Secondary Structure in Amyloids in Relation to Their Wild Type Forms
Abstract
1. Introduction
2. Results
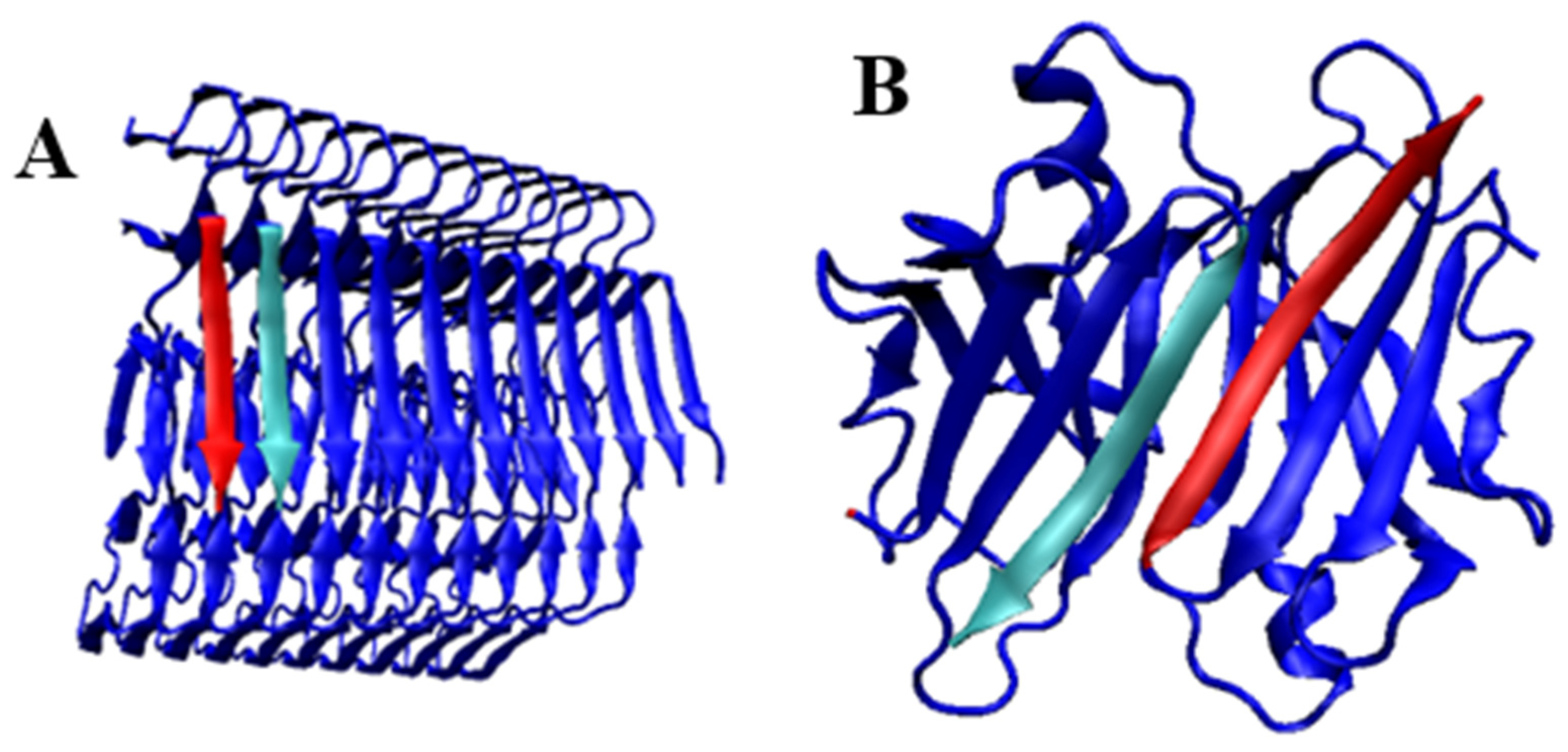
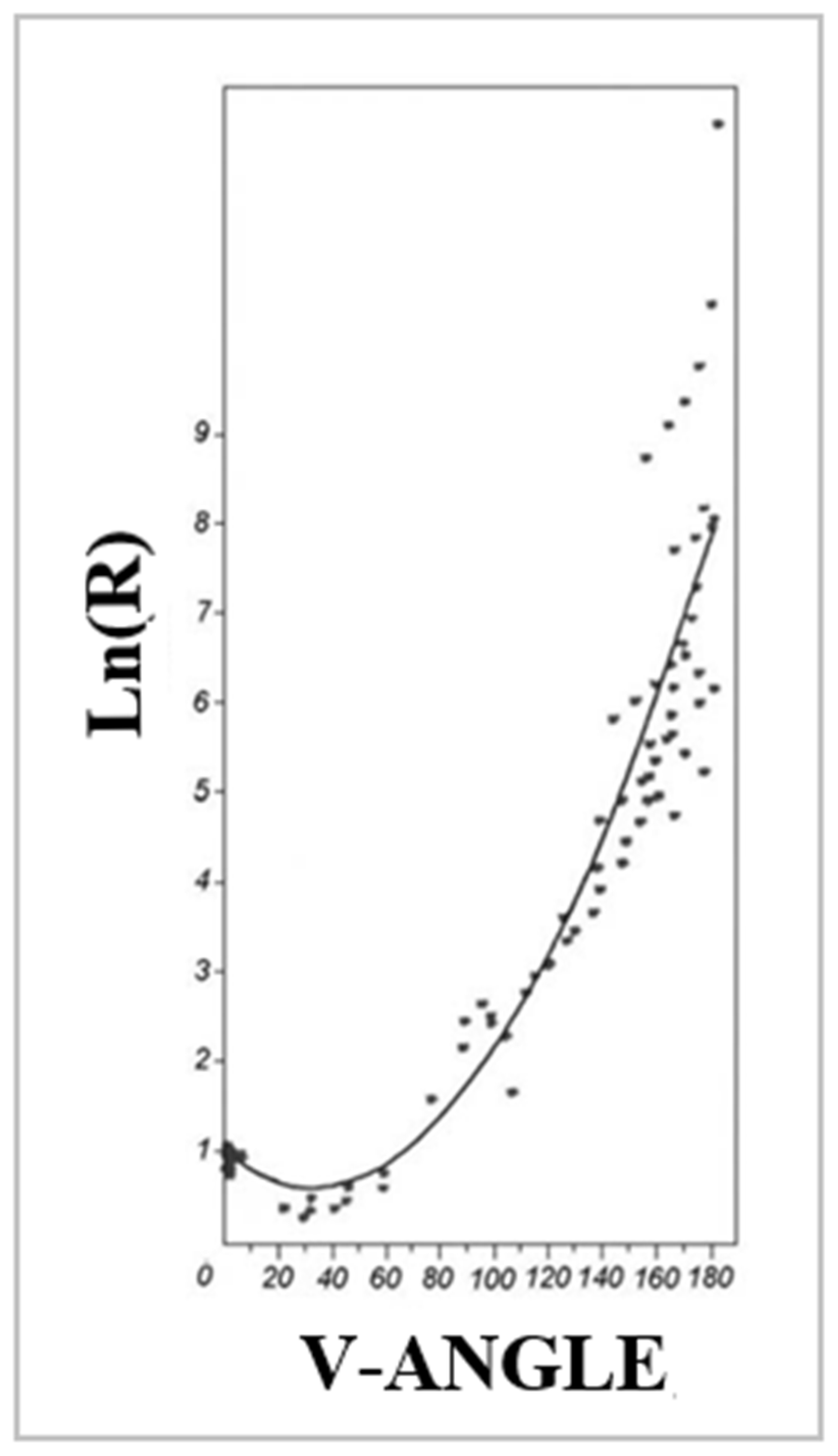
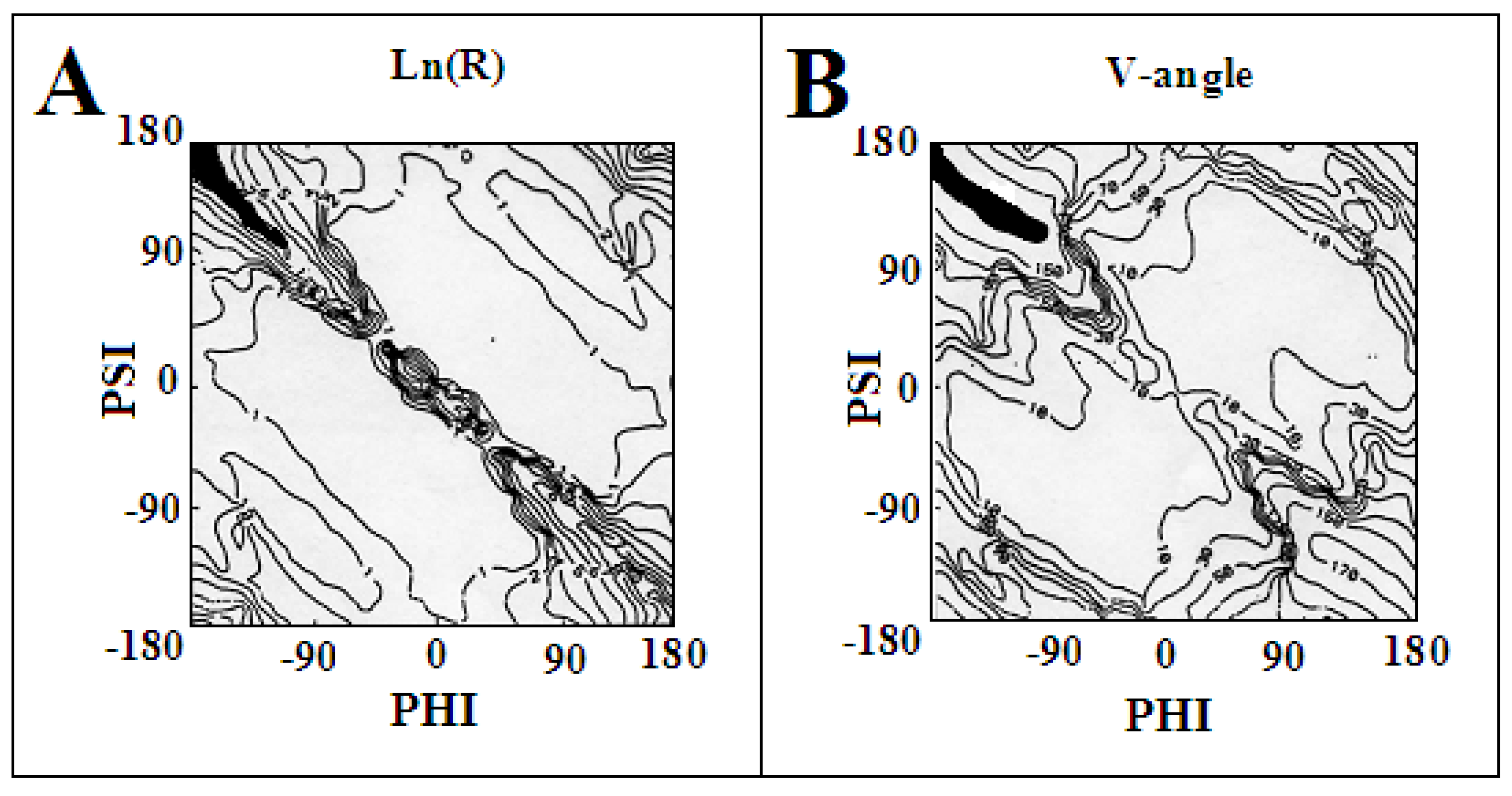
2.1. Models of an Idealised Amyloid Form of a Polypeptide Chain
2.2. Analysis of the Amyloid Structure Transthyretin
2.3. Analysis of the Structure Present in Fibrils in Relation to Their WT Forms
2.4. ABeta Amyloids
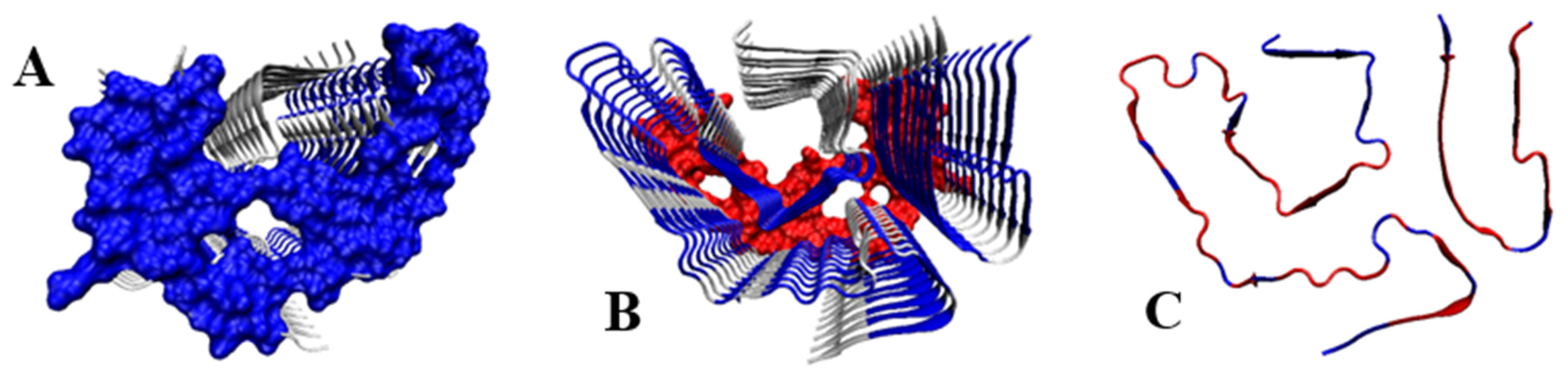
2.5. V Domain of Immunoglobulin Light Chain
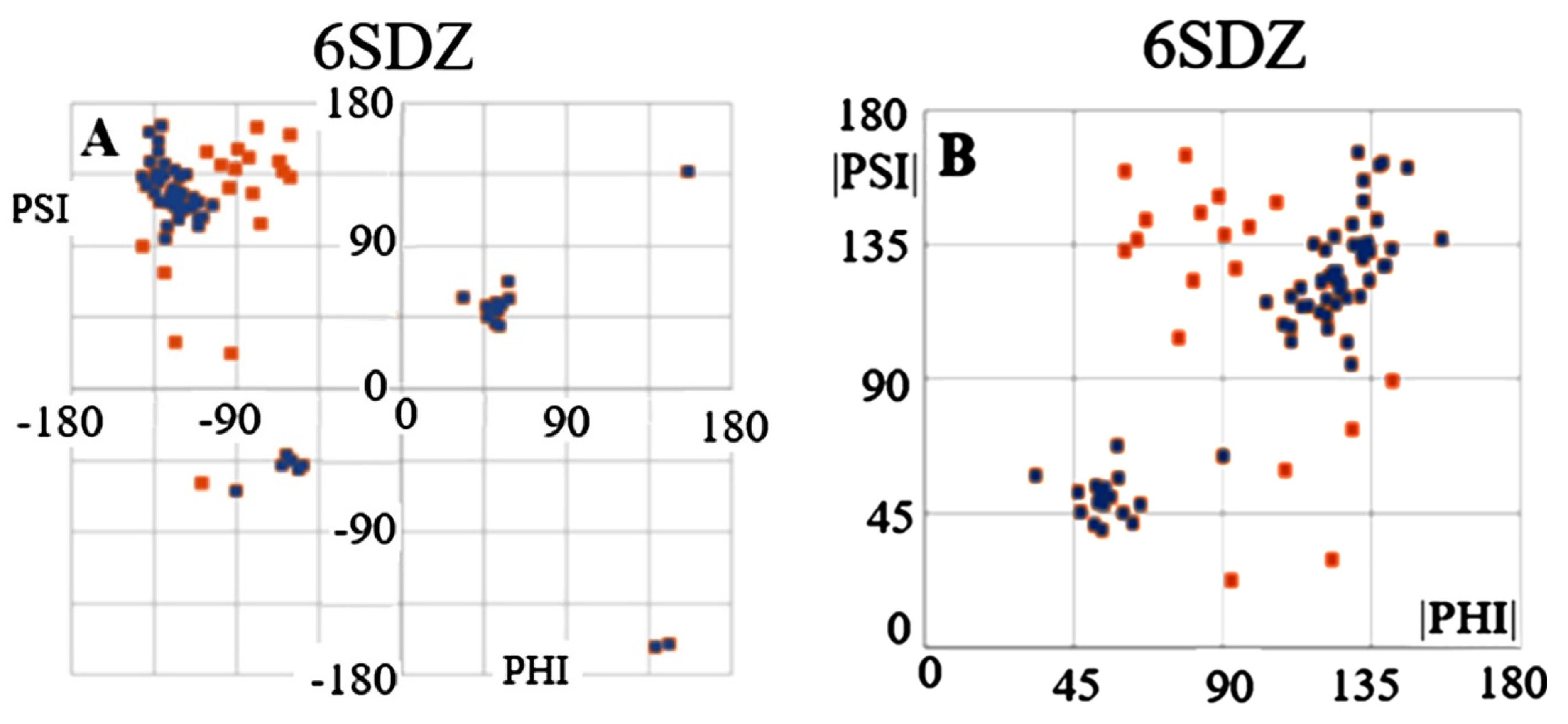
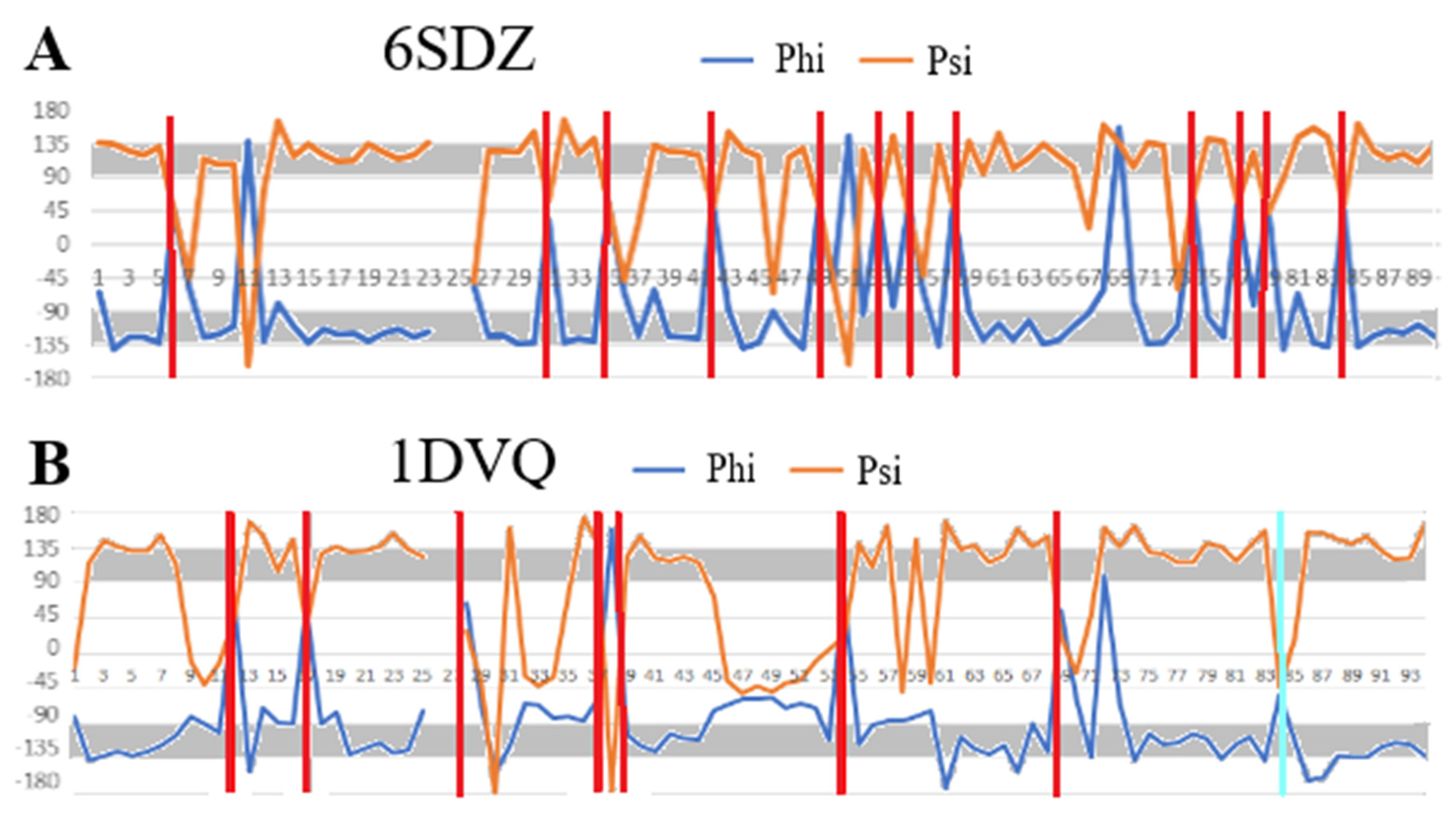
2.6. Transthyretin
2.7. Alpha-Synuclein (aSyn)
3. Discussion
4. Materials and Methods
4.1. Data
4.2. Model—Description
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Picken, M.M. The Pathology of Amyloidosis in Classification: A Review. Acta Haematol. 2020, 143, 322–334. [Google Scholar] [CrossRef]
- Hazenberg, B.P. Amyloidosis: A clinical overview. Rheum. Dis. Clin. N. Am. 2013, 39, 323–345. [Google Scholar] [CrossRef]
- Koike, H.; Iguchi, Y.; Sahashi, K.; Katsuno, M. Significance of Oligomeric and Fibrillar Species in Amyloidosis: Insights into Pathophysiology and Treatment. Molecules 2021, 26, 5091. [Google Scholar] [CrossRef]
- Soto, C.; Pritzkow, S. Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat. Neurosci. 2018, 21, 1332–1340. [Google Scholar] [CrossRef]
- Kovacs, G.G. Concepts and classification of neurodegenerative diseases. Handb. Clin. Neurol. 2017, 145, 301–307. [Google Scholar] [CrossRef]
- Khan, F.; Oloketuyi, S.F. A future perspective on neurodegenerative diseases: Nasopharyngeal and gut microbiota. J. Appl. Microbiol. 2017, 122, 306–320. [Google Scholar] [CrossRef]
- Hardy, J.A.; Higgins, G.A. Alzheimer’s disease: The amyloid cascade hypothesis. Science 1992, 256, 184–185. [Google Scholar] [CrossRef]
- Adrogue, H.E. Amyloidosis of the Heart and Kidney. Methodist DeBakey Cardiovasc. J. 2022, 18, 27–33. [Google Scholar] [CrossRef]
- Sudhakar, V.; Richardson, R.M. Gene Therapy for Neurodegenerative Diseases. Neurotherapeutics 2019, 16, 166–175. [Google Scholar] [CrossRef]
- Sugaya, K.; Vaidya, M. Stem Cell Therapies for Neurodegenerative Diseases. Adv. Exp. Med. Biol. 2018, 1056, 61–84. [Google Scholar] [CrossRef]
- Horvath, I.; Kumar, R.; Wittung-Stafshede, P. Macromolecular crowding modulates α-synuclein amyloid fiber growth. Biophys. J. 2021, 120, 3374–3381. [Google Scholar] [CrossRef]
- Zhou, J.; Ruggeri, F.S.; Zimmermann, M.R.; Meisl, G.; Longo, G.; Sekatskii, S.K.; Knowles, T.P.J.; Dietler, G. Effects of sedimentation, microgravity, hydrodynamic mixing and air-water interface on α-synuclein amyloid formation. Chem. Sci. 2020, 11, 3687–3693. [Google Scholar] [CrossRef]
- Cook, N.P.; Martí, A.A. Facile methodology for monitoring amyloid-β fibrillization. ACS Chem. Neurosci. 2012, 3, 896–899. [Google Scholar] [CrossRef][Green Version]
- Ben-Amotz, D. Electric buzz in a glass of pure water. Science 2022, 376, 800–801. [Google Scholar] [CrossRef]
- Schutzius, T.M.; Jung, S.; Maitra, T.; Graeber, G.; Köhme, M.; Poulikakos, D. Spontaneous droplet trampolining on rigid superhydrophobic surfaces. Nature 2015, 527, 82–85. [Google Scholar] [CrossRef]
- Zuo, Y.-J.; Qu, J. How Does Aqueous Solubility of Organic Reactant Affect a Water-Promoted Reaction? J. Org. Chem. 2014, 79, 6832–6839. [Google Scholar] [CrossRef]
- Eremin, D.B.; Fokin, V.V. On-Water Selectivity Switch in Microdroplets in the 1,2,3-Triazole Synthesis from Bromoethenesulfonyl Fluoride. J. Am. Chem. Society 2021, 143, 18374–18379. [Google Scholar] [CrossRef]
- Kaplaneris, N.; Vilches-Herrera, M.; Wu, J.; Ackermann, L. Sustainable Ruthenium(II)-Catalyzed C–H Activations in and on H2O. ACS Sustain. Chem. Eng. 2022, 10, 6871–6888. [Google Scholar] [CrossRef]
- Ishiyama, T.; Tahara, T.; Morita, A. Why the Photochemical Reaction of Phenol Becomes Ultrafast at the Air–Water Interface: The Effect of Surface Hydration. J. Am. Chem. Soc. 2022, 144, 6321–6325. [Google Scholar] [CrossRef]
- Gupta, A.; Vankar, J.K.; Jadav, J.P.; Gururaja, G.N. Water Mediated Direct Thioamidation of Aldehydes at Room Temperature. J. Org. Chem. 2022, 87, 2410–2420. [Google Scholar] [CrossRef]
- Cannalire, R.; Santoro, F.; Russo, C.; Graziani, G.; Tron, G.C.; Carotenuto, A.; Brancaccio, D.; Giustiniano, M. Photomicellar Catalyzed Synthesis of Amides from Isocyanides: Optimization, Scope, and NMR Studies of Photocatalyst/Surfactant Interactions. ACS Org. Inorg. Au 2022, 2, 66–74. [Google Scholar] [CrossRef]
- Jayawardena, N.; Kaur, M.; Nair, S.; Malmstrom, J.; Goldstone, D.; Negron, L.; Gerrard, J.A.; Domigan, L.J. Amyloid Fibrils from Hemoglobin. Biomolecules 2017, 7, 37. [Google Scholar] [CrossRef]
- Tuttle, M.D.; Comellas, G.; Nieuwkoop, A.J.; Covell, D.J.; Berthold, D.A.; Kloepper, K.D.; Courtney, J.M.; Kim, J.K.; Barclay, A.M.; Kendall, A.; et al. Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein. Nat. Struct. Mol. Biol. 2016, 23, 409–415. [Google Scholar] [CrossRef]
- Ulmer, T.S.; Bax, A.; Cole, N.B.; Nussbaum, R.L. Structure and dynamics of micelle-bound human alpha-synuclein. J. Biol. Chem. 2005, 280, 9595–9603. [Google Scholar] [CrossRef]
- Sawaya, M.R.; Hughes, M.P.; Rodriguez, J.A.; Riek, R.; Eisenberg, D.S. The expanding amyloid family: Structure, stability, function, and pathogenesis. Cell 2021, 184, 4857–4873. [Google Scholar] [CrossRef]
- Goedert, M.; Eisenberg, D.S.; Crowther, R.A. Propagation of Tau Aggregates and Neurodegeneration. Annu. Rev. Neurosci. 2017, 40, 189–210. [Google Scholar] [CrossRef]
- Eisenberg, D.S.; Sawaya, M.R. Structural Studies of Amyloid Proteins at the Molecular Level. Annu. Rev. Biochem. 2017, 86, 69–95. [Google Scholar] [CrossRef]
- Wei, Z.; Li, Y.; Cooks, R.G.; Yan, X. Accelerated Reaction Kinetics in Microdroplets: Overview and Recent Developments. Annu. Rev. Phys. Chem. 2020, 71, 31–51. [Google Scholar] [CrossRef]
- Roterman, I.; Stapor, K.; Dułak, D.; Konieczny, L. The Possible Mechanism of Amyloid Transformation Based on the Geometrical Parameters of Early-Stage Intermediate in Silico Model for Protein Folding. Int. J. Mol. Sci. 2022, 23, 9502. [Google Scholar] [CrossRef]
- Roterman, I. Modelling the optimal simulation path in the peptide chain folding--studies based on geometry of alanine heptapeptide. J. Theor. Biol. 1995, 177, 283–288. [Google Scholar] [CrossRef]
- Schmidt, M.; Wiese, S.; Adak, V.; Engler, J.; Agarwal, S.; Fritz, G.; Westermark, P.; Zacharias, M.; Fändrich, M. Cryo-EM structure of a transthyretin-derived amyloid fibril from a patient with hereditary ATTR amyloidosis. Nat. Commun. 2019, 10, 5008. [Google Scholar] [CrossRef]
- Sgourakis, N.G.; Yau, W.-M.; Qiang, W. Modeling an in-register, parallel “iowa” aβ fibril structure using solid-state NMR data from labeled samples with rosetta. Structure 2015, 23, 216–227. [Google Scholar] [CrossRef]
- Xiao, Y.; Ma, B.; McElheny, D.; Parthasarathy, S.; Long, F.; Hoshi, M.; Nussinov, R.; Ishii, Y. Aβ(1-42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat. Struct. Mol. Biol. 2015, 22, 499–505. [Google Scholar] [CrossRef]
- Schütz, A.K.; Vagt, T.; Huber, M.; Ovchinnikova, O.Y.; Cadalbert, R.; Wall, J.; Güntert, P.; Böckmann, A.; Glockshuber, R.; Meier, B.H. Atomic-resolution three-dimensional structure of amyloid β fibrils bearing the Osaka mutation. Angew. Chem. Int. Ed. Engl. 2015, 54, 331–335. [Google Scholar] [CrossRef]
- Klabunde, T.; Petrassi, H.M.; Oza, V.B.; Raman, P.; Kelly, J.W.; Sacchettini, J.C. Rational design of potent human transthyretin amyloid disease inhibitors. Nat. Struct. Biol. 2000, 7, 312–321. [Google Scholar] [CrossRef]
- Jiang, X.; Smith, C.S.; Petrassi, H.M.; Hammarström, P.; White, J.T.; Sacchettini, J.C.; Kelly, J.W. An engineered transthyretin monomer that is nonamyloidogenic, unless it is partially denatured. Biochemistry 2001, 40, 11442–11452. [Google Scholar] [CrossRef]
- Huang, D.B.; Ainsworth, C.F.; Stevens, F.J.; Schiffer, M. Three quaternary structures for a single protein. Proc. Natl. Acad. Sci. USA 1996, 93, 7017–7021. [Google Scholar] [CrossRef]
- Eneqvis, T.; Andersson, K.; Olofsson, A.; Lundgren, E.; Sauer-Eriksson, A.E. The beta-slip: A novel concept in transthyretin amyloidosis. Mol. Cell 2000, 6, 1207–1218. [Google Scholar] [CrossRef]
- Swuec, P.; Lavatelli, F.; Tasaki, M.; Paissoni, C.; Rognoni, P.; Maritan, M.; Brambilla, F.; Milani, P.; Mauri, P.; Camilloni, C.; et al. Cryo-EM structure of cardiac amyloid fibrils from an immunoglobulin light chain AL amyloidosis patient. Nat. Commun. 2019, 10, 1269. [Google Scholar] [CrossRef]
- Roterman, I.; Stapor, K.; Konieczny, L. Structural specificity of α-synuclein amyloid. submitted.
- Serpell, L.C. Alzheimer’s amyloid fibrils: Structure and assembly. Biochim. Biophys. Acta 2000, 1502, 16–30. [Google Scholar] [CrossRef]

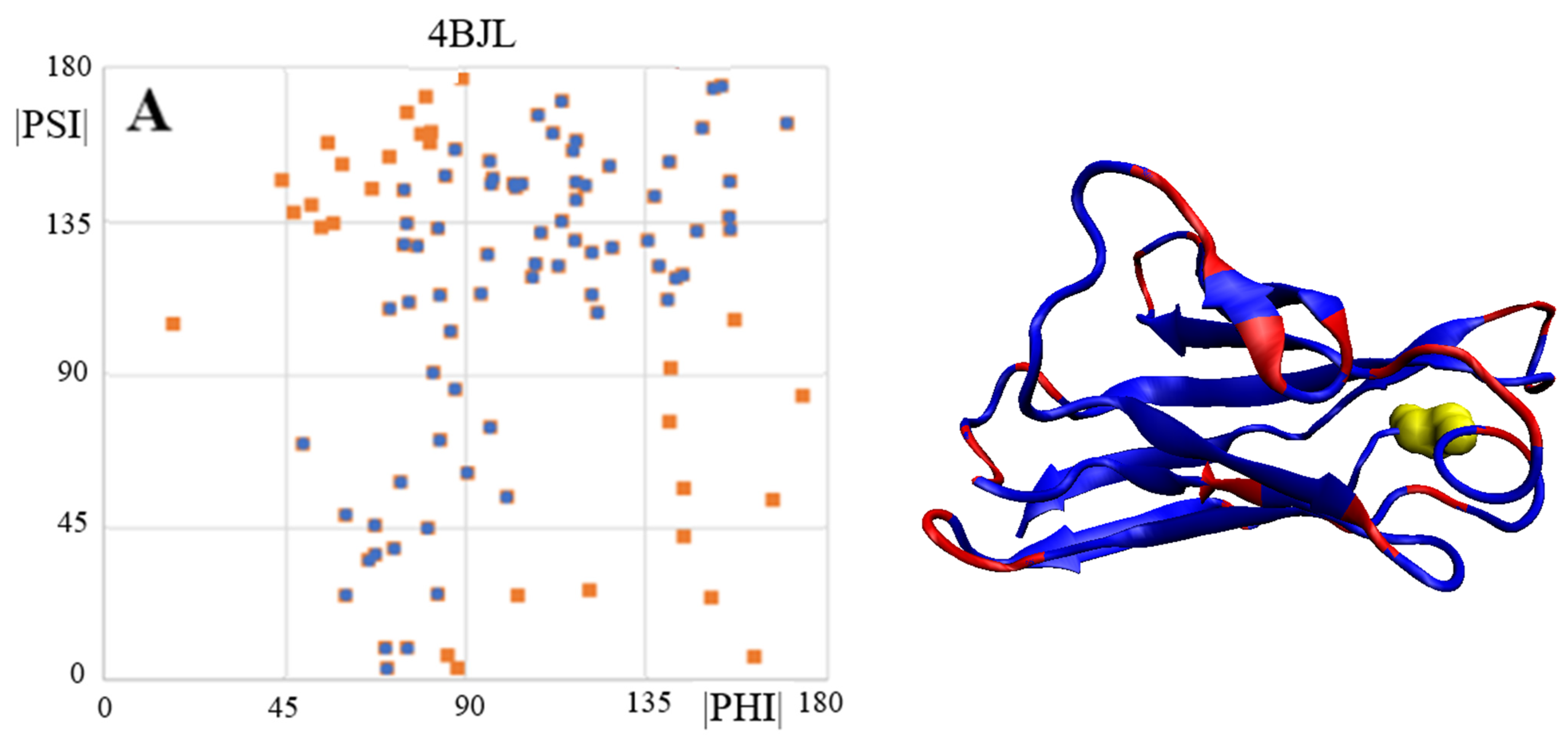
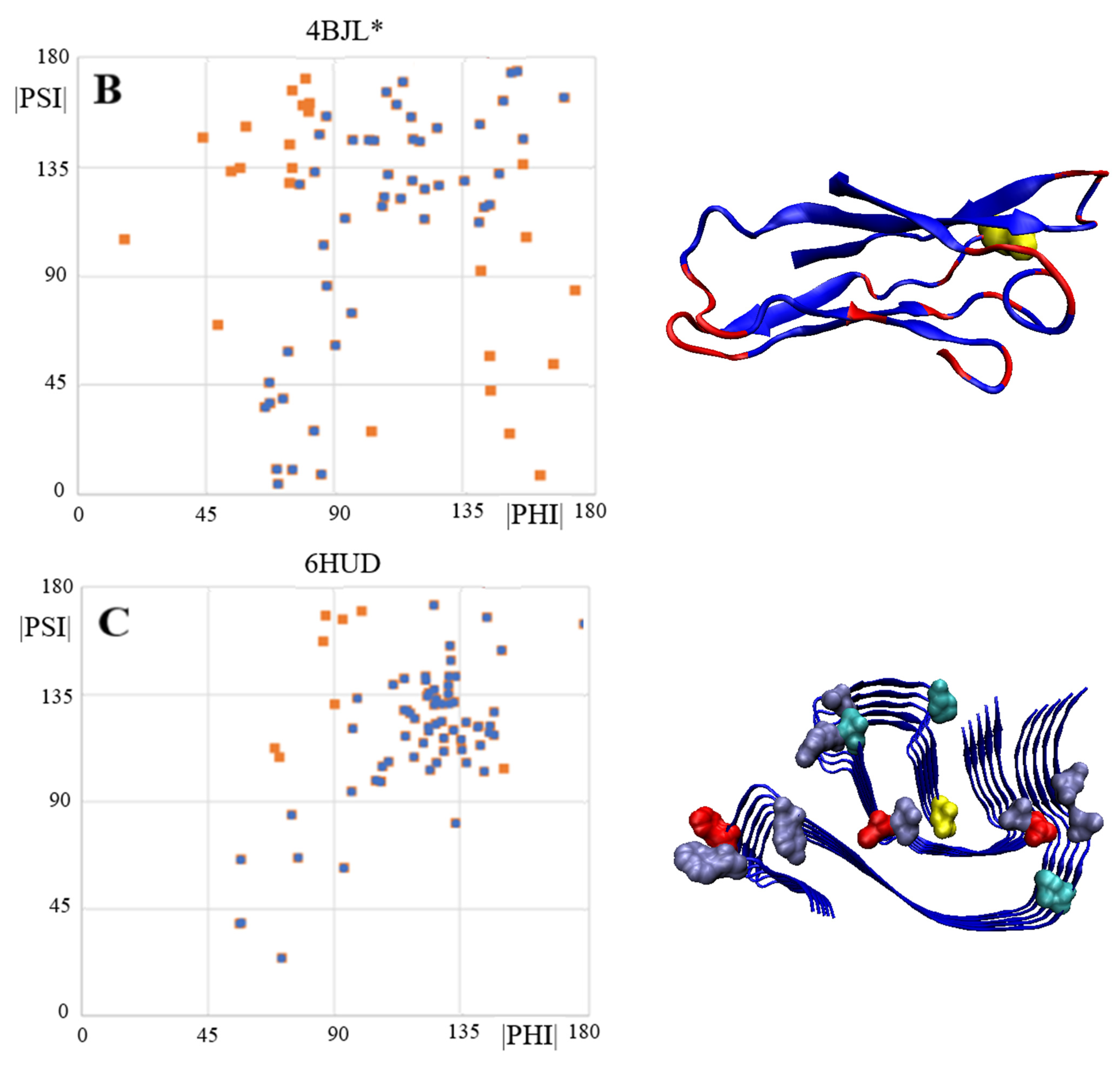
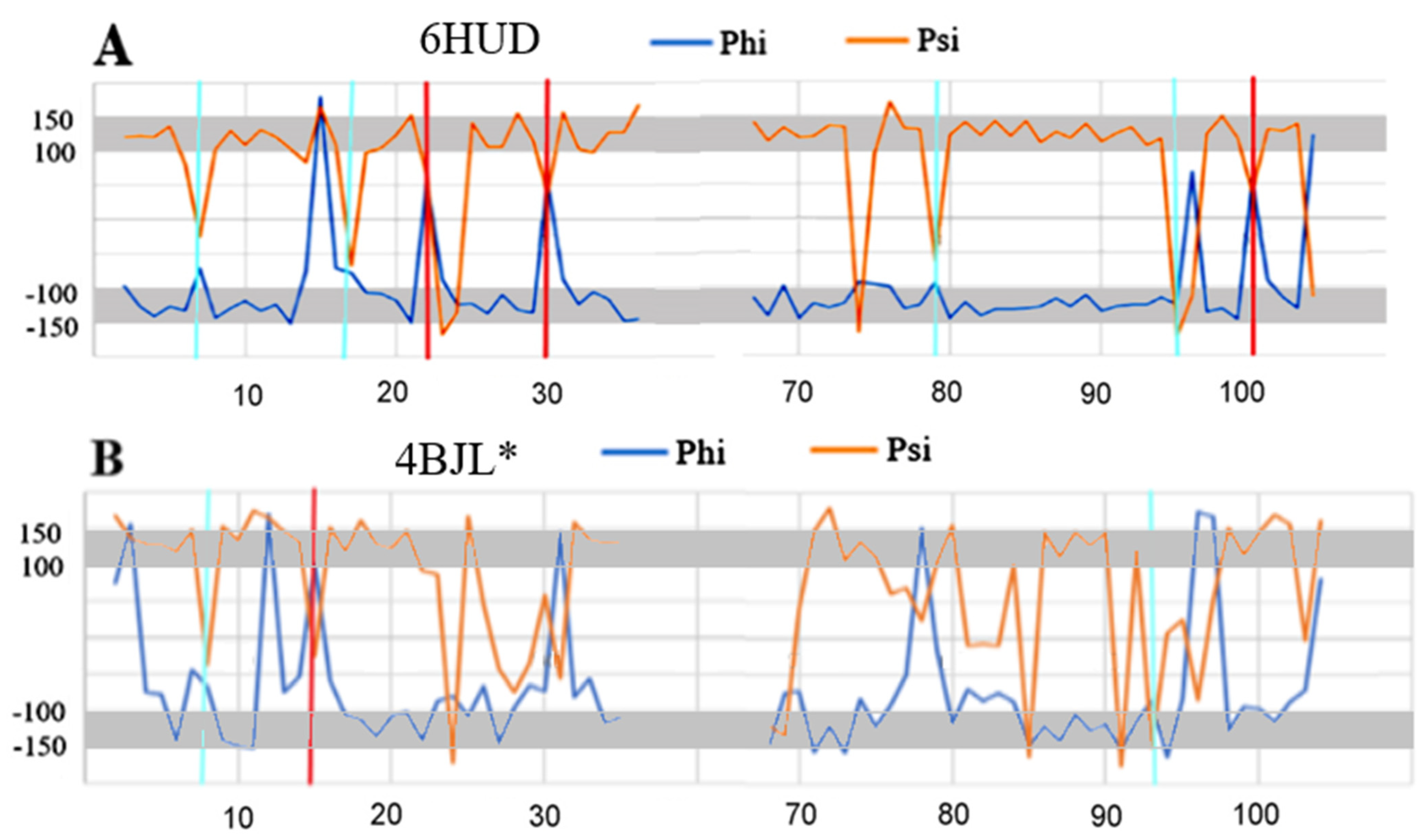
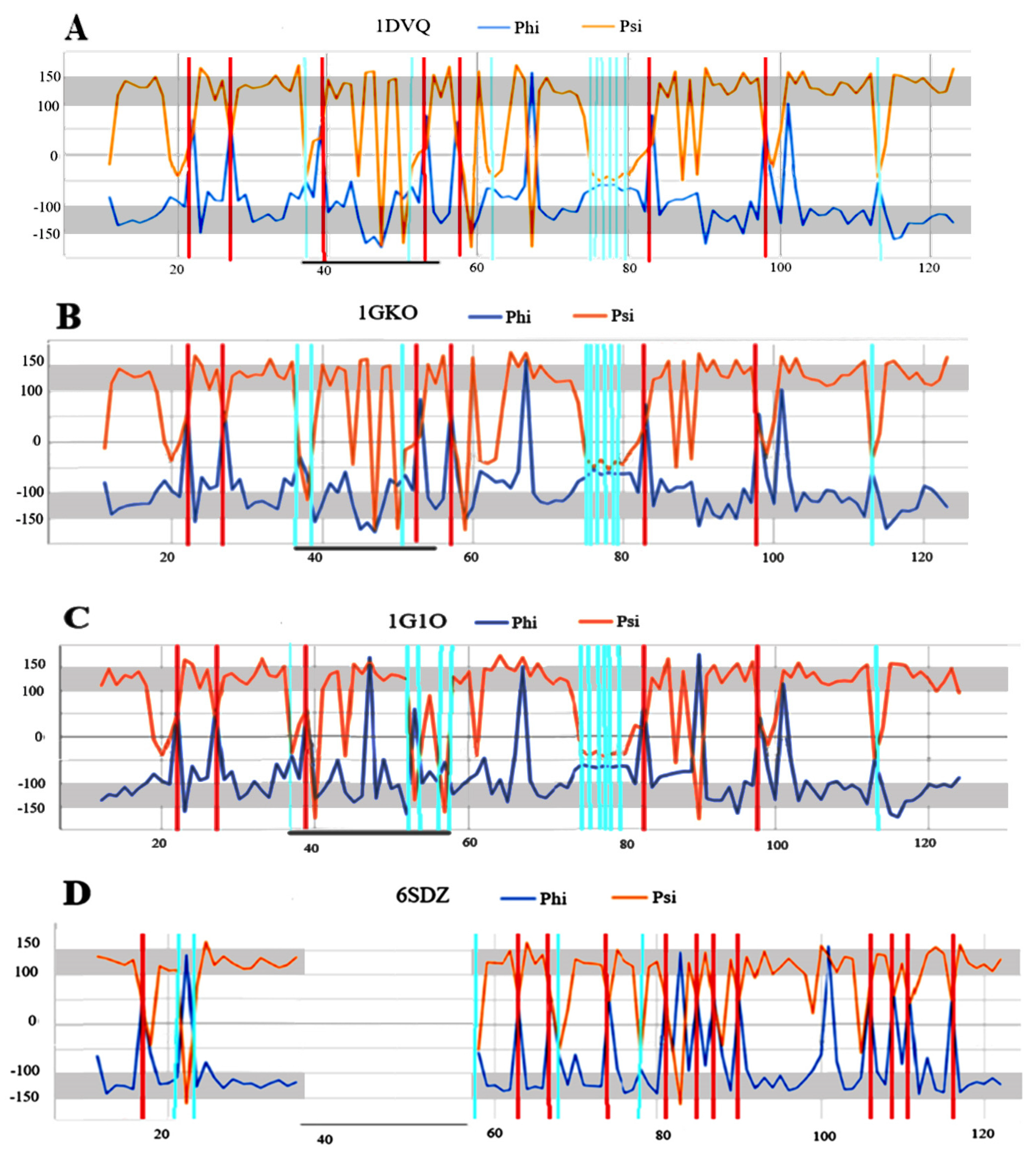

| PDB ID | CC-0 | RESIDUES ELIMINATED | CC-F |
|---|---|---|---|
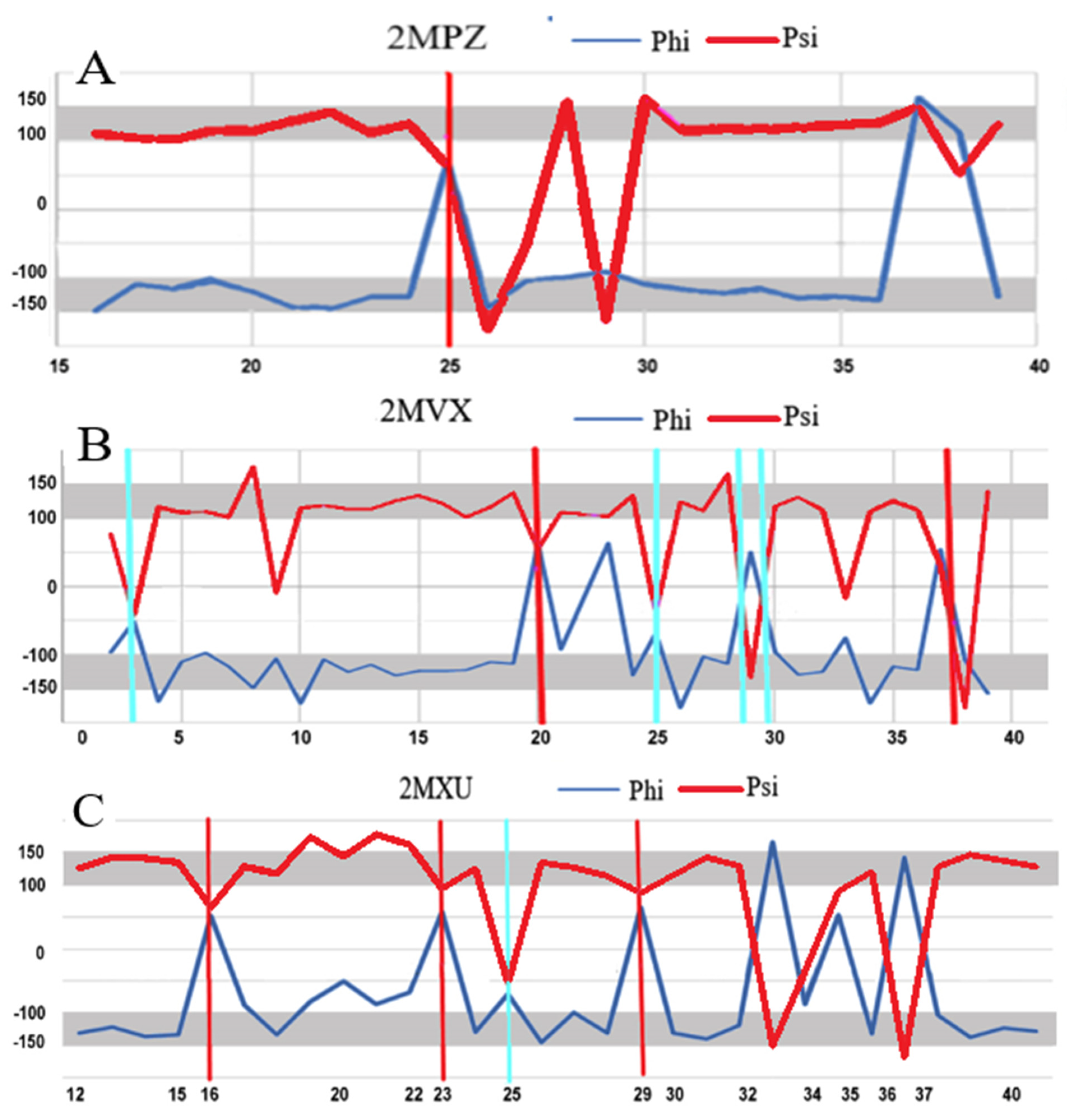
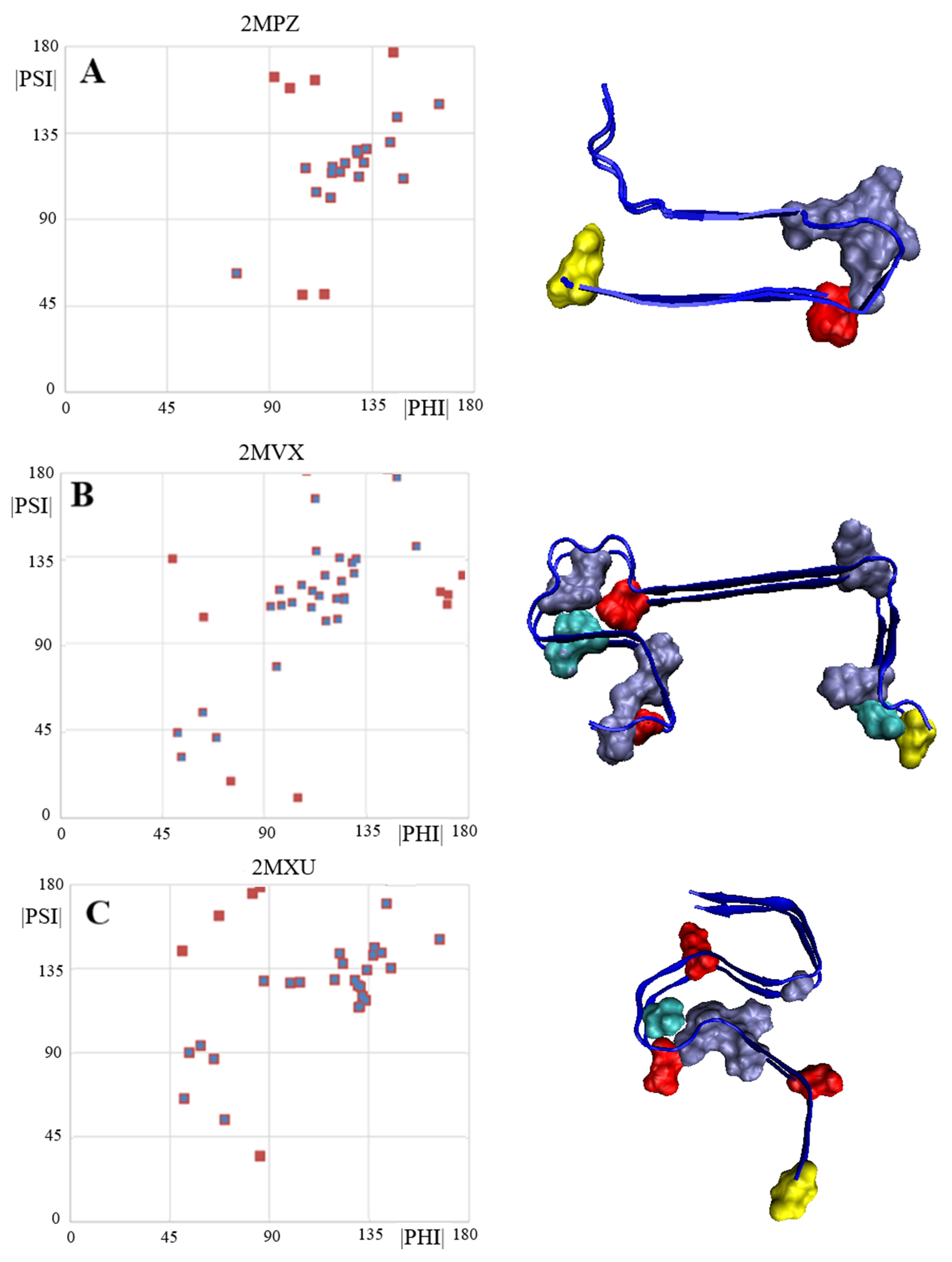
| 2MPZ [32] | 0.364 | 26, 27, 28, 29, 30 | 0.860 |
| 2MXU [33] | 0.403 | 19, 20, 21, 22, 34 | 0.820 |
| 2MVX [34] | 0.521 | 4, 9, 10, 23, 26, 29, 33, 34, 38 | 0.860 |
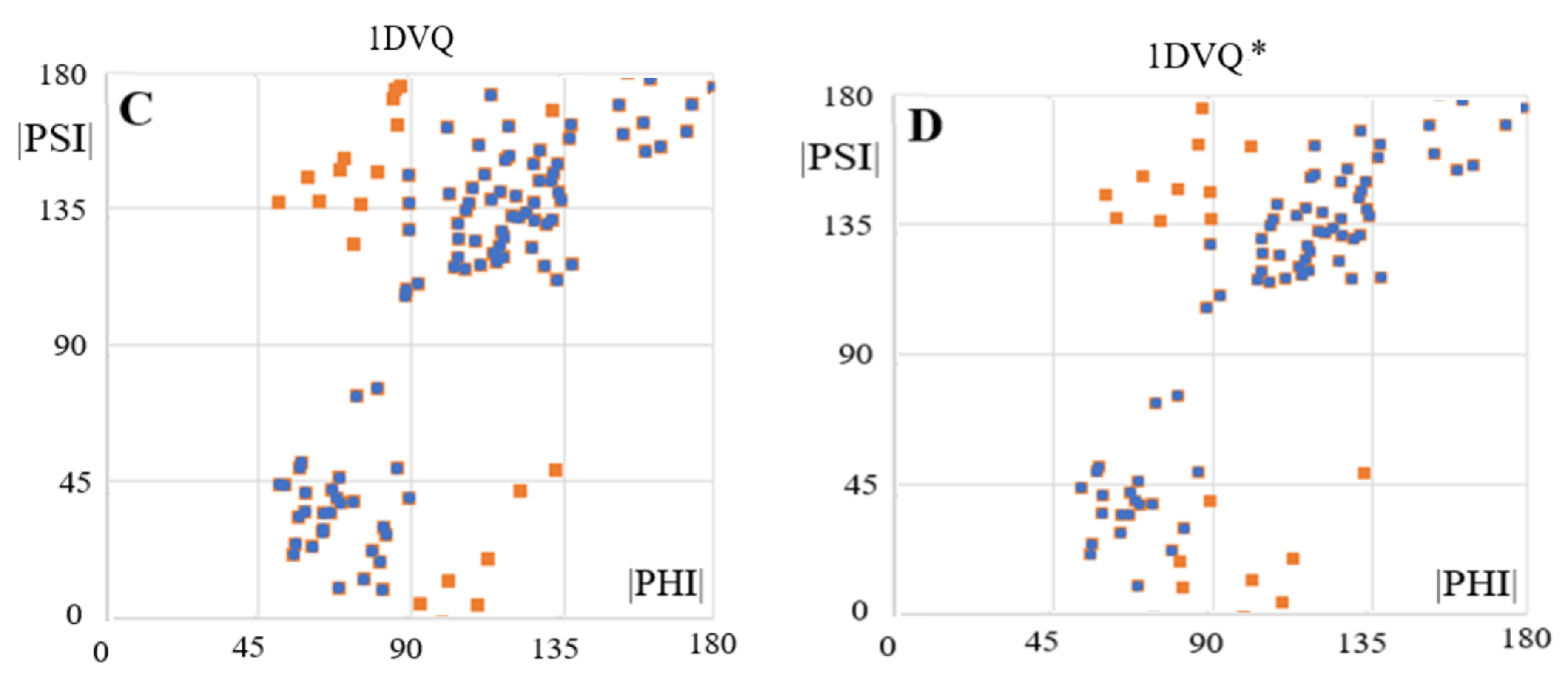
| 1DVQ [35] | 0.619 | 21, 24, 29, 35, 36, 43, 44, 49, 50, 52, 65, 66, 82, 86, 88, 100, 102, 114 | 0.862 |
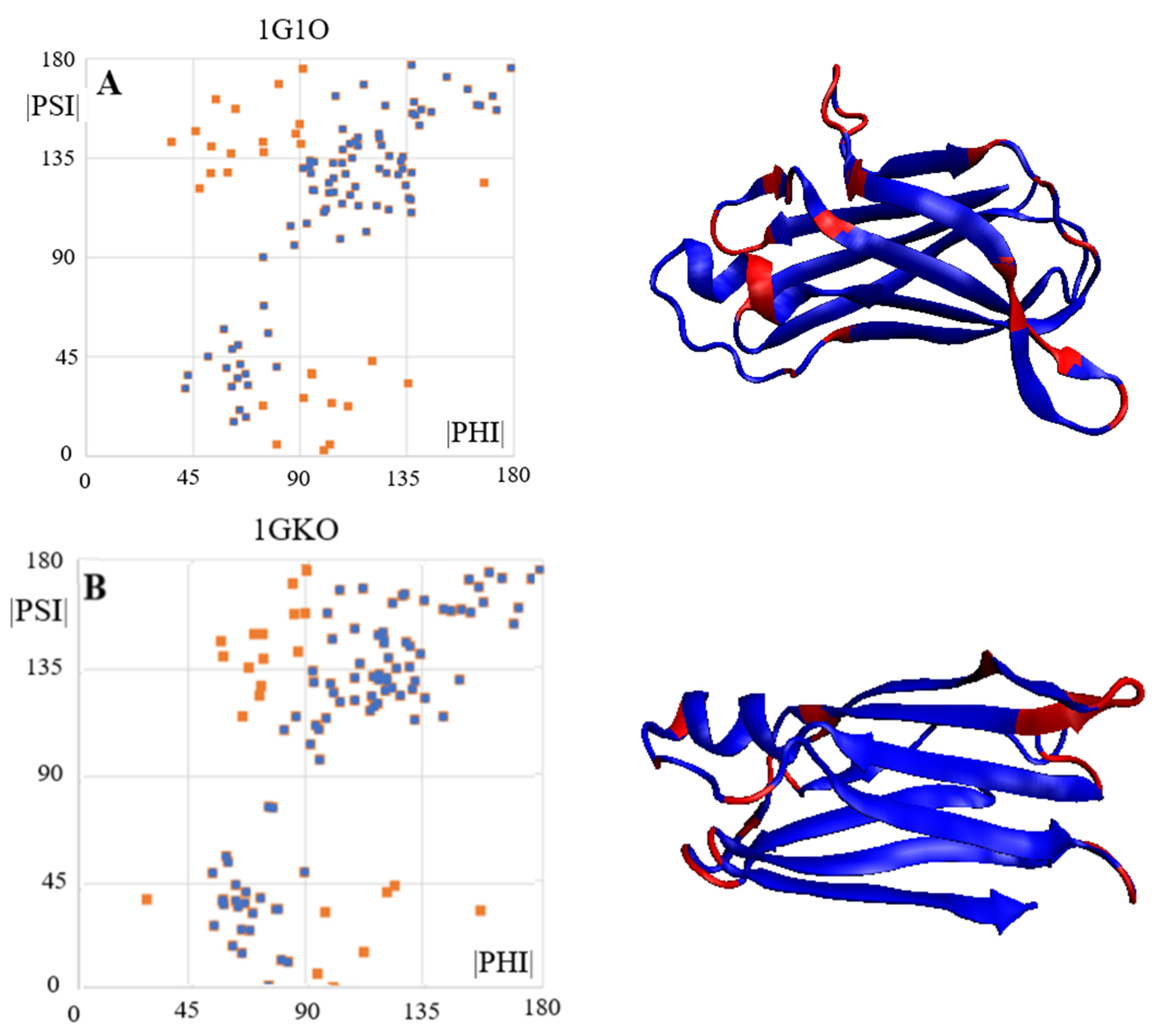
| 1GKO [36] | 0.570 | 20, 21, 24, 26, 29, 35–39, 43, 44, 49, 50, 52, 65, 66, 82, 85, 88, 100, 102, 114 | 0.852 |
| 1G1O [37] | 0.477 | 19–21, 24, 26, 29, 35, 36, 38, 43, 44, 49, 52–54, 56, 57, 62, 64, 66, 82, 88, 89, 100, 102, 114 | 0.855 |
| 1DVQ * [35] | 0.663 | 11, 19–21, 24, 26, 29, 65, 66, 82, 86, 88, 96, 100, 101, 102, 114 | 0.913 |
| 1GKO * [36] | 0.657 | 12, 19–21, 24, 26, 29, 65, 66, 82, 85–88, 93, 100–102, 117 | 0.918 |
| 1G1O * [36] | 0.508 | 19–21, 24, 26, 29, 62, 64, 66, 82, 86, 88, 100, 102, 114 | 0.842 |
| 6SDZ [31] | 0.622 | 12, 23, 24, 69, 70, 75, 84, 86, 91, 93, 99, 100, 102, 105, 110, 112, 113 | 0.938 |
| 4BJL-V [38] | 0.108 | 7, 14, 15, 16, 22, 24, 27, 31–33, 41, 42, 44, 45, 51, 53, 55, 56, 67, 73, 78, 79, 82, 94, 96, 97 | 0.635 |
| 4BJL-V * [38] | 0.120 | 2–4, 7, 13, 14, 15, 16, 22, 24, 27, 31–33, 67, 69, 73, 77–79, 94, 96, 97 | 0.694 |
| 6HUD [39] | 0.495 | 13, 16, 23, 31, 74, 76, 96, 101 | 0.730 |
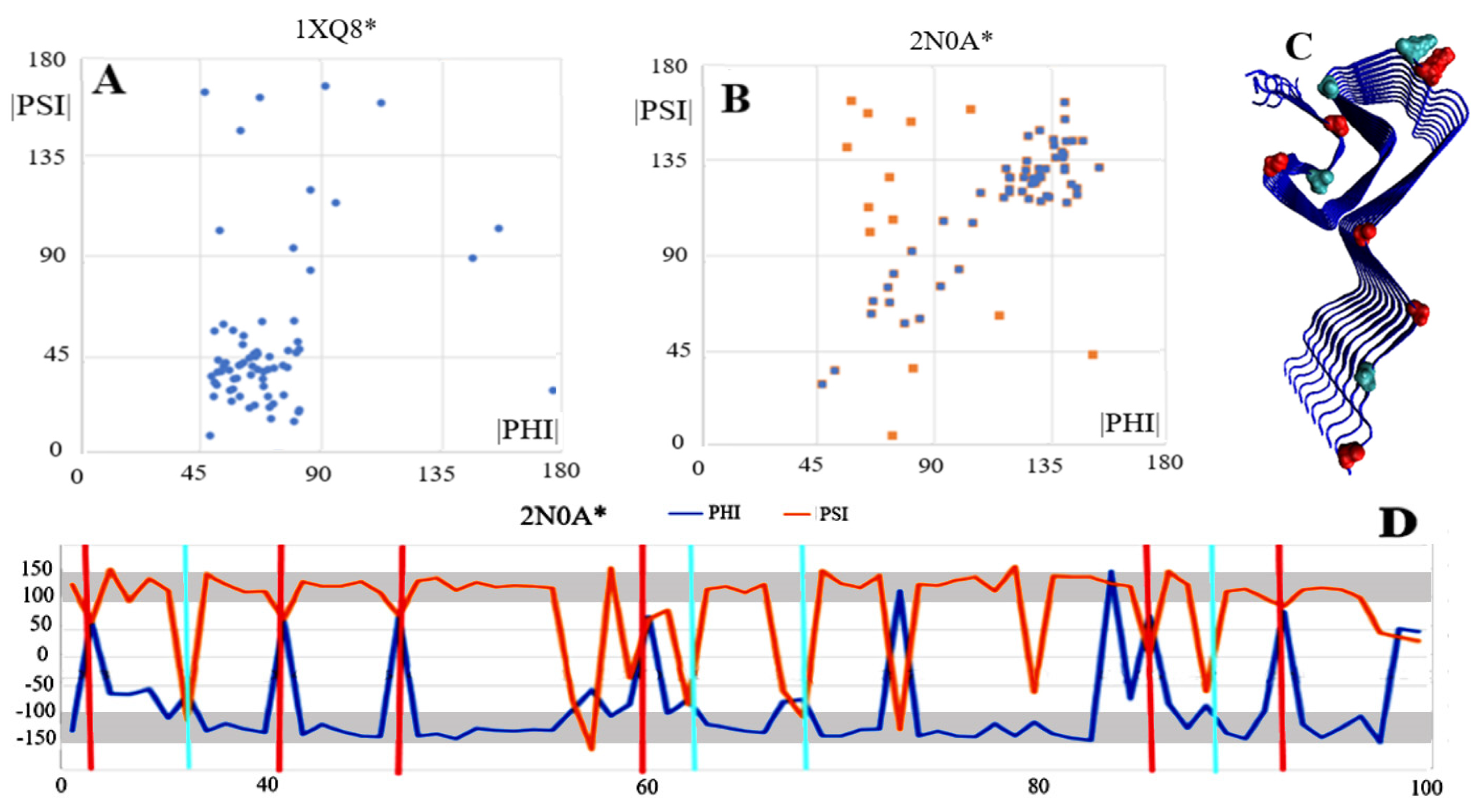
| 1XQ8 * [23] | 0.256 | ||
| 2N0A * [24] | 0.512 | 32–34, 36, 57–59, 68, 80, 85–87, 98 | 0.913 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roterman, I.; Stapor, K.; Konieczny, L. Secondary Structure in Amyloids in Relation to Their Wild Type Forms. Int. J. Mol. Sci. 2023, 24, 154. https://doi.org/10.3390/ijms24010154
Roterman I, Stapor K, Konieczny L. Secondary Structure in Amyloids in Relation to Their Wild Type Forms. International Journal of Molecular Sciences. 2023; 24(1):154. https://doi.org/10.3390/ijms24010154
Chicago/Turabian StyleRoterman, Irena, Katarzyna Stapor, and Leszek Konieczny. 2023. "Secondary Structure in Amyloids in Relation to Their Wild Type Forms" International Journal of Molecular Sciences 24, no. 1: 154. https://doi.org/10.3390/ijms24010154
APA StyleRoterman, I., Stapor, K., & Konieczny, L. (2023). Secondary Structure in Amyloids in Relation to Their Wild Type Forms. International Journal of Molecular Sciences, 24(1), 154. https://doi.org/10.3390/ijms24010154







