A Comprehensive Biomarker Analysis of Microsatellite Unstable/Mismatch Repair Deficient Colorectal Cancer Cohort Treated with Immunotherapy
Abstract
1. Introduction
2. Results
2.1. Cohort Description
2.2. Cohort Genomic Profile
2.3. TMB Scores and Indels
2.4. Immune Microenvironment
2.5. Diagnosis Techniques of MSI-H/dMMR Phenotype
3. Discussion
4. Materials and Methods
4.1. Patients and Samples
4.2. Library Preparation and Bioinformatic Analysis
4.3. Immune Microenvironment Analysis
4.4. MSI/MMR Status Analysis
4.5. Statistical Analysis
5. Conclusions
- -
- Biallelic ARID1A mutation and CTNNB1 mutation are associated with high and low T cell immune infiltrates, respectively, in MSI-H/dMMR CRC.
- -
- Both ARID1A and PTEN biallelic mutations should be further investigated as immunotherapy biomarkers in MSI-H/dMMR CRC.
- -
- TMB does not correlate with immunotherapy benefits based on PD-1/PD-L1 inhibitors in MSI-H/dMMR CRC.
- -
- Discordances in MSI-H/dMMR assessment between IHC and PCR are associated with limited benefit to immunotherapy.
- -
- Immunotherapy biomarkers in MSI-H/dMMR mCRC should rely on tumour genomic profiles and their dominant immune-contexture.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Kim, T.W.; Van Cutsem, E.; Geva, R.; Jäger, D.; Hara, H.; Burge, M.; O’Neil, B.; Kavan, P.; Yoshino, T.; et al. Phase II open-label study of pembrolizumab in treatment-refractory, microsatellite instability–high/mismatch repair–deficient metastatic colorectal cancer: KEYNOTE-164. J. Clin. Oncol. 2020, 38, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Overman, M.J.; Lonardi, S.; Wong, K.Y.M.; Lenz, H.-J.; Gelsomino, F.; Aglietta, M.; Morse, M.A.; Van Cutsem, E.; McDermott, R.; Hill, A.; et al. Durable Clinical Benefit with Nivolumab Plus Ipilimumab in DNA Mismatch Repair–Deficient/Microsatellite Instability–High Metastatic Colorectal Cancer. J. Clin. Oncol. 2018, 36, 773–779. [Google Scholar] [CrossRef]
- Overman, M.J.; McDermott, R.; Leach, J.L.; Lonardi, S.; Lenz, H.-J.; Morse, M.A.; Desai, J.; Hill, A.; Axelson, M.; Moss, R.A.; et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): An open-label, multicentre, phase 2 study. Lancet Oncol. 2017, 18, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- André, T.; Shiu, K.-K.; Kim, T.W.; Jensen, B.V.; Jensen, L.H.; Punt, C.; Smith, D.; Garcia-Carbonero, R.; Benavides, M.; Gibbs, P.; et al. Pembrolizumab in Microsatellite-Instability–High Advanced Colorectal Cancer. N. Engl. J. Med. 2020, 383, 2207–2218. [Google Scholar] [CrossRef]
- Lenz, H.-J.; Lonardi, S.; Zagonel, V.; Van Cutsem, E.; Limon, M.L.; Wong, K.Y.M.; Hendlisz, A.; Aglietta, M.; Garcia-Alfonso, P.; Neyns, B.; et al. Nivolumab plus low-dose ipilimumab as first-line therapy in microsatellite instability-high/DNA mismatch repair deficient metastatic colorectal cancer: Clinical update. J. Clin. Oncol. 2020, 38, 11. [Google Scholar] [CrossRef]
- Muzny, D.M.; Bainbridge, M.N.; Chang, K. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar]
- Grasso, C.S.; Giannakis, M.; Wells, D.K.; Hamada, T.; Mu, X.J.; Quist, M.; Nowak, J.A.; Nishihara, R.; Qian, Z.R.; Inamura, K.; et al. Genetic Mechanisms of Immune Evasion in Colorectal Cancer. Cancer Discov. 2018, 8, 730–749. [Google Scholar] [CrossRef]
- Turajlic, S.; Litchfield, K.; Xu, H.; Rosenthal, R.; McGranahan, N.; Reading, J.L.; Wong, Y.N.S.; Rowan, A.; Kanu, N.; Al Bakir, M.; et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: A pan-cancer analysis. Lancet Oncol. 2017, 18, 1009–1021. [Google Scholar] [CrossRef]
- Guinney, J.; Dienstmann, R.; Wang, X.; De Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Becht, E.; de Reyniès, A.; Giraldo, N.A.; Pilati, C.; Buttard, B.; Lacroix, L.; Selves, J.; Sautès-Fridman, C.; Laurent-Puig, P.; Fridman, W.H. Immune and Stromal Classification of Colorectal Cancer Is Associated with Molecular Subtypes and Relevant for Precision Immunotherapy. Clin. Cancer Res. 2016, 22, 4057–4066. [Google Scholar] [CrossRef]
- Llosa, N.J.; Cruise, M.; Tam, A.; Wicks, E.C.; Hechenbleikner, E.M.; Taube, J.M.; Blosser, R.L.; Fan, H.; Wang, H.; Luber, B.S.; et al. The Vigorous Immune Microenvironment of Microsatellite Instable Colon Cancer Is Balanced by Multiple Counter-Inhibitory Checkpoints. Cancer Discov. 2015, 5, 43–51. [Google Scholar] [CrossRef]
- Lal, N.; Beggs, A.; E Willcox, B.; Middleton, G.W. An immunogenomic stratification of colorectal cancer: Implications for development of targeted immunotherapy. OncoImmunology 2015, 4, e976052. [Google Scholar] [CrossRef]
- Ozcan, M.; Janikovits, J.; von Knebel Doeberitz, M.; Kloor, M. Complex pattern of immune evasion in MSI colorectal cancer. OncoImmunology 2018, 7, e1445453. [Google Scholar] [CrossRef]
- Shin, D.S.; Zaretsky, J.M.; Escuin-Ordinas, H.; Garcia-Diaz, A.; Hu-Lieskovan, S.; Kalbasi, A.; Grasso, C.S.; Hugo, W.; Sandoval, S.; Torrejon, D.Y.; et al. Primary Resistance to PD-1 Blockade Mediated by JAK1/2 MutationsPrimary Resistance to PD-1 Blockade. Cancer Discov. 2017, 7, 188–201. [Google Scholar] [CrossRef]
- Gurjao, C.; Liu, D.; Hofree, M.; AlDubayan, S.H.; Wakiro, I.; Su, M.-J.; Felt, K.; Gjini, E.; Brais, L.K.; Rotem, A.; et al. Intrinsic Resistance to Immune Checkpoint Blockade in a Mismatch Repair–Deficient Colorectal Cancer. Cancer Immunol. Res. 2019, 7, 1230–1236. [Google Scholar] [CrossRef]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Lu, S.; Kemberling, H.; Wilt, C.; Luber, B.S.; et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef]
- Llosa, N.J.; Luber, B.; Siegel, N.; Awan, A.H.; Oke, T.; Zhu, Q.; Bartlett, B.R.; Aulakh, L.K.; Thompson, E.D.; Jaffee, E.M.; et al. Immunopathologic Stratification of Colorectal Cancer for Checkpoint Blockade Immunotherapy. Cancer Immunol. Res. 2019, 7, 1574–1579. [Google Scholar] [CrossRef]
- Yaeger, R.; Chatila, W.K.; Lipsyc, M.D.; Hechtman, J.F.; Cercek, A.; Sanchez-Vega, F.; Jayakumaran, G.; Middha, S.; Zehir, A.; Donoghue, M.T.A.; et al. Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell 2018, 33, 125–136.e3. [Google Scholar] [CrossRef]
- cBioPortal for Cancer Genomics. Available online: www.cbioportal.org (accessed on 5 July 2021).
- Pich, O.; Muiños, F.; Lolkema, M.P.; Steeghs, N.; Gonzalez-Perez, A.; Lopez-Bigas, N. The mutational footprints of cancer therapies. Nat. Genet. 2019, 51, 1732–1740. [Google Scholar] [CrossRef]
- COSMIC. Catalogue of Somatic Mutations in Cancer. Available online: cancer.sanger.ac.uk/cosmic (accessed on 5 July 2021).
- Middha, S.; Yaeger, R.; Shia, J.; Stadler, Z.K.; King, S.; Guercio, S.; Paroder, V.; Bates, D.D.; Rana, S.; Diaz, L.A.; et al. Majority of B2M-Mutant and -Deficient Colorectal Carcinomas Achieve Clinical Benefit from Immune Checkpoint Inhibitor Therapy and Are Microsatellite Instability-High. JCO Precis. Oncol. 2019, 3, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Luke, J.J.; Bao, R.; Sweis, R.F.; Spranger, S.; Gajewski, T.F. WNT/β-catenin Pathway Activation Correlates with Immune Exclusion across Human Cancers. Clin. Cancer Res. 2019, 25, 3074–3083. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Lavrijsen, M.; Bakker, A.; Magierowski, M.; Magierowska, K.; Liu, P.; Wang, W.; Peppelenbosch, M.P.; Smits, R. Commonly observed RNF43 mutations retain functionality in attenuating Wnt/β-catenin signaling and unlikely confer Wnt-dependency onto colorectal cancers. Oncogene 2020, 39, 3458–3472. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Ju, Z.; Zhao, W.; Wang, L.; Peng, Y.; Ge, Z.; Nagel, Z.D.; Zou, J.; Wang, C.; Kapoor, P.; et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat. Med. 2018, 24, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Tokunaga, R.; Xiu, J.; Goldberg, R.M.; Philip, P.A.; Seeber, A.; Battaglin, F.; Arai, H.; Lo, J.H.; Puccini, A.; Naseem, M.; et al. Gene mutations of SWI/SNF complex and molecular profile in colorectal cancer. J. Clin. Oncol. 2019, 37, 3600. [Google Scholar] [CrossRef]
- Pan, D.; Kobayashi, A.; Jiang, P.; de Andrade, L.F.; Tay, R.E.; Luoma, A.M.; Tsoucas, D.; Qiu, X.; Lim, K.; Rao, P.; et al. A major chromatin regulator determines resistance of tumor cells to T cell–mediated killing. Science 2018, 359, 770–775. [Google Scholar] [CrossRef]
- Okamura, R.; Kato, S.; Lee, S.; Jimenez, R.E.; Sicklick, J.K.; Kurzrock, R. ARID1A alterations function as a biomarker for longer progression-free survival after anti-PD-1/PD-L1 immunotherapy. J. Immunother. Cancer 2020, 8, e000438. [Google Scholar] [CrossRef]
- Abou Alaiwi, S.; Nassar, A.H.; Xie, W.; Bakouny, Z.; Berchuck, J.E.; Braun, D.A.; Baca, S.C.; Nuzzo, P.V.; Flippot, R.; Mouhieddine, T.H.; et al. Mammalian SWI/SNF Complex Genomic Alterations and Immune Checkpoint Blockade in Solid TumorsmSWI/SNF Genomic Alterations and ICIs in Solid Tumors. Cancer Immunol. Res. 2020, 8, 1075–1084. [Google Scholar] [CrossRef]
- Piro, G.; Carbone, C.; Carbognin, L.; Pilotto, S.; Ciccarese, C.; Iacovelli, R.; Milella, M.; Bria, E.; Tortora, G. Revising PTEN in the Era of Immunotherapy: New Perspectives for an Old Story. Cancers 2019, 11, 1525. [Google Scholar] [CrossRef]
- Conciatori, F.; Bazzichetto, C.; Falcone, I.; Ciuffreda, L.; Ferretti, G.; Vari, S.; Ferraresi, V.; Cognetti, F.; Milella, M. PTEN Function at the Interface between Cancer and Tumor Microenvironment: Implications for Response to Immunotherapy. Int. J. Mol. Sci. 2020, 21, 5337. [Google Scholar] [CrossRef]
- Vidotto, T.; Melo, C.M.; Castelli, E.; Koti, M.; dos Reis, R.B.; Squire, J.A. Emerging role of PTEN loss in evasion of the immune response to tumours. Br. J. Cancer 2020, 122, 1732–1743. [Google Scholar] [CrossRef]
- Chida, K.; Kawazoe, A.; Kawazu, M.; Suzuki, T.; Nakamura, Y.; Nakatsura, T.; Kuwata, T.; Ueno, T.; Kuboki, Y.; Kotani, D.; et al. A Low Tumor Mutational Burden and PTEN Mutations Are Predictors of a Negative Response to PD-1 Blockade in MSI-H/dMMR Gastrointestinal TumorsLow TMB and PTEN Mutations Predict ICI Response in MSI-H GI Tumors. Clin. Cancer Res. 2021, 27, 3714–3724. [Google Scholar] [CrossRef]
- Schrock, A.B.; Ouyang, C.; Sandhu, J.; Sokol, E.; Jin, D.; Ross, J.S.; Miller, V.A.; Lim, D.; Amanam, I.; Chao, J.; et al. Tumor mutational burden is predictive of response to immune checkpoint inhibitors in MSI-high metastatic colorectal cancer. Ann. Oncol. 2019, 30, 1096–1103. [Google Scholar] [CrossRef]
- McGrail, D.; Pilié, P.; Rashid, N.; Voorwerk, L.; Slagter, M.; Kok, M.; Jonasch, E.; Khasraw, M.; Heimberger, A.; Lim, B.; et al. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann. Oncol. 2021, 32, 661–672. [Google Scholar] [CrossRef]
- Rousseau, B.; Foote, M.B.; Maron, S.B.; Diplas, B.H.; Lu, S.; Argilés, G.; Cercek, A.; Diaz, L.A., Jr. The spectrum of benefit from checkpoint blockade in hypermutated tumors. N. Engl. J. Med. 2021, 384, 1168–1170. [Google Scholar] [CrossRef]
- Cohen, R.; Hain, E.; Buhard, O.; Guilloux, A.; Bardier, A.; Kaci, R.; Bertheau, P.; Renaud, F.; Bibeau, F.; Fléjou, J.-F.; et al. Association of Primary Resistance to Immune Checkpoint Inhibitors in Metastatic Colorectal Cancer with Misdiagnosis of Microsatellite Instability or Mismatch Repair Deficiency Status. JAMA Oncol. 2019, 5, 551–555. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, S.Y.; Baek, J.Y.; Cha, Y.J.; Ahn, J.B.; Kim, H.S.; Lee, K.-W.; Kim, J.-W.; Kim, T.-Y.; Chang, W.J.; et al. A Phase II Study of Avelumab Monotherapy in Patients with Mismatch Repair-Deficient/Microsatellite Instability-High or POLE-Mutated Metastatic or Unresectable Colorectal Cancer. Cancer Res. Treat. 2020, 52, 1135–1144. [Google Scholar] [CrossRef]
- Luchini, C.; Bibeau, F.; Ligtenberg, M.J.L.; Singh, N.; Nottegar, A.; Bosse, T.; Miller, R.; Riaz, N.; Douillard, J.Y.; Andre, F.; et al. ESMO recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: A systematic review-based approach. Ann. Oncol. 2019, 30, 1232–1243. [Google Scholar] [CrossRef]
- Cristescu, R.; Mogg, R.; Ayers, M.; Albright, A.; Murphy, E.; Yearley, J.; Sher, X.; Liu, X.Q.; Lu, H.; Nebozhyn, M.; et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade–based immunotherapy. Science 2018, 362, eaar3593. [Google Scholar] [CrossRef]
- Schwartz, L.H.; Litière, S.; De Vries, E.; Ford, R.; Gwyther, S.; Mandrekar, S.; Shankar, L.; Bogaerts, J.; Chen, A.; Dancey, J.; et al. RECIST 1.1-Update and clarification: From the RECIST committee. Eur. J. Cancer 2016, 62, 132–137. [Google Scholar] [CrossRef]
- Cohen, R.; Bennouna, J.; Meurisse, A.; Tournigand, C.; De La Fouchardière, C.; Tougeron, D.; Borg, C.; Mazard, T.; Chibaudel, B.; Garcia-Larnicol, M.-L.; et al. RECIST and iRECIST criteria for the evaluation of nivolumab plus ipilimumab in patients with microsatellite instability-high/mismatch repair-deficient metastatic colorectal cancer: The GERCOR NIPICOL phase II study. J. Immunother. Cancer 2020, 8, e001499. [Google Scholar] [CrossRef] [PubMed]
- Altshuler, D.M.; Durbin, R.M.; Abecasis, G.R. An integrated map of genetic variation from 1092 human genomes. Nature 2012, 491, 56–65. [Google Scholar]
- The Exome Variant Server Database. Available online: https://esp.gs.washington.edu/drupal (accessed on 5 July 2021).
- Lek, M.; Karczewski, K.J.; Minikel, E.V.; Samocha, K.E.; Banks, E.; Fennell, T.; O’Donnell-Luria, A.H.; Ware, J.S.; Hill, A.J.; Cummings, B.B.; et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016, 536, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- COSMIC. Catalogue Of Somatic Mutations In Cancer. 2021. Available online: https://cancer.sanger.ac.uk (accessed on 5 July 2021).
- cBioPortal for Cancer Genomics. 2021. Available online: https://cbioportal.org (accessed on 5 July 2021).
- ClinVar -NCBI-NIH. 2021. Available online: https://ncbi.nlm.nih.gov/clinvar/ (accessed on 5 July 2021).
- OncoKB. 2021. Available online: https://oncokb.org/ (accessed on 5 July 2021).
- The Human Genomic Variant. 2021. Available online: https://varsome.com (accessed on 5 July 2021).
- Kulangara, K.; Zhang, N.; Corigliano, E.; Guerrero, L.; Waldroup, S.; Jaiswal, D.; Ms, M.J.; Shah, S.; Hanks, D.; Wang, J.; et al. Clinical utility of the combined positive score for programmed death ligand-1 expression and the approval of pembrolizumab for treatment of gastric cancer. Arch. Pathol. Lab. Med. 2019, 143, 330–337. [Google Scholar] [CrossRef]
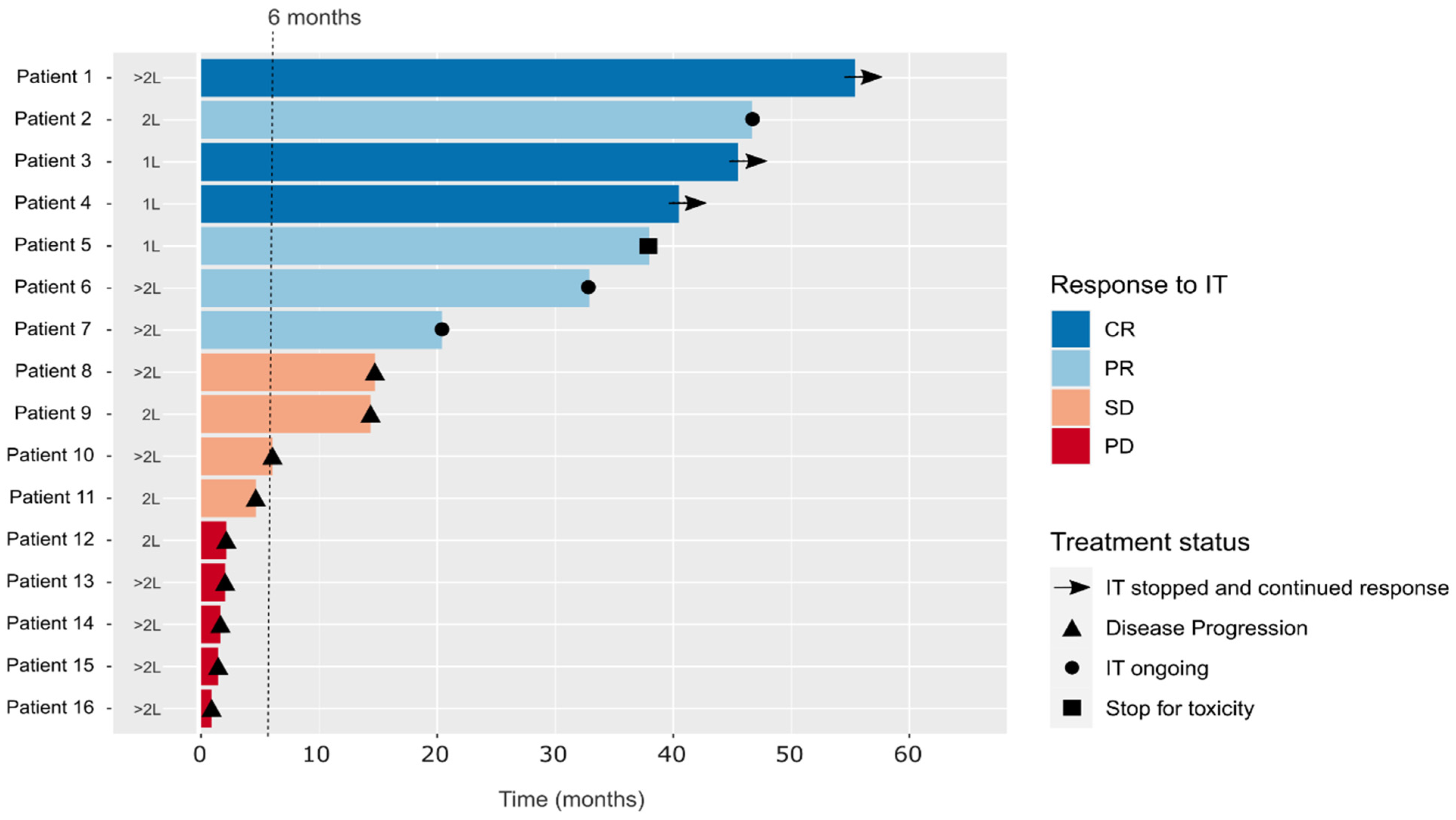
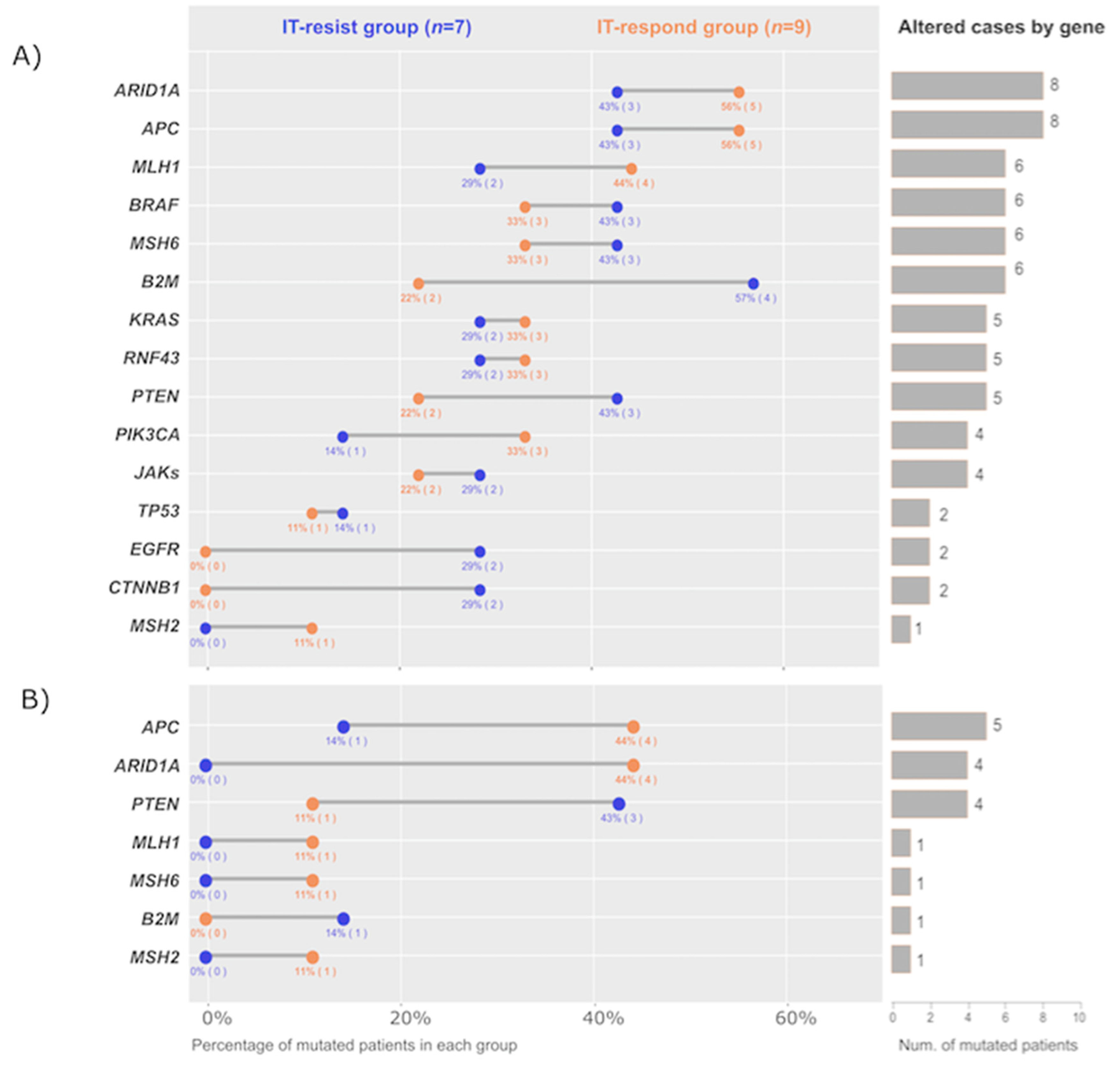
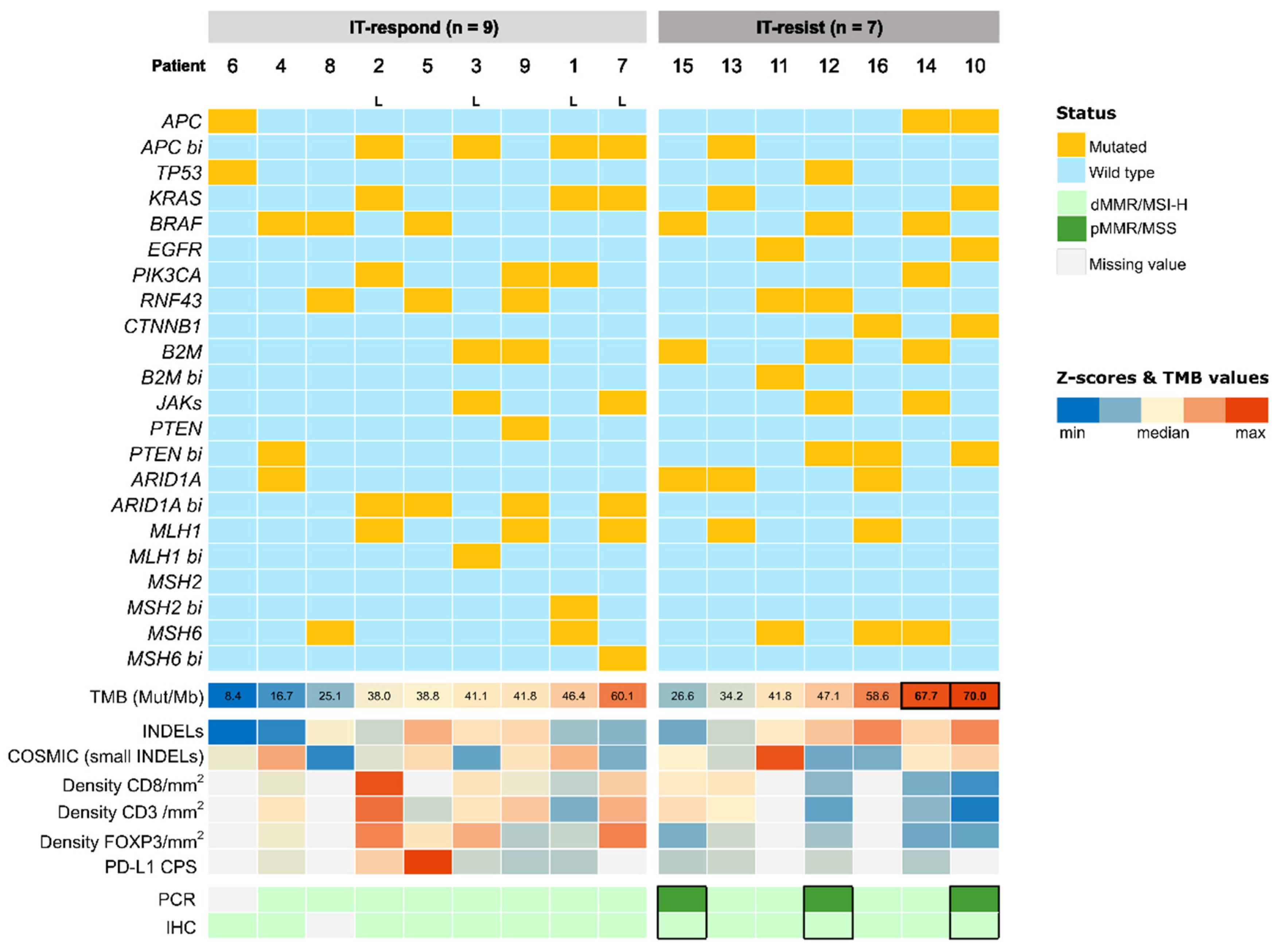
| Number of Patients n = 16 | % | IT-Respond n = 9 | % | IT-Resist n = 7 | % | |
|---|---|---|---|---|---|---|
| Median age at diagnosis and range | 55.4 | (30–90) | 57.22 | (31–90) | 53.14 | (30–71) |
| Gender | ||||||
| Male | 7 | 43.7% | 4 | 44.4% | 3 | 42.8% |
| Female | 9 | 56.3% | 5 | 55.5% | 4 | 57.1% |
| Stage at diagnosis | ||||||
| II | 1 | 6.2% | 1 | 11.1% | 0 | 0% |
| III | 8 | 50% | 3 | 33.3% | 5 | 71.4% |
| IV | 7 | 43.7% | 5 | 55.5% | 2 | 28.6% |
| Primary tumor location | ||||||
| Right | 9 | 56.2% | 5 | 55.5% | 4 | 57.1% |
| Transverse | 3 | 18.7% | 1 | 11.1% | 2 | 28.6% |
| Left-rectum | 4 | 25% | 3 | 33.3% | 1 | 14.3% |
| Number of metastatic sites | ||||||
| 1 | 6 | 37.5% | 4 | 44.4% | 2 | 28.6% |
| 2 | 7 | 43.7% | 4 | 44.4% | 3 | 42.8% |
| 3 | 3 | 18.7% | 1 | 11.1% | 2 | 28.6% |
| Anti-PD-1/PD-L1 setting | ||||||
| 1st line | 3 | 18.7% | 3 | 33.3% | 0 | 0% |
| 2nd line | 4 | 25% | 2 | 22.2% | 2 | 28.6% |
| >2nd line | 9 | 56.2% | 4 | 44.4% | 5 | 71.42% |
| KRASmutation | ||||||
| Yes | 5 | 31.2% | 3 | 33.3% | 2 | 28.6% |
| No | 11 | 68.7% | 6 | 66.6% | 5 | 71.4% |
| NRASmutation | ||||||
| Yes | 0 | 0% | 0 | 0% | 0 | 0% |
| No | 0 | 0% | 0 | 0% | 0 | 0% |
| BRAFV600E | ||||||
| Yes | 6 | 37.5% | 3 | 33.3% | 3 | 42.8% |
| No | 10 | 62.5% | 6 | 66.6% | 4 | 57.1% |
| Lynch syndrome | ||||||
| Yes | 4 | 25% | 4 | 44.4% | 0 | 0% |
| No | 12 | 75% | 5 | 55.5% | 7 | 100% |
| Anti-PD-1/PD-L1 regimen | ||||||
| Anti-PD1 | 7 | 43.7% | 5 | 55.5% | 2 | 28.6% |
| Anti-PD-L1 | 3 | 18.7% | 1 | 11.1% | 2 | 28.6% |
| Anti-PD-L1 combo: | 6 | 37.5% | 3 | 33.3% | 3 | 42.8% |
| +Bevacizumab | 3 | 18.7% | 2 | 22.2% | 1 | 14.3% |
| +CD40 agonist | 1 | 6.2% | 0 | 0% | 1 | 14.3% |
| +anti-CEA-CD3 antibody | 2 | 12.5% | 1 | 11.1% | 1 | 14.3% |
| Anti-PD-1/PD-L1 response | ||||||
| CR | 3 | 18.7% | 3 | 33.3% | 0 | 0% |
| PR | 4 | 25% | 4 | 44.4% | 0 | 0% |
| SD | 4 | 25% | 2 | 22.2% | 2 | 28.6% |
| PD | 5 | 31.2% | 0 | 0% | 5 | 71.4% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Élez, E.; Mulet-Margalef, N.; Sanso, M.; Ruiz-Pace, F.; Mancuso, F.M.; Comas, R.; Ros, J.; Argilés, G.; Martini, G.; Sanz-Garcia, E.; et al. A Comprehensive Biomarker Analysis of Microsatellite Unstable/Mismatch Repair Deficient Colorectal Cancer Cohort Treated with Immunotherapy. Int. J. Mol. Sci. 2023, 24, 118. https://doi.org/10.3390/ijms24010118
Élez E, Mulet-Margalef N, Sanso M, Ruiz-Pace F, Mancuso FM, Comas R, Ros J, Argilés G, Martini G, Sanz-Garcia E, et al. A Comprehensive Biomarker Analysis of Microsatellite Unstable/Mismatch Repair Deficient Colorectal Cancer Cohort Treated with Immunotherapy. International Journal of Molecular Sciences. 2023; 24(1):118. https://doi.org/10.3390/ijms24010118
Chicago/Turabian StyleÉlez, Elena, Núria Mulet-Margalef, Miriam Sanso, Fiorella Ruiz-Pace, Francesco M. Mancuso, Raquel Comas, Javier Ros, Guillem Argilés, Giulia Martini, Enrique Sanz-Garcia, and et al. 2023. "A Comprehensive Biomarker Analysis of Microsatellite Unstable/Mismatch Repair Deficient Colorectal Cancer Cohort Treated with Immunotherapy" International Journal of Molecular Sciences 24, no. 1: 118. https://doi.org/10.3390/ijms24010118
APA StyleÉlez, E., Mulet-Margalef, N., Sanso, M., Ruiz-Pace, F., Mancuso, F. M., Comas, R., Ros, J., Argilés, G., Martini, G., Sanz-Garcia, E., Baraibar, I., Salvà, F., Noguerido, A., Cuadra-Urteaga, J. L., Fasani, R., Garcia, A., Jimenez, J., Aguilar, S., Landolfi, S., ... Vivancos, A. (2023). A Comprehensive Biomarker Analysis of Microsatellite Unstable/Mismatch Repair Deficient Colorectal Cancer Cohort Treated with Immunotherapy. International Journal of Molecular Sciences, 24(1), 118. https://doi.org/10.3390/ijms24010118










