.; Ghassemi, S.; Milone, M.C. CAR T Cell Immunotherapy for Human Cancer. Science 2018, 359, 1361–1365. [Google Scholar] [CrossRef] [PubMed]
- Restifo, N.P.; Smyth, M.J.; Snyder, A. Acquired Resistance to Immunotherapy and Future Challenges. Nat. Rev. Cancer 2016, 16, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Subramanian, S. Intrinsic Resistance of Solid Tumors to Immune Checkpoint Blockade Therapy. Cancer Res. 2017, 77, 817–822. [Google Scholar] [CrossRef] [PubMed]
- Pardoll, D.M. The Blockade of Immune Checkpoints in Cancer Immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef]
- Ahmad, S.; Khan, H.; Siddiqui, Z.; Khan, M.Y.; Rehman, S.; Shahab, U.; Godovikova, T.; Silnikov, V. AGEs, RAGEs and s-RAGE; Friend or Foe for Cancer. Semin. Cancer Biol. 2018, 49, 44–55. [Google Scholar] [CrossRef]
- Gangemi, S.; Allegra, A.; Aguennouz, M.; Alonci, A.; Speciale, A.; Cannavò, A.; Cristani, M.; Russo, S.; Spatari, G.; Alibrandi, A. Relationship between Advanced Oxidation Protein Products, Advanced Glycation End Products, and S-Nitrosylated Proteins with Biological Risk and MDR-1 Polymorphisms in Patients Affected by B-Chronic Lymphocytic Leukemia. Cancer Investig. 2012, 30, 20–26. [Google Scholar] [CrossRef]
- Chhipa, A.S.; Borse, S.P.; Baksi, R.; Lalotra, S.; Nivsarkar, M. Targeting Receptors of Advanced Glycation End Products (RAGE): Preventing Diabetes Induced Cancer and Diabetic Complications. Pathol.-Res. Pract. 2019, 215, 152643. [Google Scholar] [CrossRef]
- Abe, R.; Yamagishi, S. AGE-RAGE System and Carcinogenesis. Curr. Pharm. Des. 2008, 14, 940–945. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Park, H.K.; Yoon, J.S.; Kim, S.J.; Kim, E.S.; Ahn, K.S.; Kim, D.-S.; Yoon, S.S.; Kim, B.K.; Lee, Y.Y. Advanced Glycation End Product (AGE)-Induced Proliferation of HEL Cells via Receptor for AGE-Related Signal Pathways. Int. J. Oncol. 2008, 33, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Riehl, A.; Németh, J.; Angel, P.; Hess, J. The Receptor RAGE: Bridging Inflammation and Cancer. Cell Commun. Signal. 2009, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.-Y.; Ko, H.-A.; Shieh, T.-M.; Chang, W.-C.; Chen, H.-I.; Chang, S.-S.; Lin, I.-H. Cell Migration Is Regulated by AGE-RAGE Interaction in Human Oral Cancer Cells in Vitro. PLoS ONE 2014, 9, e110542. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Khan, M.Y.; Rafi, Z.; Khan, H.; Siddiqui, Z.; Rehman, S.; Shahab, U.; Khan, M.S.; Saeed, M.; Alouffi, S. Oxidation, Glycation and Glycoxidation—The Vicious Cycle and Lung Cancer. Semin. Cancer Biol. 2018, 49, 29–36. [Google Scholar] [CrossRef]
- Park, J.S.; Arcaroli, J.; Yum, H.-K.; Yang, H.; Wang, H.; Yang, K.-Y.; Choe, K.-H.; Strassheim, D.; Pitts, T.M.; Tracey, K.J. Activation of Gene Expression in Human Neutrophils by High Mobility Group Box 1 Protein. Am. J. Physiol.-Cell Physiol. 2003, 284, C870–C879. [Google Scholar] [CrossRef] [PubMed]
- Rowell, J.P.; Simpson, K.L.; Stott, K.; Watson, M.; Thomas, J.O. HMGB1-Facilitated P53 DNA Binding Occurs via HMG-Box/P53 Transactivation Domain Interaction, Regulated by the Acidic Tail. Structure 2012, 20, 2014–2024. [Google Scholar] [CrossRef]
- Smolarczyk, R.; Cichoń, T.; Jarosz, M.; Szala, S. HMGB1-Rola w Progresji i Terapii Przeciwnowotworowej. Adv. Hyg. Exp. Med. /Postep. Hig. Med. Dosw. 2012, 66, 913–920. [Google Scholar] [CrossRef]
- Zhang, Q.-B.; Jia, Q.; Wang, H.; Hu, C.-X.; Sun, D.; Jiang, R.-D.; Zhang, Z.-L. High-Mobility Group Protein Box1 Expression Correlates with Peritumoral Macrophage Infiltration and Unfavorable Prognosis in Patients with Hepatocellular Carcinoma and Cirrhosis. BMC Cancer 2016, 16, 880. [Google Scholar] [CrossRef]
- Choi, Y.R.; Kim, H.; Kang, H.J.; Kim, N.-G.; Kim, J.J.; Park, K.-S.; Paik, Y.-K.; Kim, H.O.; Kim, H. Overexpression of High Mobility Group Box 1 in Gastrointestinal Stromal Tumors with KIT Mutation. Cancer Res. 2003, 63, 2188–2193. [Google Scholar]
- Völp, K.; Brezniceanu, M.-L.; Bösser, S.; Brabletz, T.; Kirchner, T.; Göttel, D.; Joos, S.; Zörnig, M. Increased Expression of High Mobility Group Box 1 (HMGB1) Is Associated with an Elevated Level of the Antiapoptotic c-IAP2 Protein in Human Colon Carcinomas. Gut 2006, 55, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Chang, Y.; Liang, X.; Cardinal, J.S.; Huang, H.; Thorne, S.H.; Monga, S.P.S.; Geller, D.A.; Lotze, M.T.; Tsung, A. High-mobility Group Box 1 Activates Caspase-1 and Promotes Hepatocellular Carcinoma Invasiveness and Metastases. Hepatology 2012, 55, 1863–1875. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Zhong, K.; Sun, Z.; Wu, G.; Ding, G. Receptor for Advanced Glycation End Products (RAGE) Partially Mediates HMGB1-ERKs Activation in Clear Cell Renal Cell Carcinoma. J. Cancer Res. Clin. Oncol. 2012, 138, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-Y.; Wu, L.-Q.; Zhang, T.; Han, Y.-F.; Lin, X. Autophagy-Mediated HMGB1 Release Promotes Gastric Cancer Cell Survival via RAGE Activation of Extracellular Signal-Regulated Kinases 1/2. Oncol. Rep. 2015, 33, 1630–1638. [Google Scholar] [CrossRef]
- Pusterla, T.; Nèmeth, J.; Stein, I.; Wiechert, L.; Knigin, D.; Marhenke, S.; Longerich, T.; Kumar, V.; Arnold, B.; Vogel, A. Receptor for Advanced Glycation Endproducts (RAGE) Is a Key Regulator of Oval Cell Activation and Inflammation-associated Liver Carcinogenesis in Mice. Hepatology 2013, 58, 363–373. [Google Scholar] [CrossRef]
- Sharma, S.; Evans, A.; Hemers, E. Mesenchymal-Epithelial Signalling in Tumour Microenvironment: Role of High-Mobility Group Box 1. Cell Tissue Res. 2016, 365, 357–366. [Google Scholar] [CrossRef]
- Van Beijnum, J.R.; Nowak-Sliwinska, P.; Van den Boezem, E.; Hautvast, P.; Buurman, W.A.; Griffioen, A.W. Tumor Angiogenesis Is Enforced by Autocrine Regulation of High-Mobility Group Box 1. Oncogene 2013, 32, 363–374. [Google Scholar] [CrossRef]
- Tang, D.; Kang, R.; Livesey, K.M.; Cheh, C.-W.; Farkas, A.; Loughran, P.; Hoppe, G.; Bianchi, M.E.; Tracey, K.J.; Zeh III, H.J. Endogenous HMGB1 Regulates Autophagy. J. Cell Biol. 2010, 190, 881–892. [Google Scholar] [CrossRef]
- Salmivirta, M.; Rauvala, H.; Elenius, K.; Jalkanen, M. Neurite Growth-Promoting Protein (Amphoterin, P30) Binds Syndecan. Exp. Cell Res. 1992, 200, 444–451. [Google Scholar] [CrossRef]
- Park, J.S.; Svetkauskaite, D.; He, Q.; Kim, J.-Y.; Strassheim, D.; Ishizaka, A.; Abraham, E. Involvement of Toll-like Receptors 2 and 4 in Cellular Activation by High Mobility Group Box 1 Protein. J. Biol. Chem. 2004, 279, 7370–7377. [Google Scholar] [CrossRef]
- Hori, O.; Brett, J.; Slattery, T.; Cao, R.; Zhang, J.; Chen, J.X.; Nagashima, M.; Lundh, E.R.; Vijay, S.; Nitecki, D. The Receptor for Advanced Glycation End Products (RAGE) Is a Cellular Binding Site for Amphoterin: Mediation of neurite outgrowth and co-expression of rage and amphoterin in the developing nervous system (∗). J. Biol. Chem. 1995, 270, 25752–25761. [Google Scholar] [CrossRef] [PubMed]
- Youn, J.H.; Oh, Y.J.; Kim, E.S.; Choi, J.E.; Shin, J.-S. High Mobility Group Box 1 Protein Binding to Lipopolysaccharide Facilitates Transfer of Lipopolysaccharide to CD14 and Enhances Lipopolysaccharide-Mediated TNF-α Production in Human Monocytes. J. Immunol. 2008, 180, 5067–5074. [Google Scholar] [CrossRef] [PubMed]
- Štros, M.; Muselíková-Polanská, E.; Pospíšilová, Š.; Strauss, F. High-Affinity Binding of Tumor-Suppressor Protein P53 and HMGB1 to Hemicatenated DNA Loops. Biochemistry 2004, 43, 7215–7225. [Google Scholar] [CrossRef] [PubMed]
- Sheflin, L.G.; Fucile, N.W.; Spaulding, S.W. The Specific Interactions of HMG 1 and 2 with Negatively Supercoiled DNA Are Modulated by Their Acidic C-Terminal Domains and Involve Cysteine Residues in Their HMG 1/2 Boxes. Biochemistry 1993, 32, 3238–3248. [Google Scholar] [CrossRef]
- Catez, F.; Yang, H.; Tracey, K.J.; Reeves, R.; Misteli, T.; Bustin, M. Network of Dynamic Interactions between Histone H1 and High-Mobility-Group Proteins in Chromatin. Mol. Cell. Biol. 2004, 24, 4321–4328. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Clarke, M.F. Regulation of P53 Localization. Eur. J. Biochem. 2001, 268, 2779–2783. [Google Scholar] [CrossRef] [PubMed]
- McKinney, K.; Prives, C. Efficient Specific DNA Binding by P53 Requires Both Its Central and C-Terminal Domains as Revealed by Studies with High-Mobility Group 1 Protein. Mol. Cell. Biol. 2002, 22, 6797–6808. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Mi, Y.; Yang, H.; Hu, A.; Zhang, Q.; Shang, C. The Activation of HMGB1 as a Progression Factor on Inflammation Response in Normal Human Bronchial Epithelial Cells through RAGE/JNK/NF-ΚB Pathway. Mol. Cell. Biochem. 2013, 380, 249–257. [Google Scholar] [CrossRef]
- Chen, R.-C.; Yi, P.-P.; Zhou, R.-R.; Xiao, M.-F.; Huang, Z.-B.; Tang, D.-L.; Huang, Y.; Fan, X.-G. The Role of HMGB1-RAGE Axis in Migration and Invasion of Hepatocellular Carcinoma Cell Lines. Mol. Cell. Biochem. 2014, 390, 271–280. [Google Scholar] [CrossRef]
- Dhumale, S.S.; Waghela, B.N.; Pathak, C. Quercetin Protects Necrotic Insult and Promotes Apoptosis by Attenuating the Expression of RAGE and Its Ligand HMGB1 in Human Breast Adenocarcinoma Cells. IUBMB Life 2015, 67, 361–373. [Google Scholar] [CrossRef]
- Huang, Z.; Zhong, Z.; Zhang, L.; Wang, X.; Xu, R.; Zhu, L.; Wang, Z.; Hu, S.; Zhao, X. Down-Regulation of HMGB1 Expression by ShRNA Constructs Inhibits the Bioactivity of Urothelial Carcinoma Cell Lines via the NF-ΚB Pathway. Sci. Rep. 2015, 5, 12807. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Li, X.; Chen, Y.; Fang, J.; Ge, Z. High-Mobility Group Box 1: A Novel Inducer of the Epithelial–Mesenchymal Transition in Colorectal Carcinoma. Cancer Lett. 2015, 357, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Zhang, Y.; Zhang, S. High-Mobility Group Box 1 Is Overexpressed in Cervical Carcinoma and Promotes Cell Invasion and Migration in Vitro. Oncol. Rep. 2017, 37, 831–840. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Geng, Y.; Deng, Q.; Li, R.; Shao, X.; Zhang, Z.; Xu, W.; Wu, Y.; Ma, Q. Translationally Controlled Tumor Protein Affects Colorectal Cancer Metastasis through the High Mobility Group Box 1-Dependent Pathway. Int. J. Oncol. 2018, 53, 1481–1492. [Google Scholar] [CrossRef]
- Kuniyasu, H.; Chihara, Y.; Kondo, H. Differential Effects between Amphoterin and Advanced Glycation End Products on Colon Cancer Cells. Int. J. Cancer 2003, 104, 722–727. [Google Scholar] [CrossRef]
- Sajithlal, G.; Huttunen, H.; Rauvala, H.; Münch, G. Receptor for Advanced Glycation End Products Plays a More Important Role in Cellular Survival than in Neurite Outgrowth during Retinoic Acid-Induced Differentiation of Neuroblastoma Cells. J. Biol. Chem. 2002, 277, 6888–6897. [Google Scholar] [CrossRef]
- Tang, C.-H.; Keng, Y.-T.; Liu, J.-F. HMGB-1 Induces Cell Motility and A5β1 Integrin Expression in Human Chondrosarcoma Cells. Cancer Lett. 2012, 322, 98–106. [Google Scholar] [CrossRef]
- Peng, T.; Hu, M.; Wu, T.; Chen, Z.; Zhang, C.; Huang, S.; Zhou, X. Effects of High-mobility Group Box 1 Knockdown on Proliferation, Migration and Invasion of the HONE-1 Human Nasopharyngeal Carcinoma Cell Line. Mol. Med. Rep. 2015, 12, 7531–7537. [Google Scholar] [CrossRef][Green Version]
- Arumugam, T.; Ramachandran, V.; Gomez, S.B.; Schmidt, A.M.; Logsdon, C.D. S100P-Derived RAGE Antagonistic Peptide Reduces Tumor Growth and Metastasis. Clin. Cancer Res. 2012, 18, 4356–4364. [Google Scholar] [CrossRef]
- Yaser, A.-M.; Huang, Y.; Zhou, R.-R.; Hu, G.-S.; Xiao, M.-F.; Huang, Z.-B.; Duan, C.-J.; Tian, W.; Tang, D.-L.; Fan, X.-G. The Role of Receptor for Advanced Glycation End Products (RAGE) in the Proliferation of Hepatocellular Carcinoma. Int. J. Mol. Sci. 2012, 13, 5982–5997. [Google Scholar] [CrossRef]
- Gong, W.; Wang, Z.-Y.; Chen, G.-X.; Liu, Y.-Q.; Gu, X.-Y.; Liu, W.-W. Invasion Potential of H22 Hepatocarcinoma Cells Is Increased by HMGB1-Induced Tumor NF-ΚB Signaling via Initiation of HSP70. Oncol. Rep. 2013, 30, 1249–1256. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, A.; Yamamoto, Y.; Munesue, S.; Harashima, A.; Watanabe, T.; Yonekura, H.; Yamamoto, H.; Tsuchiya, H. Low Molecular Weight Heparin Suppresses Receptor for Advanced Glycation End Products-mediated Expression of Malignant Phenotype in Human Fibrosarcoma Cells. Cancer Sci. 2013, 104, 740–749. [Google Scholar] [CrossRef]
- Berridge, M.J.; Lipp, P.; Bootman, M.D. The Versatility and Universality of Calcium Signalling. Nat. Rev. Mol. Cell Biol. 2000, 1, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Kretsinger, R.H. Why Cells Must Export Calcium; Wiley-Liss: New York, NY, USA, 1990; pp. 439–457. [Google Scholar]
- Kretsinger, R.H.; Wasserman, R.H. Structure and Evolution of Calcium-Modulated Protein. Crit. Rev. Biochem. 1980, 8, 119–174. [Google Scholar] [CrossRef] [PubMed]
- Kretsinger, R.H.; Tolbert, D.; Nakayama, S.; Pearson, W. The EF-Hand, Homologs and Analogs. In Novel Calcium-Binding Proteins; Springer: Berlin/Heidelberg, Germany, 1991; pp. 17–37. [Google Scholar]
- Heizmann, C.W.; Fritz, G.; Schafer, B.W. S100 Proteins: Structure, Functions and Pathology. Front. Biosci. 2002, 7, 1356–1368. [Google Scholar]
- Donato, R. Intracellular and Extracellular Roles of S100 Proteins. Microsc. Res. Tech. 2003, 60, 540–551. [Google Scholar] [CrossRef]
- Ostendorp, T.; Leclerc, E.; Galichet, A.; Koch, M.; Demling, N.; Weigle, B.; Heizmann, C.W.; Kroneck, P.M.H.; Fritz, G. Structural and Functional Insights into RAGE Activation by Multimeric S100B. EMBO J. 2007, 26, 3868–3878. [Google Scholar] [CrossRef]
- Chaudhuri, D.; Horrocks, W.D.; Amburgey, J.C.; Weber, D.J. Characterization of Lanthanide Ion Binding to the EF-Hand Protein S100β by Luminescence Spectroscopy. Biochemistry 1997, 36, 9674–9680. [Google Scholar] [CrossRef]
- Mishra, S.K.; Siddique, H.R.; Saleem, M. S100A4 Calcium-Binding Protein Is Key Player in Tumor Progression and Metastasis: Preclinical and Clinical Evidence. Cancer Metastasis Rev. 2012, 31, 163–172. [Google Scholar] [CrossRef]
- Boye, K.; Mælandsmo, G.M. S100A4 and Metastasis: A Small Actor Playing Many Roles. Am. J. Pathol. 2010, 176, 528–535. [Google Scholar] [CrossRef]
- Yammani, R.R.; Carlson, C.S.; Bresnick, A.R.; Loeser, R.F. Increase in Production of Matrix Metalloproteinase 13 by Human Articular Chondrocytes Due to Stimulation with S100A4: Role of the Receptor for Advanced Glycation End Products. Arthritis Rheum. 2006, 54, 2901–2911. [Google Scholar] [CrossRef] [PubMed]
- Garrett, S.C.; Varney, K.M.; Weber, D.J.; Bresnick, A.R. S100A4, a Mediator of Metastasis. J. Biol. Chem. 2006, 281, 677–680. [Google Scholar] [CrossRef] [PubMed]
- Dahlmann, M.; Okhrimenko, A.; Marcinkowski, P.; Osterland, M.; Herrmann, P.; Smith, J.; Heizmann, C.W.; Schlag, P.M.; Stein, U. RAGE Mediates S100A4-Induced Cell Motility via MAPK/ERK and Hypoxia Signaling and Is a Prognostic Biomarker for Human Colorectal Cancer Metastasis. Oncotarget 2014, 5, 3220. [Google Scholar] [CrossRef]
- Grotterød, I.; Mælandsmo, G.M.; Boye, K. Signal Transduction Mechanisms Involved in S100A4-Induced Activation of the Transcription Factor NF-ΚB. BMC Cancer 2010, 10, 241. [Google Scholar] [CrossRef] [PubMed]
- Haase-Kohn, C.; Wolf, S.; Herwig, N.; Mosch, B.; Pietzsch, J. Metastatic Potential of B16-F10 Melanoma Cells Is Enhanced by Extracellular S100A4 Derived from RAW264. 7 Macrophages. Biochem. Biophys. Res. Commun. 2014, 446, 143–148. [Google Scholar] [CrossRef]
- Haase-Kohn, C.; Wolf, S.; Lenk, J.; Pietzsch, J. Copper-Mediated Cross-Linking of S100A4, but Not of S100A2, Results in Proinflammatory Effects in Melanoma Cells. Biochem. Biophys. Res. Commun. 2011, 413, 494–498. [Google Scholar] [CrossRef]
- Herwig, N.; Belter, B.; Pietzsch, J. Extracellular S100A4 Affects Endothelial Cell Integrity and Stimulates Transmigration of A375 Melanoma Cells. Biochem. Biophys. Res. Commun. 2016, 477, 963–969. [Google Scholar] [CrossRef]
- Nikitenko, L.L.; Lloyd, B.H.; Rudland, P.S.; Fear, S.; Barraclough, R. Localisation by in Situ Hybridisation of S100A4 (P9Ka) MRNA in Primary Human Breast Tumour Specimens. Int. J. Cancer 2000, 86, 219–228. [Google Scholar] [CrossRef]
- Ebralidze, A.; Tulchinsky, E.; Grigorian, M.; Afanasyeva, A.; Senin, V.; Revazova, E.; Lukanidin, E. Isolation and Characterization of a Gene Specifically Expressed in Different Metastatic Cells and Whose Deduced Gene Product Has a High Degree of Homology to a Ca2+-Binding Protein Family. Genes Dev. 1989, 3, 1086–1093. [Google Scholar] [CrossRef]
- Boye, K.; Grotterød, I.; Aasheim, H.; Hovig, E.; Mælandsmo, G.M. Activation of NF-κB by Extracellular S100A4: Analysis of Signal Transduction Mechanisms and Identification of Target Genes. Int. J. Cancer 2008, 123, 1301–1310. [Google Scholar] [CrossRef]
- Schmidt-Hansen, B.; Örnås, D.; Grigorian, M.; Klingelhöfer, J.; Tulchinsky, E.; Lukanidin, E.; Ambartsumian, N. Extracellular S100A4 (Mts1) Stimulates Invasive Growth of Mouse Endothelial Cells and Modulates MMP-13 Matrix Metalloproteinase Activity. Oncogene 2004, 23, 5487–5495. [Google Scholar] [CrossRef] [PubMed]
- Novitskaya, V.; Grigorian, M.; Kriajevska, M.; Tarabykina, S.; Bronstein, I.; Berezin, V.; Bock, E.; Lukanidin, E. Oligomeric Forms of the Metastasis-Related Mts1 (S100A4) Protein Stimulate Neuronal Differentiation in Cultures of Rat Hippocampal Neurons. J. Biol. Chem. 2000, 275, 41278–41286. [Google Scholar] [CrossRef] [PubMed]
- Semov, A.; Moreno, M.J.; Onichtchenko, A.; Abulrob, A.; Ball, M.; Ekiel, I.; Pietrzynski, G.; Stanimirovic, D.; Alakhov, V. Metastasis-Associated Protein S100A4 Induces Angiogenesis through Interaction with Annexin II and Accelerated Plasmin Formation. J. Biol. Chem. 2005, 280, 20833–20841. [Google Scholar] [CrossRef] [PubMed]
- Kiryushko, D.; Novitskaya, V.; Soroka, V.; Klingelhofer, J.; Lukanidin, E.; Berezin, V.; Bock, E. Molecular Mechanisms of Ca2+ Signaling in Neurons Induced by the S100A4 Protein. Mol. Cell. Biol. 2006, 26, 3625–3638. [Google Scholar] [CrossRef]
- Baudier, J.; Glasser, N.; Gerard, D. Ions Binding to S100 Proteins. I. Calcium-and Zinc-Binding Properties of Bovine Brain S100 Alpha Alpha, S100a (Alpha Beta), and S100b (Beta Beta) Protein: Zn2+ Regulates Ca2+ Binding on S100b Protein. J. Biol. Chem. 1986, 261, 8192–8203. [Google Scholar] [CrossRef]
- Kligman, D.; Hilt, D.C. The S100 Protein Family. Trends Biochem. Sci. 1988, 13, 437–443. [Google Scholar] [CrossRef]
- Vallely, K.M.; Rustandi, R.R.; Ellis, K.C.; Varlamova, O.; Bresnick, A.R.; Weber, D.J. Solution Structure of Human Mts1 (S100A4) as Determined by NMR Spectroscopy. Biochemistry 2002, 41, 12670–12680. [Google Scholar] [CrossRef]
- Malashkevich, V.N.; Varney, K.M.; Garrett, S.C.; Wilder, P.T.; Knight, D.; Charpentier, T.H.; Ramagopal, U.A.; Almo, S.C.; Weber, D.J.; Bresnick, A.R. Structure of Ca2+-Bound S100A4 and Its Interaction with Peptides Derived from Nonmuscle Myosin-IIA. Biochemistry 2008, 47, 5111–5126. [Google Scholar] [CrossRef]
- Gingras, A.R.; Basran, J.; Prescott, A.; Kriajevska, M.; Bagshaw, C.R.; Barsukov, I.L. Crystal Structure of the Ca2+-Form and Ca2+-Binding Kinetics of Metastasis-Associated Protein, S100A4. FEBS Lett. 2008, 582, 1651–1656. [Google Scholar] [CrossRef]
- Leclerc, E.; Fritz, G.; Weibel, M.; Heizmann, C.W.; Galichet, A. S100B and S100A6 Differentially Modulate Cell Survival by Interacting with Distinct RAGE (Receptor for Advanced Glycation End Products) Immunoglobulin Domains. J. Biol. Chem. 2007, 282, 31317–31331. [Google Scholar] [CrossRef]
- Leśniak, W.; Słomnicki, Ł.P.; Filipek, A. S100A6–New Facts and Features. Biochem. Biophys. Res. Commun. 2009, 390, 1087–1092. [Google Scholar] [CrossRef]
- Yan, S.F.; Ramasamy, R.; Schmidt, A.M. Receptor for AGE (RAGE) and Its Ligands—Cast into Leading Roles in Diabetes and the Inflammatory Response. J. Mol. Med. 2009, 87, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Moroz, O.V.; Wilson, K.S.; Bronstein, I.B. The Role of Zinc in the S100 Proteins: Insights from the X-Ray Structures. Amino Acids 2011, 41, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Filipek, A.; Leśniak, W. Current View on Cellular Function of S100A6 and Its Ligands, CacyBP/SIP and Sgt1. Postep. Biochem. 2018, 64, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Ma, Q.; Wang, Y.; Ma, G.; Zhai, H. CacyBP/SIP Expression Is Involved in the Clinical Progression of Breast Cancer. World J. Surg. 2010, 34, 2545–2552. [Google Scholar] [CrossRef]
- Topolska-Woś, A.M.; Chazin, W.J.; Filipek, A. CacyBP/SIP—Structure and Variety of Functions. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2016, 1860, 79–85. [Google Scholar] [CrossRef]
- Leong, S.; Christopherson, R.; Baxter, R. Profiling of Apoptotic Changes in Human Breast Cancer Cells Using SELDI-TOF Mass Spectrometry. Cell. Physiol. Biochem. 2007, 20, 579–590. [Google Scholar] [CrossRef]
- Mohan, S.K.; Gupta, A.A.; Yu, C. Interaction of the S100A6 Mutant (C3S) with the V Domain of the Receptor for Advanced Glycation End Products (RAGE). Biochem. Biophys. Res. Commun. 2013, 434, 328–333. [Google Scholar] [CrossRef]
- Li, F.; Men, X.; Zhang, W. S100 Protein in Breast Tumor. Indian J. Cancer 2014, 51, 67. [Google Scholar]
- Cross, S.S.; Hamdy, F.C.; Deloulme, J.-C.; Rehman, I. Expression of S100 Proteins in Normal Human Tissues and Common Cancers Using Tissue Microarrays: S100A6, S100A8, S100A9 and S100A11 Are All Overexpressed in Common Cancers. Histopathology 2005, 46, 256–269. [Google Scholar] [CrossRef]
- Ito, Y.; Yoshida, H.; Tomoda, C.; Uruno, T.; Miya, A.; Kobayashi, K.; Matsuzuka, F.; Kakudo, K.; Kuma, K.; Miyauchi, A. Expression of S100A2 and S100A6 in Thyroid Carcinomas. Histopathology 2005, 46, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wang, K.; Tan, Z.; Zheng, C.; Liang, Z.; Zhao, J. Identification of Potential Therapeutic Targets for Papillary Thyroid Carcinoma by Bioinformatics Analysis. Oncol. Lett. 2016, 11, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liu, X.; Lang, H.; Zhang, S.; Luo, Y.; Zhang, J. S100 Calcium-Binding Protein A6 Promotes Epithelial-Mesenchymal Transition through β-Catenin in Pancreatic Cancer Cell Line. PLoS ONE 2015, 10, e0121319. [Google Scholar] [CrossRef] [PubMed]
- Lyu, X.-J.; Li, H.-Z.; Ma, X.; Li, X.-T.; Gao, Y.; Ni, D.; Shen, D.-L.; Gu, L.-Y.; Wang, B.-J.; Zhang, Y. Elevated S100A6 (Calcyclin) Enhances Tumorigenesis and Suppresses CXCL14-Induced Apoptosis in Clear Cell Renal Cell Carcinoma. Oncotarget 2015, 6, 6656. [Google Scholar] [CrossRef] [PubMed]
- Tamai, H.; Miyake, K.; Yamaguchi, H.; Shimada, T.; Dan, K.; Inokuchi, K. Inhibition of S100A6 Induces GVL Effects in MLL/AF4-Positive ALL in Human PBMC-SCID Mice. Bone Marrow Transplant. 2014, 49, 699–703. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wolf, R.; Howard, O.M.Z.; Dong, H.-F.; Voscopoulos, C.; Boeshans, K.; Winston, J.; Divi, R.; Gunsior, M.; Goldsmith, P.; Ahvazi, B. Chemotactic Activity of S100A7 (Psoriasin) Is Mediated by the Receptor for Advanced Glycation End Products and Potentiates Inflammation with Highly Homologous but Functionally Distinct S100A15. J. Immunol. 2008, 181, 1499–1506. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, Q.; Chen, Y.; Wang, Y.; Gao, S.; Mao, Y.; Li, M.; Peng, A.; He, D.; Xiao, X. Selective Expression of S100A7 in Lung Squamous Cell Carcinomas and Large Cell Carcinomas but Not in Adenocarcinomas and Small Cell Carcinomas. Thorax 2008, 63, 352–359. [Google Scholar] [CrossRef][Green Version]
- Guilloteau, K.; Paris, I.; Pedretti, N.; Boniface, K.; Juchaux, F.; Huguier, V.; Guillet, G.; Bernard, F.-X.; Lecron, J.-C.; Morel, F. Skin Inflammation Induced by the Synergistic Action of IL-17A, IL-22, Oncostatin M, IL-1α, and TNF-α Recapitulates Some Features of Psoriasis. J. Immunol. 2010, 184, 5263–5270. [Google Scholar] [CrossRef]
- Brodersen, D.E.; Etzerodt, M.; Madsen, P.; Celis, J.E.; Thøgersen, H.C.; Nyborg, J.; Kjeldgaard, M. EF-Hands at Atomic Resolution: The Structure of Human Psoriasin (S100A7) Solved by MAD Phasing. Structure 1998, 6, 477–489. [Google Scholar] [CrossRef]
- Celis, J.E.; Rasmussen, H.H.; Vorum, H.; Madsen, P.; Honore, B.; Wolf, H.; Orntoft, T.F. Bladder Squamous Cell Carcinomas Express Psoriasin and Externalize It to the Urine. J. Urol. 1996, 155, 2105–2112. [Google Scholar] [CrossRef]
- Ralhan, R.; DeSouza, L.V.; Matta, A.; Tripathi, S.C.; Ghanny, S.; Gupta, S.D.; Bahadur, S.; Siu, K.W.M. Discovery and Verification of Head-and-Neck Cancer Biomarkers by Differential Protein Expression Analysis Using ITRAQ Labeling, Multidimensional Liquid Chromatography, and Tandem Mass Spectrometry. Mol. Cell. Proteom. 2008, 7, 1162–1173. [Google Scholar] [CrossRef] [PubMed]
- Webb, M.; Emberley, E.D.; Lizardo, M.; Alowami, S.; Qing, G.; Alfia’ar, A.; Snell-Curtis, L.J.; Niu, Y.; Civetta, A.; Myal, Y. Expression Analysis of the Mouse S100A7/Psoriasin Gene in Skin Inflammation and Mammary Tumorigenesis. BMC Cancer 2005, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Brouard, M.C.; Saurat, J.-H.; Ghanem, G.; Siegenthaler, G. Urinary Excretion of Epidermal-Type Fatty Acid-Binding Protein and S100A7 Protein in Patients with Cutaneous Melanoma. Melanoma Res. 2002, 12, 627–631. [Google Scholar] [CrossRef] [PubMed]
- Moog-Lutz, C.; Bouillet, P.; Régnier, C.H.; Tomasetto, C.; Mattei, M.; Chenard, M.; Anglard, P.; Rio, M.; Basset, P. Comparative Expression of the Psoriasin (S100A7) and S100C Genes in Breast Carcinoma and Co-localization to Human Chromosome 1q21-q22. Int. J. Cancer 1995, 63, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Enerbäck, C.; Porter, D.A.; Seth, P.; Sgroi, D.; Gaudet, J.; Weremowicz, S.; Morton, C.C.; Schnitt, S.; Pitts, R.L.; Stampl, J. Psoriasin Expression in Mammary Epithelial Cells in Vitro and in Vivo. Cancer Res. 2002, 62, 43–47. [Google Scholar]
- León, R.; Murray, J.I.; Cragg, G.; Farnell, B.; West, N.R.; Pace, T.C.S.; Watson, P.H.; Bohne, C.; Boulanger, M.J.; Hof, F. Identification and Characterization of Binding Sites on S100A7, a Participant in Cancer and Inflammation Pathways. Biochemistry 2009, 48, 10591–10600. [Google Scholar] [CrossRef]
- Tian, T.; Li, X.; Hua, Z.; Ma, J.; Wu, X.; Liu, Z.; Chen, H.; Cui, Z. S100A7 Promotes the Migration, Invasion and Metastasis of Human Cervical Cancer Cells through Epithelial–Mesenchymal Transition. Oncotarget 2017, 8, 24964. [Google Scholar] [CrossRef]
- Yin, C.; Li, H.; Zhang, B.; Liu, Y.; Lu, G.; Lu, S.; Sun, L.; Qi, Y.; Li, X.; Chen, W. RAGE-Binding S100A8/A9 Promotes the Migration and Invasion of Human Breast Cancer Cells through Actin Polymerization and Epithelial–Mesenchymal Transition. Breast Cancer Res. Treat. 2013, 142, 297–309. [Google Scholar] [CrossRef]
- Vetter, S.W.; Leclerc, E. Novel Aspects of Calmodulin Target Recognition and Activation. Eur. J. Biochem. 2003, 270, 404–414. [Google Scholar] [CrossRef]
- Iotzova-Weiss, G.; Dziunycz, P.J.; Freiberger, S.N.; Läuchli, S.; Hafner, J.; Vogl, T.; French, L.E.; Hofbauer, G.F.L. S100A8/A9 Stimulates Keratinocyte Proliferation in the Development of Squamous Cell Carcinoma of the Skin via the Receptor for Advanced Glycation-End Products. PLoS ONE 2015, 10, e0120971. [Google Scholar] [CrossRef][Green Version]
- Hunter, M.J.; Chazin, W.J. High Level Expression and Dimer Characterization of the S100 EF-Hand Proteins, Migration Inhibitory Factor-Related Proteins 8 and 14. J. Biol. Chem. 1998, 273, 12427–12435. [Google Scholar] [CrossRef] [PubMed]
- Leukert, N.; Sorg, C.; Roth, J. Molecular Basis of the Complex Formation between the Two Calcium-Binding Proteins S100A8 (MRP8) and S100A9 (MRP14). Biol. Chem. 2005, 386, 429–434. [Google Scholar] [CrossRef]
- Pröpper, C.; Huang, X.; Roth, J.; Sorg, C.; Nacken, W. Analysis of the MRP8-MRP14 Protein-Protein Interaction by the Two-Hybrid System Suggests a Prominent Role of the C-Terminal Domain of S100 Proteins in Dimer Formation. J. Biol. Chem. 1999, 274, 183–188. [Google Scholar] [CrossRef]
- Nacken, W.; Roth, J.; Sorg, C.; Kerkhoff, C. S100A9/S100A8: Myeloid Representatives of the S100 Protein Family as Prominent Players in Innate Immunity. Microsc. Res. Tech. 2003, 60, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Stříž, I.; Trebichavský, I. Calprotectin—A Pleiotropic Molecule in Acute and Chronic Inflammation. Physiol. Res. 2004, 53, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Ghavami, S.; Rashedi, I.; Dattilo, B.M.; Eshraghi, M.; Chazin, W.J.; Hashemi, M.; Wesselborg, S.; Kerkhoff, C.; Los, M. S100A8/A9 at Low Concentration Promotes Tumor Cell Growth via RAGE Ligation and MAP Kinase-dependent Pathway. J. Leukoc. Biol. 2008, 83, 1484–1492. [Google Scholar] [CrossRef] [PubMed]
- Vogl, T.; Tenbrock, K.; Ludwig, S.; Leukert, N.; Ehrhardt, C.; Van Zoelen, M.A.D.; Nacken, W.; Foell, D.; Van Der Poll, T.; Sorg, C.; et al. Mrp8 and Mrp14 Are Endogenous Activators of Toll-like Receptor 4, Promoting Lethal, Endotoxin-Induced Shock. Nat. Med. 2007, 13, 1042–1049. [Google Scholar] [CrossRef]
- Loser, K.; Vogl, T.; Voskort, M.; Lueken, A.; Kupas, V.; Nacken, W.; Klenner, L.; Kuhn, A.; Foell, D.; Sorokin, L. The Toll-like Receptor 4 Ligands Mrp8 and Mrp14 Are Crucial in the Development of Autoreactive CD8+ T Cells. Nat. Med. 2010, 16, 713–717. [Google Scholar] [CrossRef]
- Hermani, A.; De Servi, B.; Medunjanin, S.; Tessier, P.A.; Mayer, D. S100A8 and S100A9 Activate MAP Kinase and NF-ΚB Signaling Pathways and Trigger Translocation of RAGE in Human Prostate Cancer Cells. Exp. Cell Res. 2006, 312, 184–197. [Google Scholar] [CrossRef]
- Greten, F.R.; Eckmann, L.; Greten, T.F.; Park, J.M.; Li, Z.-W.; Egan, L.J.; Kagnoff, M.F.; Karin, M. IKKβ Links Inflammation and Tumorigenesis in a Mouse Model of Colitis-Associated Cancer. Cell 2004, 118, 285–296. [Google Scholar] [CrossRef]
- Karin, M.; Greten, F.R. NF-ΚB: Linking Inflammation and Immunity to Cancer Development and Progression. Nat. Rev. Immunol. 2005, 5, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Németh, J.; Stein, I.; Haag, D.; Riehl, A.; Longerich, T.; Horwitz, E.; Breuhahn, K.; Gebhardt, C.; Schirmacher, P.; Hahn, M. S100A8 and S100A9 Are Novel Nuclear Factor Kappa B Target Genes during Malignant Progression of Murine and Human Liver Carcinogenesis. Hepatology 2009, 50, 1251–1262. [Google Scholar] [CrossRef] [PubMed]
- Itou, H.; Yao, M.; Fujita, I.; Watanabe, N.; Suzuki, M.; Nishihira, J.; Tanaka, I. The Crystal Structure of Human MRP14 (S100A9), a Ca2+-Dependent Regulator Protein in Inflammatory Process. J. Mol. Biol. 2002, 316, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Korndörfer, I.P.; Brueckner, F.; Skerra, A. The Crystal Structure of the Human (S100A8/S100A9) 2 Heterotetramer, Calprotectin, Illustrates How Conformational Changes of Interacting α-Helices Can Determine Specific Association of Two EF-Hand Proteins. J. Mol. Biol. 2007, 370, 887–898. [Google Scholar] [CrossRef]
- Santamaria-Kisiel, L.; Rintala-Dempsey, A.C.; Shaw, G.S. Calcium-Dependent and-Independent Interactions of the S100 Protein Family. Biochem. J. 2006, 396, 201–214. [Google Scholar] [CrossRef]
- Ishikawa, K.; Nakagawa, A.; Tanaka, I.; Suzuki, M.; Nishihira, J. The Structure of Human MRP8, a Member of the S100 Calcium-Binding Protein Family, by MAD Phasing at 1.9 Å Resolution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2000, 56, 559–566. [Google Scholar] [CrossRef]
- Pietas, A.; Schlüns, K.; Marenholz, I.; Schäfer, B.W.; Heizmann, C.W.; Petersen, I. Molecular Cloning and Characterization of the Human S100A14 Gene Encoding a Novel Member of the S100 Family. Genomics 2002, 79, 513–522. [Google Scholar] [CrossRef]
- Jin, Q.; Chen, H.; Luo, A.; Ding, F.; Liu, Z. S100A14 Stimulates Cell Proliferation and Induces Cell Apoptosis at Different Concentrations via Receptor for Advanced Glycation End Products (RAGE). PLoS ONE 2011, 6, e19375. [Google Scholar] [CrossRef]
- Bresnick, A.R.; Weber, D.J.; Zimmer, D.B. S100 Proteins in Cancer. Nat. Rev. Cancer 2015, 15, 96–109. [Google Scholar] [CrossRef]
- Sapkota, D.; Bruland, O.; Costea, D.E.; Haugen, H.; Vasstrand, E.N.; Ibrahim, S.O. S100A14 Regulates the Invasive Potential of Oral Squamous Cell Carcinoma Derived Cell-Lines in Vitro by Modulating Expression of Matrix Metalloproteinases, MMP1 and MMP9. Eur. J. Cancer 2011, 47, 600–610. [Google Scholar] [CrossRef]
- Sapkota, D.; Costea, D.E.; Blø, M.; Bruland, O.; Lorens, J.B.; Vasstrand, E.N.; Ibrahim, S.O. S100A14 Inhibits Proliferation of Oral Carcinoma Derived Cells through G1-Arrest. Oral Oncol. 2012, 48, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Chen, H.; Wang, X.; Gao, J.; Che, Y.; Li, Y.; Ding, F.; Luo, A.; Zhang, S.; Liu, Z. S100A14, a Member of the EF-Hand Calcium-Binding Proteins, Is Overexpressed in Breast Cancer and Acts as a Modulator of HER2 Signaling. J. Biol. Chem. 2014, 289, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Bigelow, R.L.H.; Williams, B.J.; Carroll, J.L.; Daves, L.K.; Cardelli, J.A. TIMP-1 Overexpression Promotes Tumorigenesis of MDA-MB-231 Breast Cancer Cells and Alters Expression of a Subset of Cancer Promoting Genes in Vivo Distinct from Those Observed in Vitro. Breast Cancer Res. Treat. 2009, 117, 31–44. [Google Scholar] [CrossRef]
- Zhu, M.; Wang, H.; Cui, J.; Li, W.; An, G.; Pan, Y.; Zhang, Q.; Xing, R.; Lu, Y. Calcium-Binding Protein S100A14 Induces Differentiation and Suppresses Metastasis in Gastric Cancer. Cell Death Dis. 2017, 8, e2938. [Google Scholar] [CrossRef] [PubMed]
- Lavoie, C.; Meerloo, T.; Lin, P.; Farquhar, M.G. Calnuc, an EF-Hand Ca2+-Binding Protein, Is Stored and Processed in the Golgi and Secreted by the Constitutive-like Pathway in AtT20 Cells. Mol. Endocrinol. 2002, 16, 2462–2474. [Google Scholar] [CrossRef] [PubMed]
- Meng, D.-F.; Sun, R.; Liu, G.-Y.; Peng, L.-X.; Zheng, L.-S.; Xie, P.; Lin, S.-T.; Mei, Y.; Qiang, Y.-Y.; Li, C.-Z. S100A14 Suppresses Metastasis of Nasopharyngeal Carcinoma by Inhibition of NF-KB Signaling through Degradation of IRAK1. Oncogene 2020, 39, 5307–5322. [Google Scholar] [CrossRef]
- Medapati, M.R.; Dahlmann, M.; Ghavami, S.; Pathak, K.A.; Lucman, L.; Klonisch, T.; Hoang-Vu, C.; Stein, U.; Hombach-Klonisch, S. RAGE Mediates the Pro-Migratory Response of Extracellular S100A4 in Human Thyroid Cancer Cells. Thyroid 2015, 25, 514–527. [Google Scholar] [CrossRef]
- Adam, P.J.; Boyd, R.; Tyson, K.L.; Fletcher, G.C.; Stamps, A.; Hudson, L.; Poyser, H.R.; Redpath, N.; Griffiths, M.; Steers, G. Comprehensive Proteomic Analysis of Breast Cancer Cell Membranes Reveals Unique Proteins with Potential Roles in Clinical Cancer. J. Biol. Chem. 2003, 278, 6482–6489. [Google Scholar] [CrossRef]
- Yao, R.; Lopez-Beltran, A.; Maclennan, G.T.; Montironi, R.; Eble, J.N.; Cheng, L. Expression of S100 Protein Family Members in the Pathogenesis of Bladder Tumors. Anticancer. Res. 2007, 27, 3051–3058. [Google Scholar]
- Smirnov, D.A.; Zweitzig, D.R.; Foulk, B.W.; Miller, M.C.; Doyle, G.V.; Pienta, K.J.; Meropol, N.J.; Weiner, L.M.; Cohen, S.J.; Moreno, J.G. Global Gene Expression Profiling of Circulating Tumor Cells. Cancer Res. 2005, 65, 4993–4997. [Google Scholar] [CrossRef]
- Chen, H.; Yu, D.; Luo, A.; Tan, W.; Zhang, C.; Zhao, D.; Yang, M.; Liu, J.; Lin, D.; Liu, Z. Functional Role of S100A14 Genetic Variants and Their Association with Esophageal Squamous Cell Carcinoma. Cancer Res. 2009, 69, 3451–3457. [Google Scholar] [CrossRef] [PubMed]
- Bertini, I.; Borsi, V.; Cerofolini, L.; Das Gupta, S.; Fragai, M.; Luchinat, C. Solution Structure and Dynamics of Human S100A14. JBIC J. Biol. Inorg. Chem. 2013, 18, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Nogueira-Machado, J.A.; M de Oliveira Volpe, C. HMGB-1 as a Target for Inflammation Controlling. Recent Pat. Endocr. Metab. Immune Drug Discov. 2012, 6, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Arnesano, F.; Banci, L.; Bertini, I.; Fantoni, A.; Tenori, L.; Viezzoli, M.S. Structural Interplay between Calcium (II) and Copper (II) Binding to S100A13 Protein. Angew. Chem. 2005, 117, 6499–6502. [Google Scholar] [CrossRef]
- Otterbein, L.R.; Kordowska, J.; Witte-Hoffmann, C.; Wang, C.-L.A.; Dominguez, R. Crystal Structures of S100A6 in the Ca2+-Free and Ca2+-Bound States: The Calcium Sensor Mechanism of S100 Proteins Revealed at Atomic Resolution. Structure 2002, 10, 557–567. [Google Scholar] [CrossRef][Green Version]
- Drohat, A.C.; Amburgey, J.C.; Abildgaard, F.; Starich, M.R.; Baldisseri, D.; Weber, D.J. Solution Structure of Rat Apo-S100B (Ββ) as Determined by NMR Spectroscopy. Biochemistry 1996, 35, 11577–11588. [Google Scholar] [CrossRef]
- Smith, S.P.; Shaw, G.S. A Novel Calcium-Sensitive Switch Revealed by the Structure of Human S100B in the Calcium-Bound Form. Structure 1998, 6, 211–222. [Google Scholar] [CrossRef][Green Version]
- Kiess, A.P.; Cho, S.Y.; Pomper, M.G. Translational Molecular Imaging of Prostate Cancer. Curr. Radiol. Rep. 2013, 1, 216–226. [Google Scholar] [CrossRef]
- Marenholz, I.; Heizmann, C.W. S100A16, a Ubiquitously Expressed EF-Hand Protein Which Is up-Regulated in Tumors. Biochem. Biophys. Res. Commun. 2004, 313, 237–244. [Google Scholar] [CrossRef]
- Heizmann, C.W.; Fritz, G. The Family of S100 Cell Signaling Proteins. In Handbook of Cell Signaling; Elsevier: Amsterdam, The Netherlands, 2010; pp. 983–993. [Google Scholar]
- He, M.; Kubo, H.; Morimoto, K.; Fujino, N.; Suzuki, T.; Takahasi, T.; Yamada, M.; Yamaya, M.; Maekawa, T.; Yamamoto, Y. Receptor for Advanced Glycation End Products Binds to Phosphatidylserine and Assists in the Clearance of Apoptotic Cells. EMBO Rep. 2011, 12, 358–364. [Google Scholar] [CrossRef]
- Andersen, J.S.; Lam, Y.W.; Leung, A.K.L.; Ong, S.-E.; Lyon, C.E.; Lamond, A.I.; Mann, M. Nucleolar Proteome Dynamics. Nature 2005, 433, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Sturchler, E.; Cox, J.A.; Durussel, I.; Weibel, M.; Heizmann, C.W. S100A16, a Novel Calcium-Binding Protein of the EF-Hand Superfamily. J. Biol. Chem. 2006, 281, 38905–38917. [Google Scholar] [CrossRef] [PubMed]
- Sapkota, D.; Costea, D.E.; Ibrahim, S.O.; Johannessen, A.C.; Bruland, O. S100A14 Interacts with S100A16 and Regulates Its Expression in Human Cancer Cells. PLoS ONE 2013, 8, e76058. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Pan, H.; Xia, T.; Xue, J.; Cheng, L.; Fan, P.; Zhang, Y.; Zhu, W.; Xue, Y.; Liu, X. Up-Regulation of S100A16 Expression Promotes Epithelial-Mesenchymal Transition via Notch1 Pathway in Breast Cancer. J. Biomed. Sci. 2014, 21, 97. [Google Scholar] [CrossRef] [PubMed]
- Sapkota, D.; Bruland, O.; Parajuli, H.; Osman, T.A.; Teh, M.-T.; Johannessen, A.C.; Costea, D.E. S100A16 Promotes Differentiation and Contributes to a Less Aggressive Tumor Phenotype in Oral Squamous Cell Carcinoma. BMC Cancer 2015, 15, 631. [Google Scholar] [CrossRef]
- Kobayashi, M.; Nagashio, R.; Saito, K.; Aguilar-Bonavides, C.; Ryuge, S.; Katono, K.; Igawa, S.; Tsuchiya, B.; Jiang, S.-X.; Ichinoe, M. Prognostic Significance of S100A16 Subcellular Localization in Lung Adenocarcinoma. Hum. Pathol. 2018, 74, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, T.; Ogris, C.; Sonnhammer, E.L.L. FunCoup 3.0: Database of Genome-Wide Functional Coupling Networks. Nucleic Acids Res. 2014, 42, D380–D388. [Google Scholar] [CrossRef]
- Price, D.J.; Avraham, S.; Feuerstein, J.; Fu, Y.; Avraham, H.K. The Invasive Phenotype in HMT-3522 Cells Requires Increased EGF Receptor Signaling through Both PI 3-Kinase and ERK 1, 2 Pathways. Cell Commun. Adhes. 2002, 9, 87–102. [Google Scholar] [CrossRef]
- Peng, S.; Zhang, Y.; Zhang, J.; Wang, H.; Ren, B. ERK in Learning and Memory: A Review of Recent Research. Int. J. Mol. Sci. 2010, 11, 222–232. [Google Scholar] [CrossRef]
- Zhu, W.; Xue, Y.; Liang, C.; Zhang, R.; Zhang, Z.; Li, H.; Su, D.; Liang, X.; Zhang, Y.; Huang, Q. S100A16 Promotes Cell Proliferation and Metastasis via AKT and ERK Cell Signaling Pathways in Human Prostate Cancer. Tumor Biol. 2016, 37, 12241–12250. [Google Scholar] [CrossRef]
- Babini, E.; Bertini, I.; Borsi, V.; Calderone, V.; Hu, X.; Luchinat, C.; Parigi, G. Structural Characterization of Human S100A16, a Low-Affinity Calcium Binder. JBIC J. Biol. Inorg. Chem. 2011, 16, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Steiner, J.; Bogerts, B.; Schroeter, M.L.; Bernstein, H.-G. S100B Protein in Neurodegenerative Disorders. Clin. Chem. Lab. Med. 2011, 49, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.P.; Tort, A.B.L.; Amaral, O.B.; Schmidt, A.P.; Walz, R.; Vettorazzi-Stuckzynski, J.; Martins-Costa, S.H.; Ramos, J.G.L.; Souza, D.O.; Portela, L.V.C. Serum S100B in Pregnancy-Related Hypertensive Disorders: A Case–Control Study. Clin. Chem. 2004, 50, 435–438. [Google Scholar] [CrossRef] [PubMed]
- Harpio, R.; Einarsson, R. S100 Proteins as Cancer Biomarkers with Focus on S100B in Malignant Melanoma. Clin. Biochem. 2004, 37, 512–518. [Google Scholar] [CrossRef] [PubMed]
- Hamberg, A.P.; Korse, C.M.; Bonfrer, J.M.G.; De Gast, G.C. Serum S100B Is Suitable for Prediction and Monitoring of Response to Chemoimmunotherapy in Metastatic Malignant Melanoma. Melanoma Res. 2003, 13, 45–49. [Google Scholar] [CrossRef]
- Huang, M.-Y.; Wang, H.-M.; Chang, H.-J.; Hsiao, C.-P.; Wang, J.-Y.; Lin, S.-R. Overexpression of S100B, TM4SF4, and OLFM4 Genes Is Correlated with Liver Metastasis in Taiwanese Colorectal Cancer Patients. DNA Cell Biol. 2012, 31, 43–49. [Google Scholar] [CrossRef]
- Hwang, C.-C.; Chai, H.-T.; Chen, H.-W.; Tsai, H.-L.; Lu, C.-Y.; Yu, F.-J.; Huang, M.-Y.; Wang, J.-Y. S100B Protein Expressions as an Independent Predictor of Early Relapse in UICC Stages II and III Colon Cancer Patients after Curative Resection. Ann. Surg. Oncol. 2011, 18, 139–145. [Google Scholar] [CrossRef]
- Villarreal, A.; Aviles Reyes, R.X.; Angelo, M.F.; Reines, A.G.; Ramos, A.J. S100B Alters Neuronal Survival and Dendrite Extension via RAGE-mediated NF-κB Signaling. J. Neurochem. 2011, 117, 321–332. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, L.; Zhang, I.Y.; Chen, X.; Da Fonseca, A.; Wu, S.; Ren, H.; Badie, S.; Sadeghi, S.; Ouyang, M. S100B Promotes Glioma Growth through Chemoattraction of Myeloid-Derived Macrophages. Clin. Cancer Res. 2013, 19, 3764–3775. [Google Scholar] [CrossRef]
- Yang, T.; Cheng, J.; Yang, Y.; Qi, W.; Zhao, Y.; Long, H.; Xie, R.; Zhu, B. S100B Mediates Stemness of Ovarian Cancer Stem-like Cells through Inhibiting P53. Stem Cells 2017, 35, 325–336. [Google Scholar] [CrossRef]
- Zhang, F.; Banker, G.; Liu, X.; Suwanabol, P.A.; Lengfeld, J.; Yamanouchi, D.; Kent, K.C.; Liu, B. The Novel Function of Advanced Glycation End Products in Regulation of MMP-9 Production. J. Surg. Res. 2011, 171, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Donato, R. RAGE: A Single Receptor for Several Ligands and Different Cellular Responses: The Case of Certain S100 Proteins. Curr. Mol. Med. 2007, 7, 711–724. [Google Scholar] [CrossRef] [PubMed]
- Baudier, J.; Delphin, C.; Grunwald, D.; Khochbin, S.; Lawrence, J.J. Characterization of the Tumor Suppressor Protein P53 as a Protein Kinase C Substrate and a S100b-Binding Protein. Proc. Natl. Acad. Sci. USA 1992, 89, 11627–11631. [Google Scholar] [CrossRef]
- Rustandi, R.R.; Drohat, A.C.; Baldisseri, D.M.; Wilder, P.T.; Weber, D.J. The Ca2+-Dependent Interaction of S100B (Ββ) with a Peptide Derived from P53. Biochemistry 1998, 37, 1951–1960. [Google Scholar] [CrossRef] [PubMed]
- Delphin, C.; Ronjat, M.; Deloulme, J.C.; Garin, G.; Debussche, L.; Higashimoto, Y.; Sakaguchi, K.; Baudier, J. Calcium-Dependent Interaction of S100B with the C-Terminal Domain of the Tumor Suppressor P53. J. Biol. Chem. 1999, 274, 10539–10544. [Google Scholar] [CrossRef]
- Lin, J.; Yang, Q.; Yan, Z.; Markowitz, J.; Wilder, P.T.; Carrier, F.; Weber, D.J. Inhibiting S100B Restores P53 Levels in Primary Malignant Melanoma Cancer Cells. J. Biol. Chem. 2004, 279, 34071–34077. [Google Scholar] [CrossRef]
- Lin, J.; Blake, M.; Tang, C.; Zimmer, D.; Rustandi, R.R.; Weber, D.J.; Carrier, F. Inhibition of P53 Transcriptional Activity by the S100B Calcium-Binding Protein. J. Biol. Chem. 2001, 276, 35037–35041. [Google Scholar] [CrossRef]
- Wilder, P.T.; Lin, J.; Bair, C.L.; Charpentier, T.H.; Yang, D.; Liriano, M.; Varney, K.M.; Lee, A.; Oppenheim, A.B.; Adhya, S. Recognition of the Tumor Suppressor Protein P53 and Other Protein Targets by the Calcium-Binding Protein S100B. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2006, 1763, 1284–1297. [Google Scholar] [CrossRef]
- Drohat, A.C.; Baldisseri, D.M.; Rustandi, R.R.; Weber, D.J. Solution Structure of Calcium-Bound Rat S100B (Ββ) as Determined by Nuclear Magnetic Resonance Spectroscopy. Biochemistry 1998, 37, 2729–2740. [Google Scholar] [CrossRef]
- Rust, R.R.; Baldisseri, D.M.; Weber, D.J. Structure of the Negative Regulatory Domain of P53 Bound to S100B (Ββ). Nat. Struct. Biol. 2000, 7, 570–574. [Google Scholar]
- Amburgey, J.C.; Abildgaard, F.; Starich, M.R.; Shah, S.; Hilt, D.C.; Weber, D.J. 1 H, 13 C and 15 N NMR Assignments and Solution Secondary Structure of Rat Apo-S100β. J. Biomol. NMR 1995, 6, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Fritz, G.; Heizmann, C.W. 3D Structures of the Calcium and Zinc Binding S100 Proteins. In Handbook of Metalloproteins; Wiley: Hoboken, NJ, USA, 2004; Volume 3, pp. 529–540. [Google Scholar]
- Xie, J.; Méndez, J.D.; Méndez-Valenzuela, V.; Aguilar-Hernández, M.M. Cellular Signalling of the Receptor for Advanced Glycation End Products (RAGE). Cell. Signal. 2013, 25, 2185–2197. [Google Scholar] [CrossRef] [PubMed]
- Ostendorp, T.; Diez, J.; Heizmann, C.W.; Fritz, G. The Crystal Structures of Human S100B in the Zinc-and Calcium-Loaded State at Three PH Values Reveal Zinc Ligand Swapping. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2011, 1813, 1083–1091. [Google Scholar] [CrossRef] [PubMed]
- Sorci, G.; Riuzzi, F.; Agneletti, A.L.; Marchetti, C.; Donato, R. S100B Causes Apoptosis in a Myoblast Cell Line in a RAGE-independent Manner. J. Cell. Physiol. 2004, 199, 274–283. [Google Scholar] [CrossRef]
- Huttunen, H.J.; Kuja-Panula, J.; Sorci, G.; Agneletti, A.L.; Donato, R.; Rauvala, H. Coregulation of Neurite Outgrowth and Cell Survival by Amphoterin and S100 Proteins through Receptor for Advanced Glycation End Products (RAGE) Activation. J. Biol. Chem. 2000, 275, 40096–40105. [Google Scholar] [CrossRef]
- Marsden, B.J.; Shaw, G.S.; Sykes, B.D. Calcium Binding Proteins. Elucidating the Contributions to Calcium Affinity from an Analysis of Species Variants and Peptide Fragments. Biochem. Cell Biol. 1990, 68, 587–601. [Google Scholar] [CrossRef]
- Averboukh, L.; Liang, P.; Kantoff, P.W.; Pardee, A.B. Regulation of S100P Expression by Androgen. Prostate 1996, 29, 350–355. [Google Scholar] [CrossRef]
- Logsdon, C.D.; Simeone, D.M.; Binkley, C.; Arumugam, T.; Greenson, J.K.; Giordano, T.J.; Misek, D.E.; Hanash, S. Molecular Profiling of Pancreatic Adenocarcinoma and Chronic Pancreatitis Identifies Multiple Genes Differentially Regulated in Pancreatic Cancer. Cancer Res. 2003, 63, 2649–2657. [Google Scholar]
- Beer, D.G.; Kardia, S.L.R.; Huang, C.-C.; Giordano, T.J.; Levin, A.M.; Misek, D.E.; Lin, L.; Chen, G.; Gharib, T.G.; Thomas, D.G. Gene-Expression Profiles Predict Survival of Patients with Lung Adenocarcinoma. Nat. Med. 2002, 8, 816–824. [Google Scholar] [CrossRef]
- Fuentes, M.K.; Nigavekar, S.S.; Arumugam, T.; Logsdon, C.D.; Schmidt, A.M.; Park, J.C.; Huang, E.H. RAGE Activation by S100P in Colon Cancer Stimulates Growth, Migration, and Cell Signaling Pathways. Dis. Colon Rectum 2007, 50, 1230–1240. [Google Scholar] [CrossRef]
- Wang, G.; Platt-Higgins, A.; Carroll, J.; de Silva Rudland, S.; Winstanley, J.; Barraclough, R.; Rudland, P.S. Induction of Metastasis by S100P in a Rat Mammary Model and Its Association with Poor Survival of Breast Cancer Patients. Cancer Res. 2006, 66, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Rehbein, G.; Simm, A.; Hofmann, H.-S.; Silber, R.-E.; Bartling, B. Molecular Regulation of S100P in Human Lung Adenocarcinomas. Int. J. Mol. Med. 2008, 22, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, Y.-N.; Lin, G.-L.; Qiu, H.-Z.; Wu, B.; Wu, H.-Y.; Zhao, Y.; Chen, Y.-J.; Lu, C.-M. S100P, a Potential Novel Prognostic Marker in Colorectal Cancer. Oncol. Rep. 2012, 28, 303–310. [Google Scholar] [PubMed]
- Tothova, V.; Gibadulinova, A. S100P, a Peculiar Member of S100 Family of Calcium-Binding Proteins Implicated in Cancer. Acta Virol. 2013, 57, 238–246. [Google Scholar] [CrossRef]
- Lee, Y.-C.; Volk, D.E.; Thiviyanathan, V.; Kleerekoper, Q.; Gribenko, A.V.; Zhang, S.; Gorenstein, D.G.; Makhatadze, G.I.; Luxon, B.A. Letter to the Editor: NMR Structure of the Apo-S100P Protein. J. Biomol. NMR 2004, 29, 399–402. [Google Scholar] [CrossRef]
- Austermann, J.; Nazmi, A.R.; Müller-Tidow, C.; Gerke, V. Characterization of the Ca2+-Regulated Ezrin-S100P Interaction and Its Role in Tumor Cell Migration∗. J. Biol. Chem. 2008, 283, 29331–29340. [Google Scholar] [CrossRef]
- Cavallaro, U.G.O.; Christofori, G. Multitasking in Tumor Progression: Signaling Functions of Cell Adhesion Molecules. Ann. New York Acad. Sci. 2004, 1014, 58–66. [Google Scholar] [CrossRef]
- Beauchemin, N.; Arabzadeh, A. Carcinoembryonic Antigen-Related Cell Adhesion Molecules (CEACAMs) in Cancer Progression and Metastasis. Cancer Metastasis Rev. 2013, 32, 643–671. [Google Scholar] [CrossRef]
- Johnson, J.P.; Stade, B.G.; Holzmann, B.; Schwäble, W.; Riethmüller, G. De Novo Expression of Intercellular-Adhesion Molecule 1 in Melanoma Correlates with Increased Risk of Metastasis. Proc. Natl. Acad. Sci. USA 1989, 86, 641–644. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Shun, C.-T.; Wu, M.-S.; Chen, C.-C. A Novel Anticancer Effect of Thalidomide: Inhibition of Intercellular Adhesion Molecule-1–Mediated Cell Invasion and Metastasis through Suppression of Nuclear Factor-ΚB. Clin. Cancer Res. 2006, 12, 7165–7173. [Google Scholar] [CrossRef]
- Schröder, C.; Witzel, I.; Müller, V.; Krenkel, S.; Wirtz, R.M.; Jänicke, F.; Schumacher, U.; Milde-Langosch, K. Prognostic Value of Intercellular Adhesion Molecule (ICAM)-1 Expression in Breast Cancer. J. Cancer Res. Clin. Oncol. 2011, 137, 1193–1201. [Google Scholar] [CrossRef] [PubMed]
- Usami, Y.; Ishida, K.; Sato, S.; Kishino, M.; Kiryu, M.; Ogawa, Y.; Okura, M.; Fukuda, Y.; Toyosawa, S. Intercellular Adhesion Molecule-1 (ICAM-1) Expression Correlates with Oral Cancer Progression and Induces Macrophage/Cancer Cell Adhesion. Int. J. Cancer 2013, 133, 568–578. [Google Scholar] [CrossRef] [PubMed]
- Farahani, E.; Patra, H.K.; Jangamreddy, J.R.; Rashedi, I.; Kawalec, M.; Rao Pariti, R.K.; Batakis, P.; Wiechec, E. Cell Adhesion Molecules and Their Relation to (Cancer) Cell Stemness. Carcinogenesis 2014, 35, 747–759. [Google Scholar] [CrossRef] [PubMed]
- Buckley, S.T.; Medina, C.; Kasper, M.; Ehrhardt, C. Interplay between RAGE, CD44, and Focal Adhesion Molecules in Epithelial-Mesenchymal Transition of Alveolar Epithelial Cells. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2011, 300, L548–L559. [Google Scholar] [CrossRef]
- Sessa, L.; Gatti, E.; Zeni, F.; Antonelli, A.; Catucci, A.; Koch, M.; Pompilio, G.; Fritz, G.; Raucci, A.; Bianchi, M.E. The Receptor for Advanced Glycation End-Products (RAGE) Is Only Present in Mammals, and Belongs to a Family of Cell Adhesion Molecules (CAMs). PLoS ONE 2014, 9, e86903. [Google Scholar] [CrossRef]
- Paschos, K.A.; Canovas, D.; Bird, N.C. The Role of Cell Adhesion Molecules in the Progression of Colorectal Cancer and the Development of Liver Metastasis. Cell. Signal. 2009, 21, 665–674. [Google Scholar] [CrossRef]
- Zecchini, S.; Cavallaro, U. Neural Cell Adhesion Molecule in Cancer: Expression and Mechanisms. In Structure and Function of the Neural Cell Adhesion Molecule NCAM; Springer: New York, NY, USA, 2010; pp. 319–333. [Google Scholar]
- Bergom, C.; Gao, C.; Newman, P.J. Mechanisms of PECAM-1-Mediated Cytoprotection and Implications for Cancer Cell Survival. Leuk. Lymphoma 2005, 46, 1409–1421. [Google Scholar] [CrossRef]
- Weichert, W.; Knösel, T.; Bellach, J.; Dietel, M.; Kristiansen, G. ALCAM/CD166 Is Overexpressed in Colorectal Carcinoma and Correlates with Shortened Patient Survival. J. Clin. Pathol. 2004, 57, 1160–1164. [Google Scholar] [CrossRef]
- Roland, C.L.; Harken, A.H.; Sarr, M.G.; Barnett Jr, C.C. ICAM-1 Expression Determines Malignant Potential of Cancer. Surgery 2007, 141, 705–707. [Google Scholar] [CrossRef]
- Donin, N.; Jurianz, K.; Ziporen, L.; Schultz, S.; Kirschfink, M.; Fishelson, Z. Complement Resistance of Human Carcinoma Cells Depends on Membrane Regulatory Proteins, Protein Kinases and Sialic Acid. Clin. Exp. Immunol. 2003, 131, 254–263. [Google Scholar] [CrossRef]
- Okroj, M.; Hsu, Y.-F.; Ajona, D.; Pio, R.; Blom, A.M. Non-Small Cell Lung Cancer Cells Produce a Functional Set of Complement Factor I and Its Soluble Cofactors. Mol. Immunol. 2008, 45, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Ajona, D.; Castano, Z.; Garayoa, M.; Zudaire, E.; Pajares, M.J.; Martinez, A.; Cuttitta, F.; Montuenga, L.M.; Pio, R. Expression of Complement Factor H by Lung Cancer Cells: Effects on the Activation of the Alternative Pathway of Complement. Cancer Res. 2004, 64, 6310–6318. [Google Scholar] [CrossRef] [PubMed]
- Ajona, D.; Ortiz-Espinosa, S.; Pio, R. Complement Anaphylatoxins C3a and C5a: Emerging Roles in Cancer Progression and Treatment. Semin. Cell Dev. Biol. 2019, 85, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Ajona, D.; Hsu, Y.-F.; Corrales, L.; Montuenga, L.M.; Pio, R. Down-Regulation of Human Complement Factor H Sensitizes Non-Small Cell Lung Cancer Cells to Complement Attack and Reduces in Vivo Tumor Growth. J. Immunol. 2007, 178, 5991–5998. [Google Scholar] [CrossRef] [PubMed]
- Reid, K. Complement Component C1q: Historical Perspective of a Functionally Versatile, and Structurally Unusual, Serum Protein. Front. Immunol. 2018, 9, 764. [Google Scholar] [CrossRef] [PubMed]
- Teh, B.K.; Yeo, J.G.; Chern, L.M.; Lu, J. C1q Regulation of Dendritic Cell Development from Monocytes with Distinct Cytokine Production and T Cell Stimulation. Mol. Immunol. 2011, 48, 1128–1138. [Google Scholar] [CrossRef] [PubMed]
- Lauvrak, V.; Brekke, O.H.; Ihle, Ø.; Lindqvist, B.H. Identification and Characterisation of C1q-Binding Phage Displayed Peptides. Biol. Chem. 1997, 378, 1509–1520. [Google Scholar] [CrossRef]
- Goodman, E.B.; Anderson, D.C.; Tenner, A.J. C1q Triggers Neutrophil Superoxide Production by a Unique CD18-dependent Mechanism. J. Leukoc. Biol. 1995, 58, 168–176. [Google Scholar] [CrossRef]
- Ma, W.; Rai, V.; Hudson, B.I.; Song, F.; Schmidt, A.M.; Barile, G.R. RAGE Binds C1q and Enhances C1q-Mediated Phagocytosis. Cell. Immunol. 2012, 274, 72–82. [Google Scholar] [CrossRef]
- Pamplona, R. Advanced Lipoxidation End-Products. Chem. -Biol. Interact. 2011, 192, 14–20. [Google Scholar] [CrossRef]
- Viedma-Poyatos, Á.; González-Jiménez, P.; Langlois, O.; Spickett, C.M.; Pérez-Sala, D. Protein Lipoxidation: Basic Concepts and Emerging Roles. Antioxidants 2021, 10, 295. [Google Scholar] [CrossRef] [PubMed]
- Dianzani, M.U. Lipid Peroxidation and Cancer: A Critical Reconsideration. Tumori J. 1989, 75, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Biasi, F.; Vizio, B.; Mascia, C.; Gaia, E.; Zarkovic, N.; Chiarpotto, E.; Leonarduzzi, G.; Poli, G. C-Jun N-Terminal Kinase Upregulation as a Key Event in the Proapoptotic Interaction between Transforming Growth Factor-Β1 and 4-Hydroxynonenal in Colon Mucosa. Free. Radic. Biol. Med. 2006, 41, 443–454. [Google Scholar] [CrossRef]
- Mol, M.; Degani, G.; Coppa, C.; Baron, G.; Popolo, L.; Carini, M.; Aldini, G.; Vistoli, G.; Altomare, A. Advanced Lipoxidation End Products (ALEs) as RAGE Binders: Mass Spectrometric and Computational Studies to Explain the Reasons Why. Redox Biol. 2019, 23, 101083. [Google Scholar] [CrossRef]
- Rakhesh, M.; Cate, M.; Vijay, R.; Shrikant, A.; Shanjana, A. A TLR4-Interacting Peptide Inhibits Lipopolysaccharide-Stimulated Inflammatory Responses, Migration and Invasion of Colon Cancer SW480 Cells. Oncoimmunology 2012, 1, 1495–1506. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, X.; Liang, J.; Li, G. Lipopolysaccharide Promotes Adhesion and Invasion of Hepatoma Cell Lines HepG2 and HepG2. 2.15. Mol. Biol. Rep. 2010, 37, 2235–2239. [Google Scholar] [CrossRef]
- Huang, T.A.O.; Chen, Z.; Fang, L. Curcumin Inhibits LPS-Induced EMT through Downregulation of NF-ΚB-Snail Signaling in Breast Cancer Cells. Oncol. Rep. 2013, 29, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Harashima, A.; Saito, H.; Tsuneyama, K.; Munesue, S.; Motoyoshi, S.; Han, D.; Watanabe, T.; Asano, M.; Takasawa, S. Septic Shock Is Associated with Receptor for Advanced Glycation End Products Ligation of LPS. J. Immunol. 2011, 186, 3248–3257. [Google Scholar] [CrossRef]
- Nakashima, H.; Nakamura, M.; Yamaguchi, H.; Yamanaka, N.; Akiyoshi, T.; Koga, K.; Yamaguchi, K.; Tsuneyoshi, M.; Tanaka, M.; Katano, M. Nuclear Factor-ΚB Contributes to Hedgehog Signaling Pathway Activation through Sonic Hedgehog Induction in Pancreatic Cancer. Cancer Res. 2006, 66, 7041–7049. [Google Scholar] [CrossRef]
- Kojima, M.; Morisaki, T.; Izuhara, K.; Uchiyama, A.; Matsunari, Y.; Katano, M.; Tanaka, M. Lipopolysaccharide Increases Cyclo-Oxygenase-2 Expression in a Colon Carcinoma Cell Line through Nuclear Factor-ΚB Activation. Oncogene 2000, 19, 1225–1231. [Google Scholar] [CrossRef]
- Wilkie, T.; Verma, A.K.; Zhao, H.; Charan, M.; Ahirwar, D.K.; Kant, S.; Pancholi, V.; Mishra, S.; Ganju, R.K. Lipopolysaccharide from the Commensal Microbiota of the Breast Enhances Cancer Growth: Role of S100A7 and TLR4. Mol. Oncol. 2021, 16, 1508–1522. [Google Scholar] [CrossRef] [PubMed]
- Nasser, M.W.; Wani, N.A.; Ahirwar, D.K.; Powell, C.A.; Ravi, J.; Elbaz, M.; Zhao, H.; Padilla, L.; Zhang, X.; Shilo, K. RAGE Mediates S100A7-Induced Breast Cancer Growth and Metastasis by Modulating the Tumor Microenvironment. Cancer Res. 2015, 75, 974–985. [Google Scholar] [CrossRef]
- You, L.; Cui, H.; Zhao, F.; Sun, H.; Zhong, H.; Zhou, G.; Chen, X. Inhibition of HMGB1/RAGE Axis Suppressed the Lipopolysaccharide (LPS)-Induced Vicious Transformation of Cervical Epithelial Cells. Bioengineered 2021, 12, 4995–5003. [Google Scholar] [CrossRef] [PubMed]
- Sharaf, H.; Matou-Nasri, S.; Wang, Q.; Rabhan, Z.; Al-Eidi, H.; Al Abdulrahman, A.; Ahmed, N. Advanced Glycation Endproducts Increase Proliferation, Migration and Invasion of the Breast Cancer Cell Line MDA-MB-231. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2015, 1852, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Allmen, E.U.; Koch, M.; Fritz, G.; Legler, D.F. V Domain of RAGE Interacts with AGEs on Prostate Carcinoma Cells. Prostate 2008, 68, 748–758. [Google Scholar] [CrossRef]
- Poser, I.; Bosserhoff, A.K. Transcription Factors Involved in Development and Progression of Malignant Melanoma. Histol. Histopathol. 2004, 19, 173–188. [Google Scholar]
- Hernandez, J.L.; Padilla, L.; Dakhel, S.; Coll, T.; Hervas, R.; Adan, J.; Masa, M.; Mitjans, F.; Martinez, J.M.; Coma, S. Therapeutic Targeting of Tumor Growth and Angiogenesis with a Novel Anti-S100A4 Monoclonal Antibody. PLoS ONE 2013, 8, e72480. [Google Scholar] [CrossRef]
- Dakhel, S.; Padilla, L.; Adan, J.; Masa, M.; Martinez, J.M.; Roque, L.; Coll, T.; Hervas, R.; Calvis, C.; Messeguer, R. S100P Antibody-Mediated Therapy as a New Promising Strategy for the Treatment of Pancreatic Cancer. Oncogenesis 2014, 3, e92. [Google Scholar] [CrossRef]
- Li, A.; Shi, D.; Xu, B.; Wang, J.; Tang, Y.; Xiao, W.; Shen, G.; Deng, W.; Zhao, C. S100A6 Promotes Cell Proliferation in Human Nasopharyngeal Carcinoma via the P38/MAPK Signaling Pathway. Mol. Carcinog. 2017, 56, 972–984. [Google Scholar] [CrossRef]
- Sakurai, M.; Miki, Y.; Takagi, K.; Suzuki, T.; Ishida, T.; Ohuchi, N.; Sasano, H. Interaction with Adipocyte Stromal Cells Induces Breast Cancer Malignancy via S100A7 Upregulation in Breast Cancer Microenvironment. Breast Cancer Res. 2017, 19, 70. [Google Scholar] [CrossRef]
- Basnet, S.; Sharma, S.; Costea, D.E.; Sapkota, D. Expression Profile and Functional Role of S100A14 in Human Cancer. Oncotarget 2019, 10, 2996. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, M.; Williams, R.; Wang, L.; Vogl, T.; Srikrishna, G. S100A8/A9 Activate Key Genes and Pathways in Colon Tumor Progression. Mol. Cancer Res. 2011, 9, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Taccioli, C.; Jiang, Y.; Chen, H.; Smalley, K.J.; Huang, K.; Liu, X.; Farber, J.L.; Croce, C.M.; Fong, L.Y.Y. Zinc Deficiency Activates S100A8 Inflammation in the Absence of COX-2 and Promotes Murine Oral-esophageal Tumor Progression. Int. J. Cancer 2011, 129, 331–345. [Google Scholar] [CrossRef]
- De Ponti, A.; Wiechert, L.; Schneller, D.; Pusterla, T.; Longerich, T.; Hogg, N.; Vogel, A.; Schirmacher, P.; Hess, J.; Angel, P. A Pro-Tumorigenic Function of S100A8/A9 in Carcinogen-Induced Hepatocellular Carcinoma. Cancer Lett. 2015, 369, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Cheng, J.; You, J.; Yan, B.; Liu, H.; Li, F. S100B Promotes Chemoresistance in Ovarian Cancer Stem Cells by Regulating P53. Oncol. Rep. 2018, 40, 1574–1582. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Shen, L.; Yang, K.; Huang, D.; Li, X.; Li, Y.; Zhao, L.; Chen, J.; Yi, Q.; Xu, H. C6 Glioma-Conditioned Medium Induces Malignant Transformation of Mesenchymal Stem Cells: Possible Role of S100B/RAGE Pathway. Biochem. Biophys. Res. Commun. 2018, 495, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Seguella, L.; Capuano, R.; Pesce, M.; Annunziata, G.; Pesce, M.; de Conno, B.; Sarnelli, G.; Aurino, L.; Esposito, G. S100B Protein Stimulates Proliferation and Angiogenic Mediators Release through RAGE/PAkt/MTOR Pathway in Human Colon Adenocarcinoma Caco-2 Cells. Int. J. Mol. Sci. 2019, 20, 3240. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, C.; Shan, X.; Wu, J.; Liu, H.; Liu, H.; Zhang, J.; Xu, W.; Sha, Z.; He, J. S100P Is Associated with Proliferation and Migration in Nasopharyngeal Carcinoma. Oncol. Lett. 2017, 14, 525–532. [Google Scholar] [CrossRef]
- Arumugam, T.; Ramachandran, V.; Logsdon, C.D. Effect of Cromolyn on S100P Interactions with RAGE and Pancreatic Cancer Growth and Invasion in Mouse Models. J. Natl. Cancer Inst. 2006, 98, 1806–1818. [Google Scholar] [CrossRef]
- Köhler, G.; Milstein, C. Derivation of Specific Antibody-producing Tissue Culture and Tumor Lines by Cell Fusion. Eur. J. Immunol. 1976, 6, 511–519. [Google Scholar] [CrossRef]
- Healey, G.D.; Pan-Castillo, B.; Garcia-Parra, J.; Davies, J.; Roberts, S.; Jones, E.; Dhar, K.; Nandanan, S.; Tofazzal, N.; Piggott, L. Antibody Drug Conjugates against the Receptor for Advanced Glycation End Products (RAGE), a Novel Therapeutic Target in Endometrial Cancer. J. Immunother. Cancer 2019, 7, 280. [Google Scholar] [CrossRef] [PubMed]
- Esumi, S.; Kitamura, Y.; Yokota-Kumasaki, H.; Ushio, S.; Yamada-Takemoto, A.; Nagai, R.; Ogawa, A.; Kawasaki, Y.; Sendo, T. Effects of Magnesium Oxide on the Serum Duloxetine Concentration and Antidepressant-like Effects of Duloxetine in Rats. Biol. Pharm. Bull. 2018, 41, 1727–1731. [Google Scholar] [CrossRef] [PubMed]
- Mizumoto, S.; Takahashi, J.; Sugahara, K. Receptor for Advanced Glycation End Products (RAGE) Functions as Receptor for Specific Sulfated Glycosaminoglycans, and Anti-RAGE Antibody or Sulfated Glycosaminoglycans Delivered in Vivo Inhibit Pulmonary Metastasis of Tumor Cells. J. Biol. Chem. 2012, 287, 18985–18994. [Google Scholar] [CrossRef] [PubMed]
- Muthyalaiah, Y.S.; Jonnalagadda, B.; John, C.M.; Arockiasamy, S. Impact of Advanced Glycation End products (AGEs) and its receptor (RAGE) on cancer metabolic signaling pathways and its progression. Glycoconj J. 2021, 38, 717–734. [Google Scholar] [CrossRef]
- Paleiron, N.; Bylicki, O.; André, M.; Rivière, E.; Grassin, F.; Robinet, G.; Chouaïd, C. Targeted Therapy for Localized Non-Small-Cell Lung Cancer: A Review. OncoTargets Ther. 2016, 9, 4099. [Google Scholar]
- Liang, H.; Zhong, Y.; Zhou, S.; Peng, L. Knockdown of RAGE Expression Inhibits Colorectal Cancer Cell Invasion and Suppresses Angiogenesis in Vitro and in Vivo. Cancer Lett. 2011, 313, 91–98. [Google Scholar] [CrossRef]
- Yu, Y.X.; Pan, W.C.; Cheng, Y.F. Silencing of Advanced Glycosylation and Glycosylation and Product-Specific Receptor (RAGE) Inhibits the Metastasis and Growth of Non-Small Cell Lung Cancer. Am. J. Transl. Res. 2017, 9, 2760. [Google Scholar]
- Finkenzeller, G.; Sparacio, A.; Technau, A.; Marmé, D.; Siemeister, G. Sp1 Recognition Sites in the Proximal Promoter of the Human Vascular Endothelial Growth Factor Gene Are Essential for Platelet-Derived Growth Factor-Induced Gene Expression. Oncogene 1997, 15, 669–676. [Google Scholar] [CrossRef]
- Li, J.; Wu, P.-W.; Zhou, Y.; Dai, B.; Zhang, P.-F.; Zhang, Y.-H.; Liu, Y.; Shi, X.-L. Rage Induces Hepatocellular Carcinoma Proliferation and Sorafenib Resistance by Modulating Autophagy. Cell Death Dis. 2018, 9, 225. [Google Scholar] [CrossRef]
- Abdelrahim, M.; Smith, R.; Burghardt, R.; Safe, S. Role of Sp Proteins in Regulation of Vascular Endothelial Growth Factor Expression and Proliferation of Pancreatic Cancer Cells. Cancer Res. 2004, 64, 6740–6749. [Google Scholar] [CrossRef]
- Matsuoka, H.; Makimura, C.; Koyama, A.; Otsuka, M.; Okamoto, W.; Fujisaka, Y.; Kaneda, H.; Tsurutani, J.; Nakagawa, K. Pilot Study of Duloxetine for Cancer Patients with Neuropathic Pain Non-Responsive to Pregabalin. Anticancer. Res. 2012, 32, 1805–1809. [Google Scholar] [PubMed]
- Voigtlaender, M.; Langer, F. Low-Molecular-Weight Heparin in Cancer Patients: Overview and Indications. Hämostaseologie 2019, 39, 67–75. [Google Scholar] [CrossRef]
- Dowling, R.J.O.; Goodwin, P.J.; Stambolic, V. Understanding the Benefit of Metformin Use in Cancer Treatment. BMC Med. 2011, 9, 33. [Google Scholar] [CrossRef] [PubMed]
- Bodmer, M.; Meier, C.; Krähenbühl, S.; Jick, S.S.; Meier, C.R. Long-Term Metformin Use Is Associated with Decreased Risk of Breast Cancer. Diabetes Care 2010, 33, 1304–1308. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Zhang, I.Y.; Zhang, L.; Song, Y.; Liu, S.; Ren, H.; Liu, H.; Zhou, H.; Su, Y.; Yang, Y. S100B Suppression Alters Polarization of Infiltrating Myeloid-Derived Cells in Gliomas and Inhibits Tumor Growth. Cancer Lett. 2018, 439, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Col, N.F.; Ochs, L.; Springmann, V.; Aragaki, A.K.; Chlebowski, R.T. Metformin and Breast Cancer Risk: A Meta-Analysis and Critical Literature Review. Breast Cancer Res. Treat. 2012, 135, 639–646. [Google Scholar] [CrossRef]
- Currie, C.J.; Poole, C.D.; Jenkins-Jones, S.; Gale, E.A.M.; Johnson, J.A.; Morgan, C.L. Mortality after Incident Cancer in People with and without Type 2 Diabetes: Impact of Metformin on Survival. Diabetes Care 2012, 35, 299–304. [Google Scholar] [CrossRef]
- Ishibashi, Y.; Matsui, T.; Takeuchi, M.; Yamagishi, S. Metformin Inhibits Advanced Glycation End Products (AGEs)-Induced Growth and VEGF Expression in MCF-7 Breast Cancer Cells by Suppressing AGEs Receptor Expression via AMP-Activated Protein Kinase. Horm. Metab. Res. 2013, 45, 387–390. [Google Scholar] [CrossRef]
- Pellegrini, L.; Xue, J.; Larson, D.; Pastorino, S.; Jube, S.; Forest, K.H.; Saad-Jube, Z.S.; Napolitano, A.; Pagano, I.; Negi, V.S. HMGB1 Targeting by Ethyl Pyruvate Suppresses Malignant Phenotype of Human Mesothelioma. Oncotarget 2017, 8, 22649. [Google Scholar] [CrossRef]
- Chen, B.; Na, F.; Yang, H.; Li, R.; Li, M.; Sun, X.; Hu, B.; Huang, G.; Lan, J.; Xu, H. Ethyl Pyruvate Alleviates Radiation-Induced Lung Injury in Mice. Biomed. Pharmacother. 2017, 92, 468–478. [Google Scholar] [CrossRef]
- Liu, Q.; Huo, Y.; Zheng, H.; Zhao, J.; Jia, L.; Wang, P. Ethyl Pyruvate Suppresses the Growth, Invasion and Migration and Induces the Apoptosis of Non-small Cell Lung Cancer Cells via the HMGB1/RAGE Axis and the NF-κB/STAT3 Pathway. Oncol. Rep. 2019, 42, 817–825. [Google Scholar] [CrossRef] [PubMed]
- Inada, M.; Shindo, M.; Kobayashi, K.; Sato, A.; Yamamoto, Y.; Akasaki, Y.; Ichimura, K.; Tanuma, S. Anticancer Effects of a Non-Narcotic Opium Alkaloid Medicine, Papaverine, in Human Glioblastoma Cells. PLoS ONE 2019, 14, e0216358. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Piao, C.; Kim, G.; Kim, J.Y.; Choi, E.; Lee, M. Production and Application of HMGB1 Derived Recombinant RAGE-Antagonist Peptide for Anti-Inflammatory Therapy in Acute Lung Injury. Eur. J. Pharm. Sci. 2018, 114, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Park, I.-H.; Chung, S.-K.; Lee, K.-B.; Yoo, Y.-C.; Kim, S.-K.; Kim, G.-S.; Song, K.-S. An Antioxidant Hispidin from the Mycelial Cultures of Phellinus Linteus. Arch. Pharmacal Res. 2004, 27, 615–618. [Google Scholar] [CrossRef] [PubMed]
- El-Far, A.H.A.M.; Munesue, S.; Harashima, A.; Sato, A.; Shindo, M.; Nakajima, S.; Inada, M.; Tanaka, M.; Takeuchi, A.; Tsuchiya, H. In Vitro Anticancer Effects of a RAGE Inhibitor Discovered Using a Structure-Based Drug Design System. Oncol. Lett. 2018, 15, 4627–4634. [Google Scholar] [CrossRef]
- Chandimali, N.; JIN, W.O.O.Y.; KWON, T. Combination Effects of Hispidin and Gemcitabine via Inhibition of Stemness in Pancreatic Cancer Stem Cells. Anticancer. Res. 2018, 38, 3967–3975. [Google Scholar] [CrossRef]
- Song, T.-Y.; Yang, N.-C.; Chen, C.-L.; Thi, T.L.V. Protective Effects and Possible Mechanisms of Ergothioneine and Hispidin against Methylglyoxal-Induced Injuries in Rat Pheochromocytoma Cells. Oxidative Med. Cell. Longev. 2017, 2017, 4824371. [Google Scholar] [CrossRef]
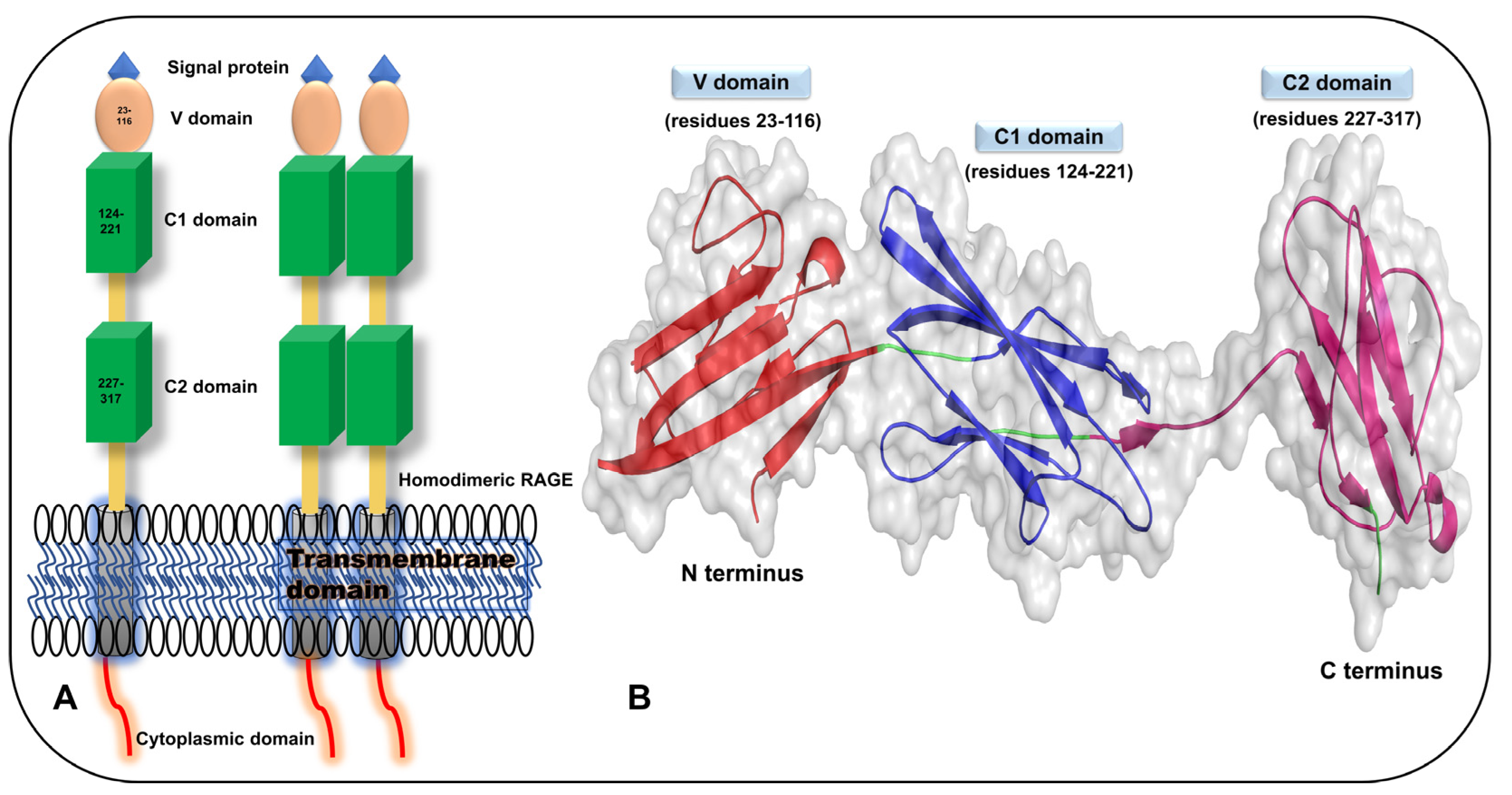
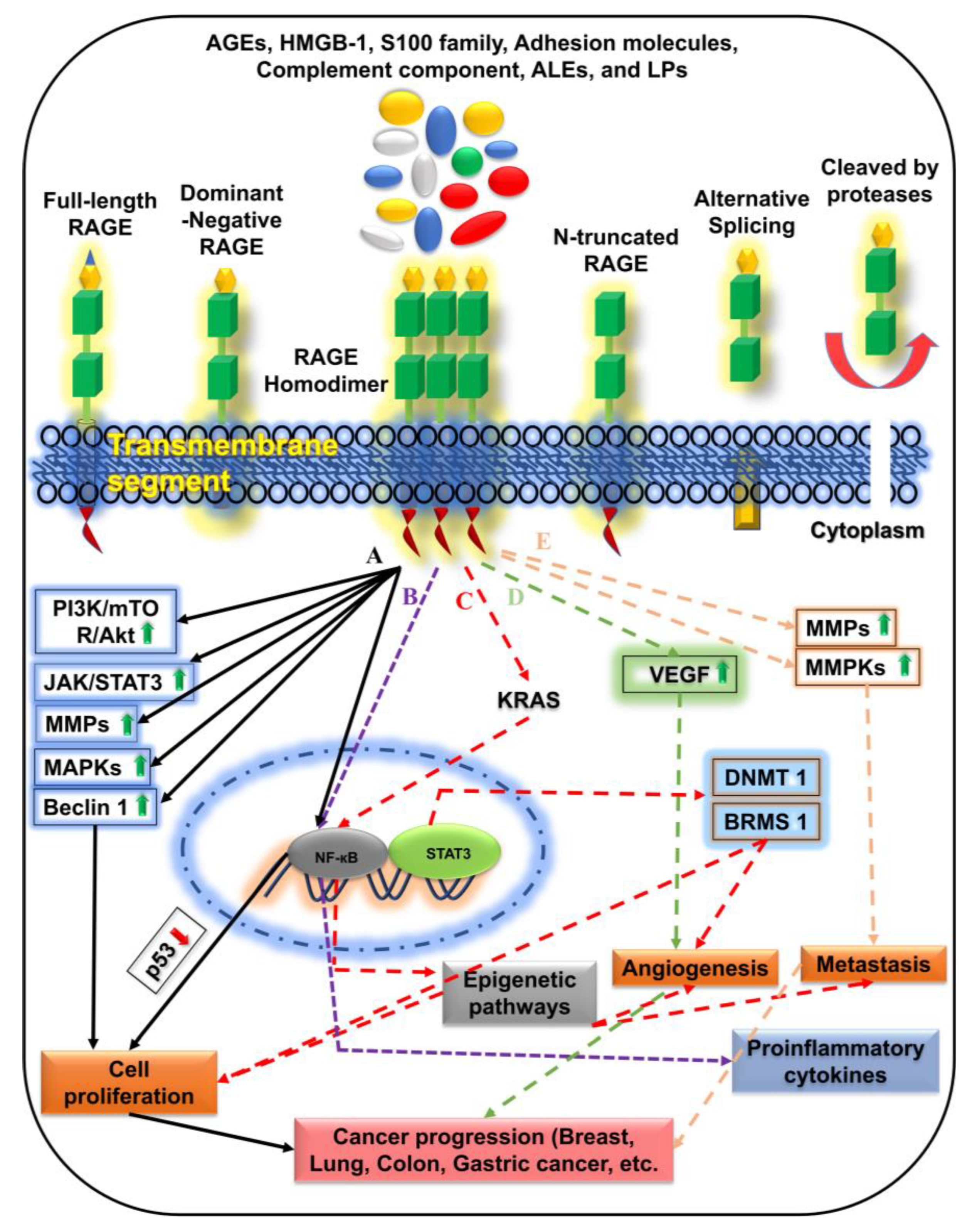
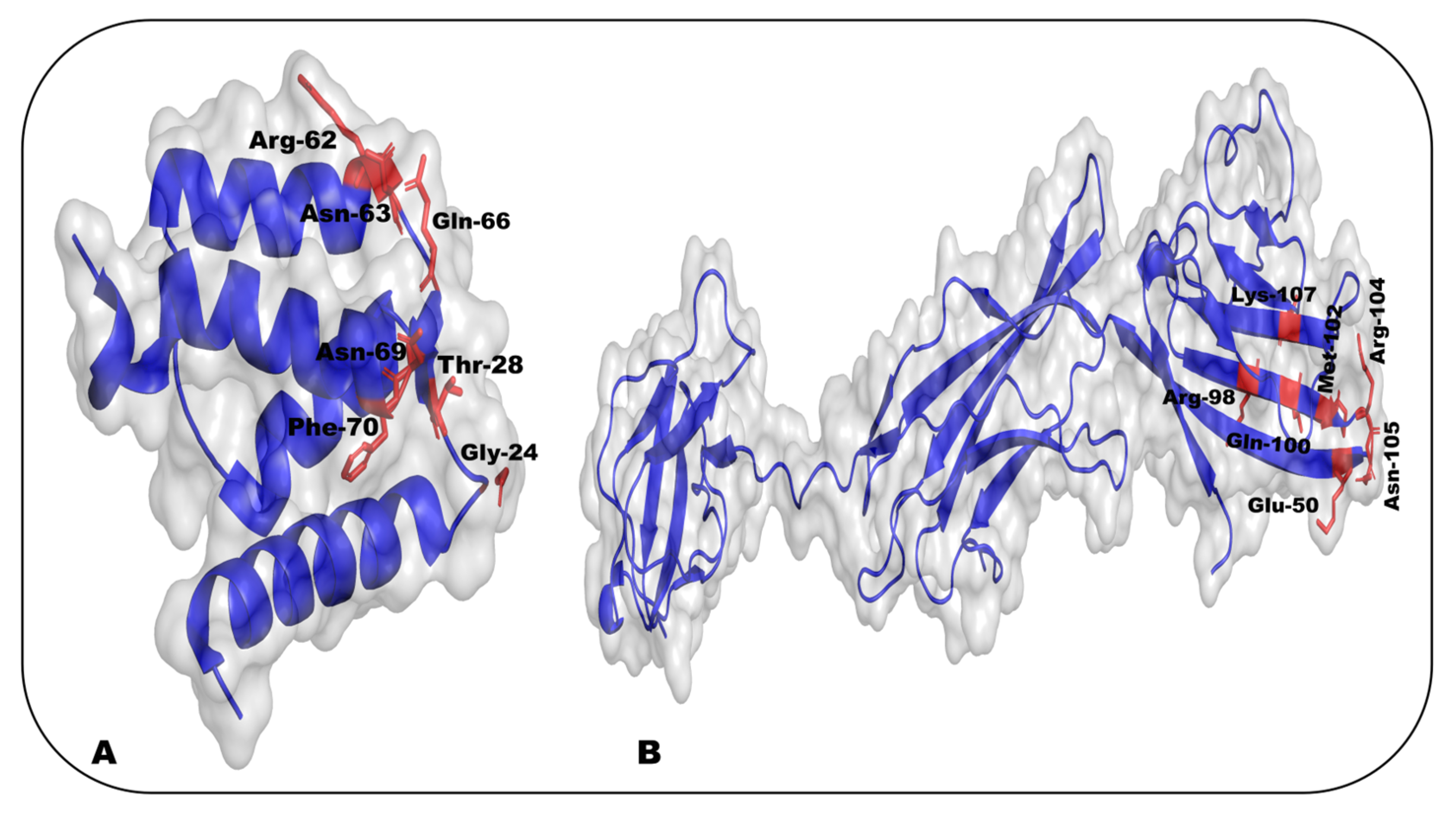
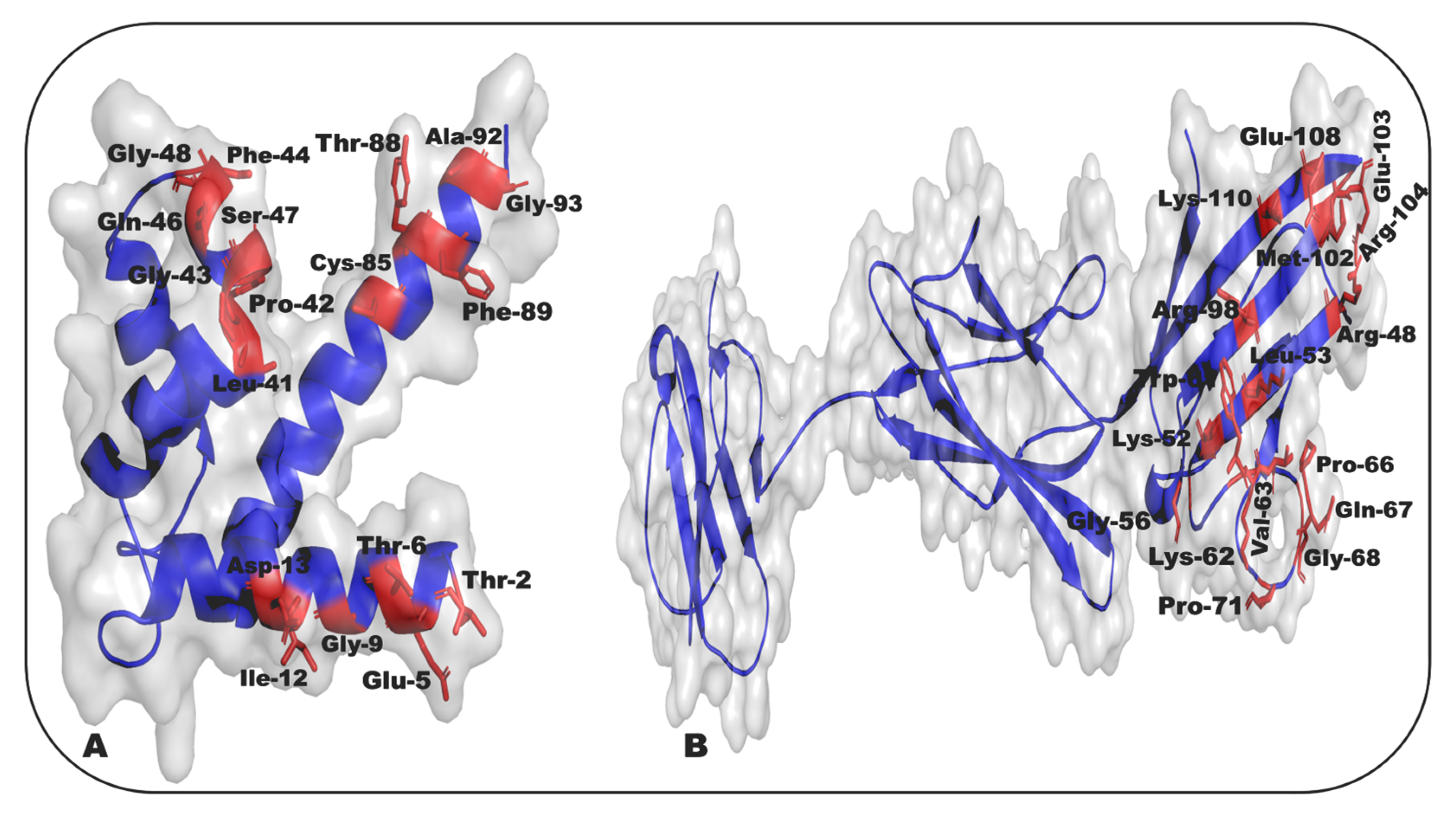
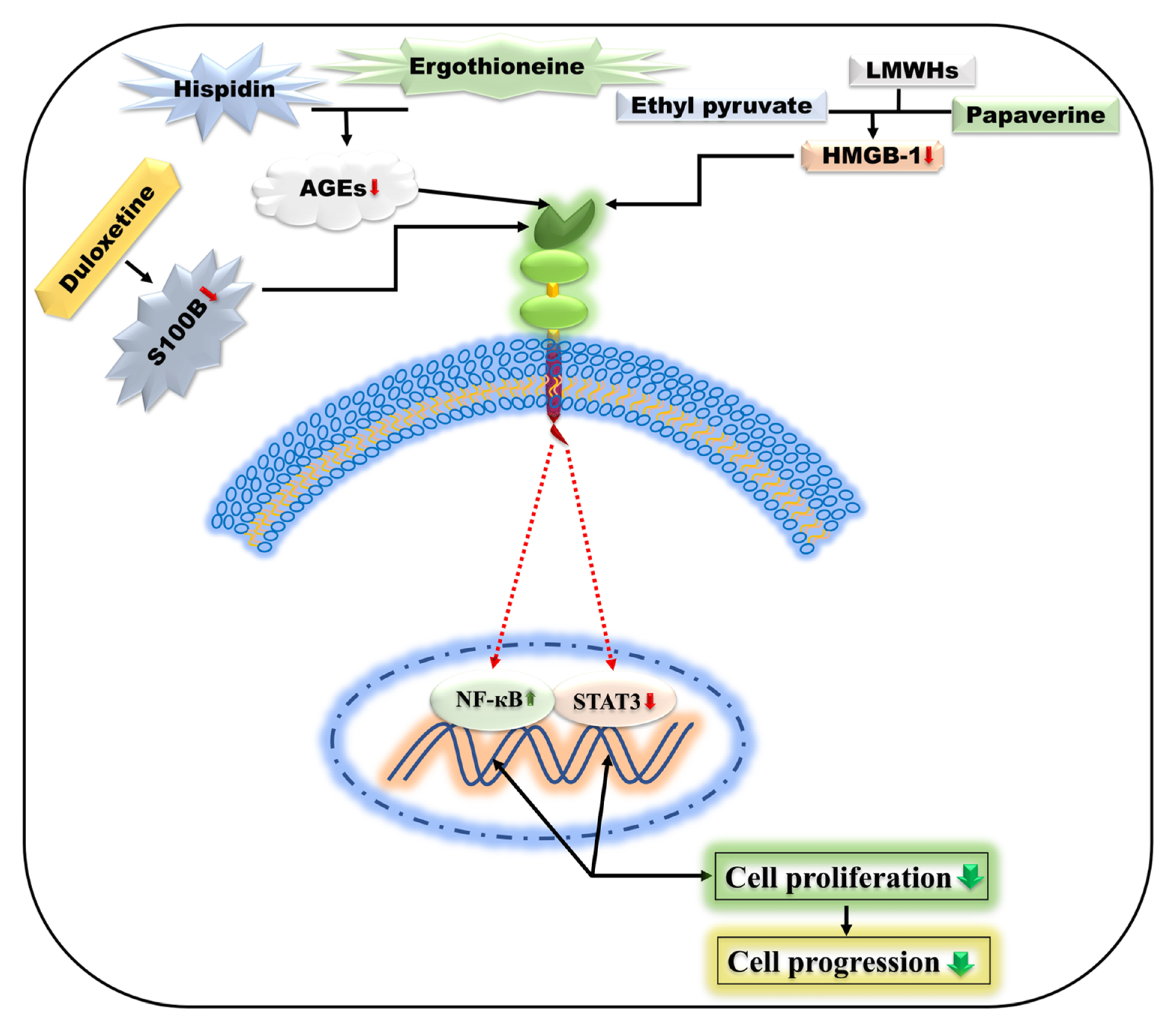
| S. No. | RAGE–Ligand Interaction | Cancer Type | In Vitro/In Vivo Study | References |
|---|---|---|---|---|
| 1. | RAGE–AGEs | Human oral cancer, breast cancer, prostate carcinoma cells | In vitro | [2,3,4,5] |
| 2. | RAGE–HMGB1 | Gastric cancer, colon cancer, breast cancer, melanoma, prostate cancer, hepatocellular carcinoma | In vitro | [2,6,7,8,9,10,11] |
| 3. | RAGE–S100A4 | Human colorectal cancer, melanoma, human osteosarcoma, human pancreatic cancer, thyroid cancer (human specimens) | In vitro/In vivo | [12,13,14,15,16] |
| 4. | RAGE–S100A6 | Nasopharyngeal carcinoma | In vitro | [10] |
| 5. | RAGE–S100A7 | Breast cancer, human cervical cancer | In vitro/In vivo | [17,18] |
| 6. | RAGE–S100A8 | Human breast cancer, human prostate cancer, colon tumor, oral-esophageal tumor, squamous cell carcinoma | In vitro/In vivo | [19,20,21,22,23] |
| 7. | RAGE–S100A9 | Human hepatocellular carcinoma | In vitro | [24] |
| 8. | RAGE–S100A14 | Esophageal squamous cell carcinoma | In vitro | [25] |
| 9. | RAGE–S100A16 | Human prostate cancer | In vitro | [26] |
| 10. | RAGE–S100B | Ovarian cancer, glioma (C6), human colon adenocarcinoma | In vitro | [27,28,29,30] |
| 11. | RAGE–S100P | Nasopharyngeal carcinoma, colon cancer, pancreatic cancer, | In vitro/In vivo | [16,31,32] |
| 12. | RAGE–LPS | Liver cancer cells, colorectal cancer, and leukemia cells | In vitro/In vivo | [33,34] |
| S. No. | RAGE Inhibitors | In Vitro/In Vivo Findings | RAGE Pathways | Studies | Cancer Types | References |
|---|---|---|---|---|---|---|
| 1. | RGBO1 | In vitro/In vivo | AKT, BCL2, and cyclin D1 | HEC1A, HEC1B, HEC50/Murine xenograft model | Endometrial cancer (EC) | [35,36] |
| 2. | Sorafenib | In vitro/In vivo | AMPK/mTOR signaling | HepG2, HCCLM3, Huh7, Bel7402/xenografts and orthotopic nude mice models | Hepatocellular carcinoma (HCC) | [37] |
| 3. | S100B inhibitor /K-Luc (Duloxetine) | In vitro/In vivo | STAT3 | GL261/Murine model | Gliomas | [38] |
| 4. | Sulfated Glycosaminoglycans (Chondroitin sulfate (CS), Heparan sulfate (HS)) | In vivo | RAGE-mediated pathway | Male C57BL/6 mice | Lung metastasis | [39,319] |
| 5. | S100P-derived inhibitor | In vitro/In vivo | via NF-κB | C6-glioma, MPanc-96 for pancreatic cancer/Murine model | Pancreatic tumors and Glioma tumor growth | [33] |
| 6. | RAGE inhibitor FPS-ZM1/ PI3K inhibitor LY294002 | In vitro/In vivo | PI3K/AKT | SiHa, CaSki, C33A, MS751/xenograft mouse model | Cervical squamous cell carcinoma (CSCC) | [40] |
| 7. | Gefitinib | In vitro | ERK1/2 MAPK | GES-1 | Gastric carcinoma cells | [41] |
| 8. | Metformin/RAGE-Ab | In vitro | AMP-activated protein kinase | MCF-7 | Breast cancer cells | [42] |
| 9. | (RAP) Potent LRP1 antagonist | In vivo | NF-κB signaling | Nude mice | Pancreatic tumors, glioma tumor growth | [43] |
| 10. | Cromolyn | In vivo | NF-κB signaling | Male CB17 SCID mice | Pancreatic cancer | [31] |
| 11. | Ethyl pyruvate | In vitro/In vivo | NF-κB/STAT3 signaling | A549, H520, and PC-9 cell lines/orthotopic MM xenograft model | Human malignant mesothelioma/non-small cell lung cancer cells | [44] |
| 12. | Papaverine | In vitro | NF-κB signaling | HT1080, U87MG, and T98G cell lines | Human fibrosarcoma, human glioblastoma | [45] |
| 13. | Ergothioneine and Hispidin | In vitro | NF-κB signaling | PC12 cell lines | Rat pheochromocytoma | [46] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Faruqui, T.; Khan, M.S.; Akhter, Y.; Khan, S.; Rafi, Z.; Saeed, M.; Han, I.; Choi, E.-H.; Yadav, D.K. RAGE Inhibitors for Targeted Therapy of Cancer: A Comprehensive Review. Int. J. Mol. Sci. 2023, 24, 266. https://doi.org/10.3390/ijms24010266
Faruqui T, Khan MS, Akhter Y, Khan S, Rafi Z, Saeed M, Han I, Choi E-H, Yadav DK. RAGE Inhibitors for Targeted Therapy of Cancer: A Comprehensive Review. International Journal of Molecular Sciences. 2023; 24(1):266. https://doi.org/10.3390/ijms24010266
Chicago/Turabian StyleFaruqui, Tabrez, Mohd Sajid Khan, Yusuf Akhter, Salman Khan, Zeeshan Rafi, Mohd Saeed, Ihn Han, Eun-Ha Choi, and Dharmendra Kumar Yadav. 2023. "RAGE Inhibitors for Targeted Therapy of Cancer: A Comprehensive Review" International Journal of Molecular Sciences 24, no. 1: 266. https://doi.org/10.3390/ijms24010266
APA StyleFaruqui, T., Khan, M. S., Akhter, Y., Khan, S., Rafi, Z., Saeed, M., Han, I., Choi, E.-H., & Yadav, D. K. (2023). RAGE Inhibitors for Targeted Therapy of Cancer: A Comprehensive Review. International Journal of Molecular Sciences, 24(1), 266. https://doi.org/10.3390/ijms24010266









