Sugar Transporters in Plasmodiophora brassicae: Genome-Wide Identification and Functional Verification
Abstract
1. Introduction
2. Results
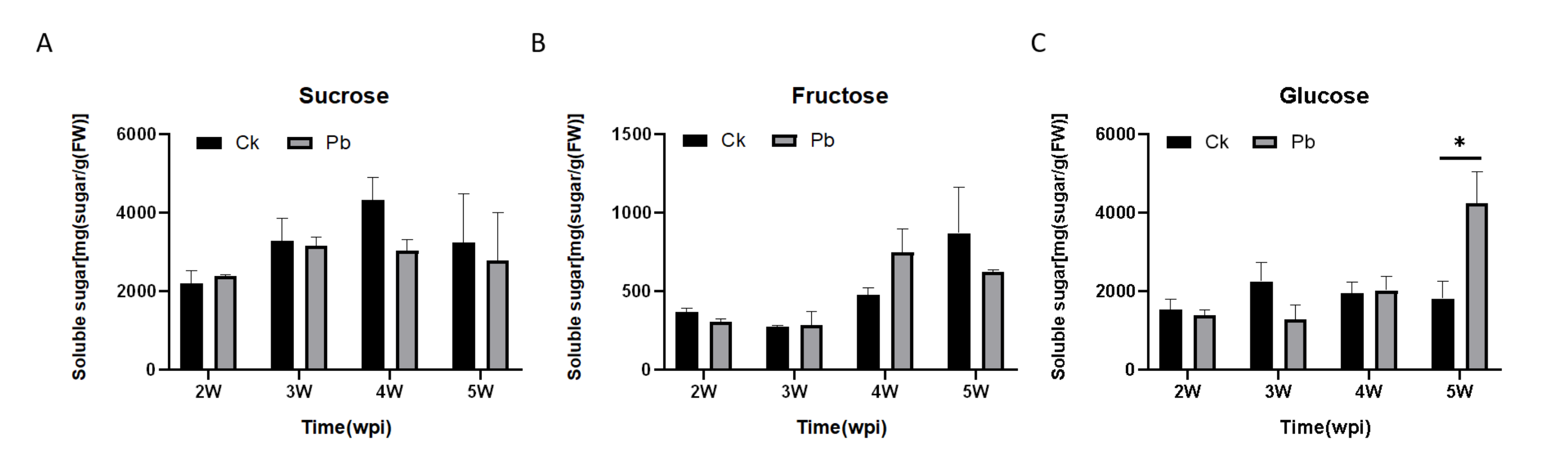
2.1. Soluble Sugar Content in Cabbage Roots after Infection with P. brassicae
2.2. Identification of Putative Sugar Transporters of P. brassicae
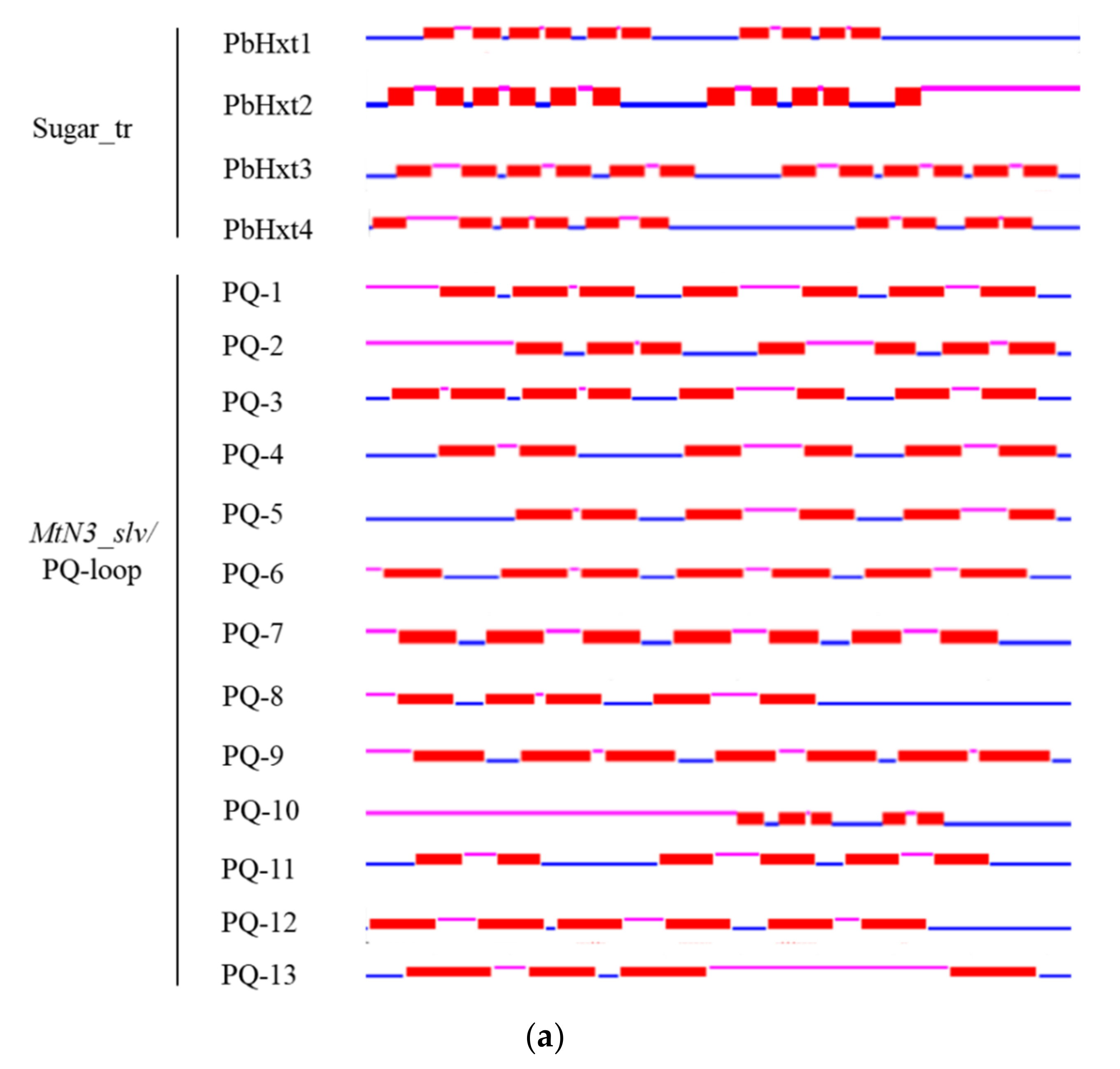
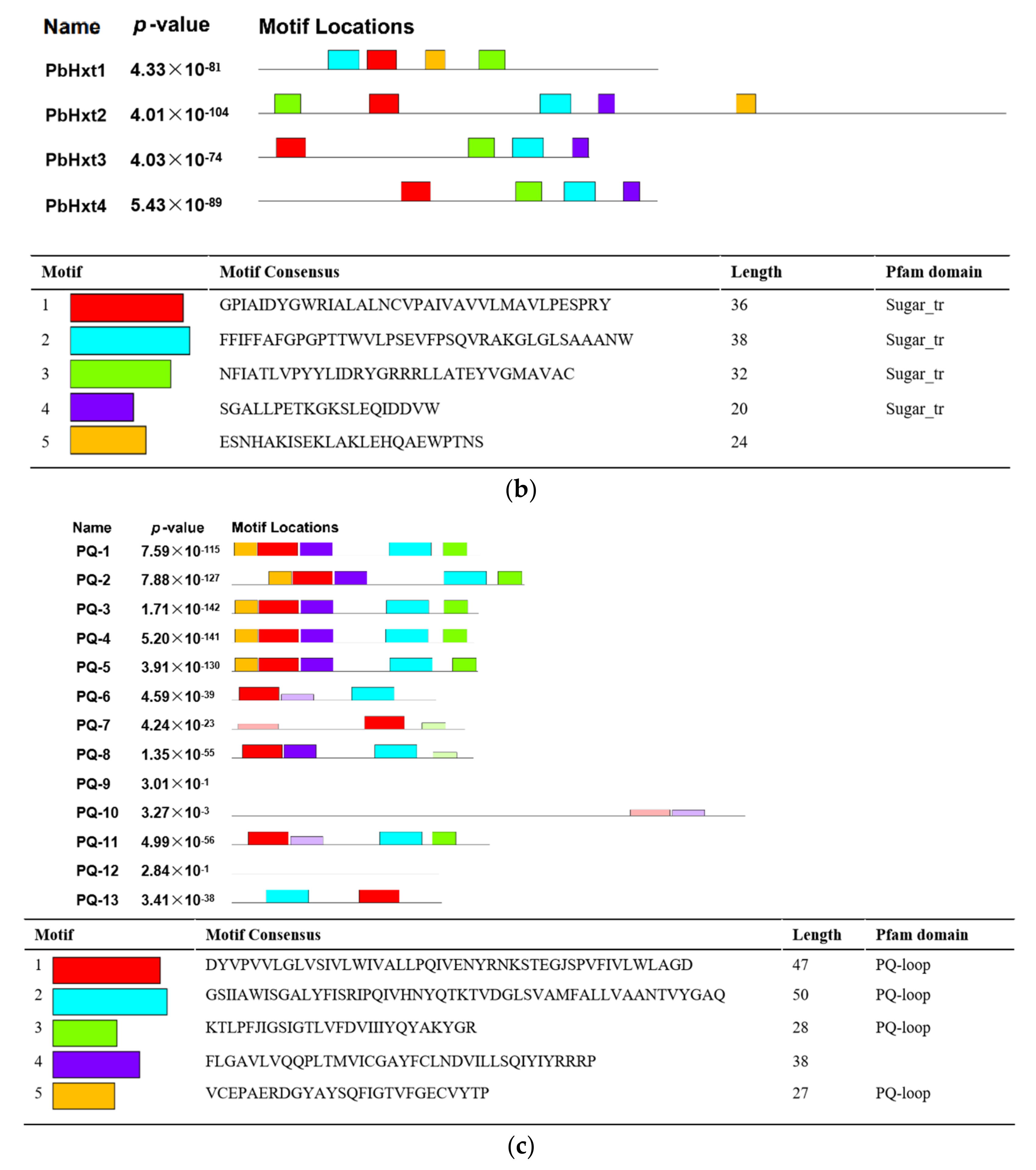
2.3. Transmembrane Helices and Motifs of Sugar Transporters in P. brassicae
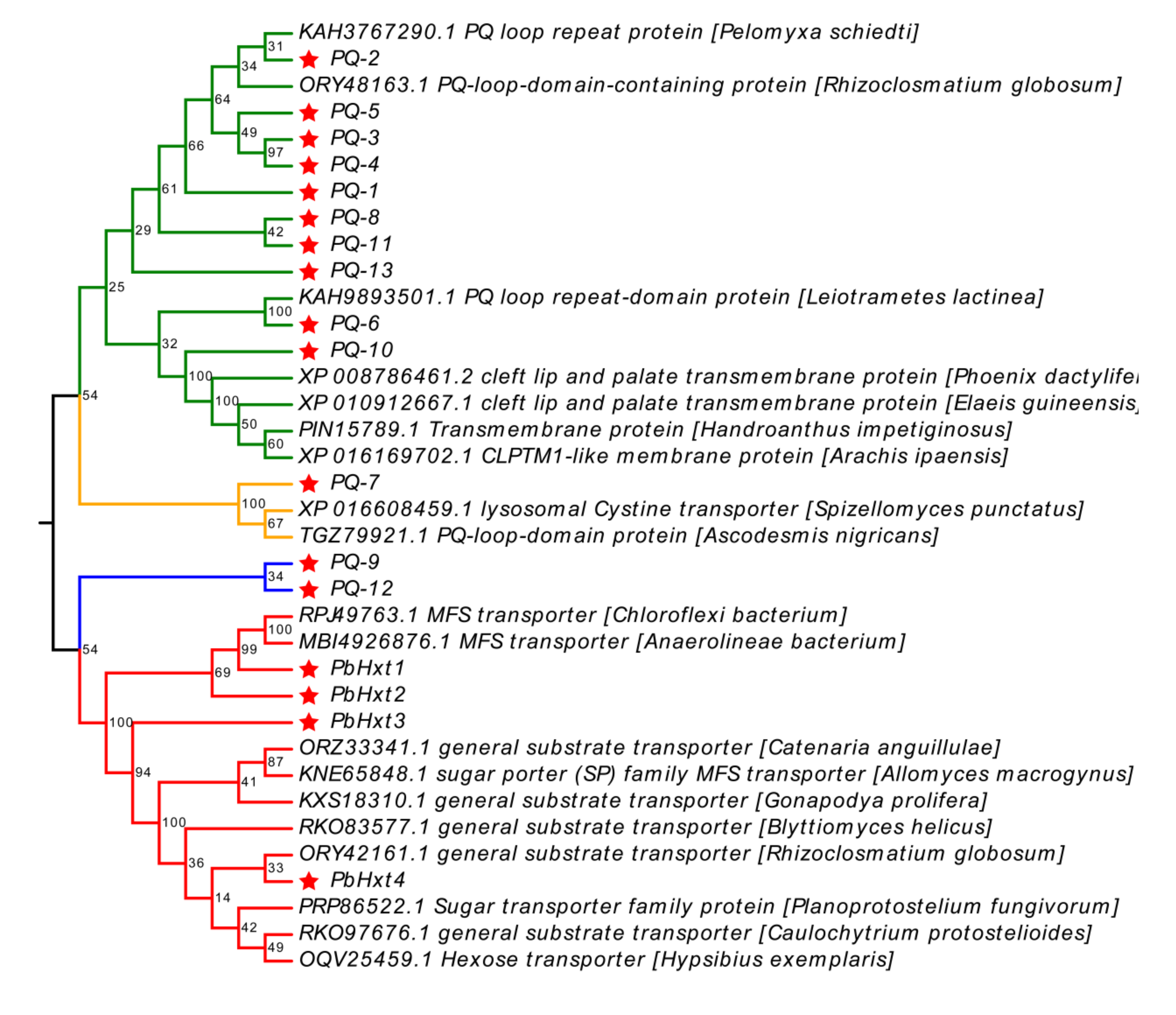
2.4. Phylogenetic Analyses of Hexose Transporters Identified in P. brassicae
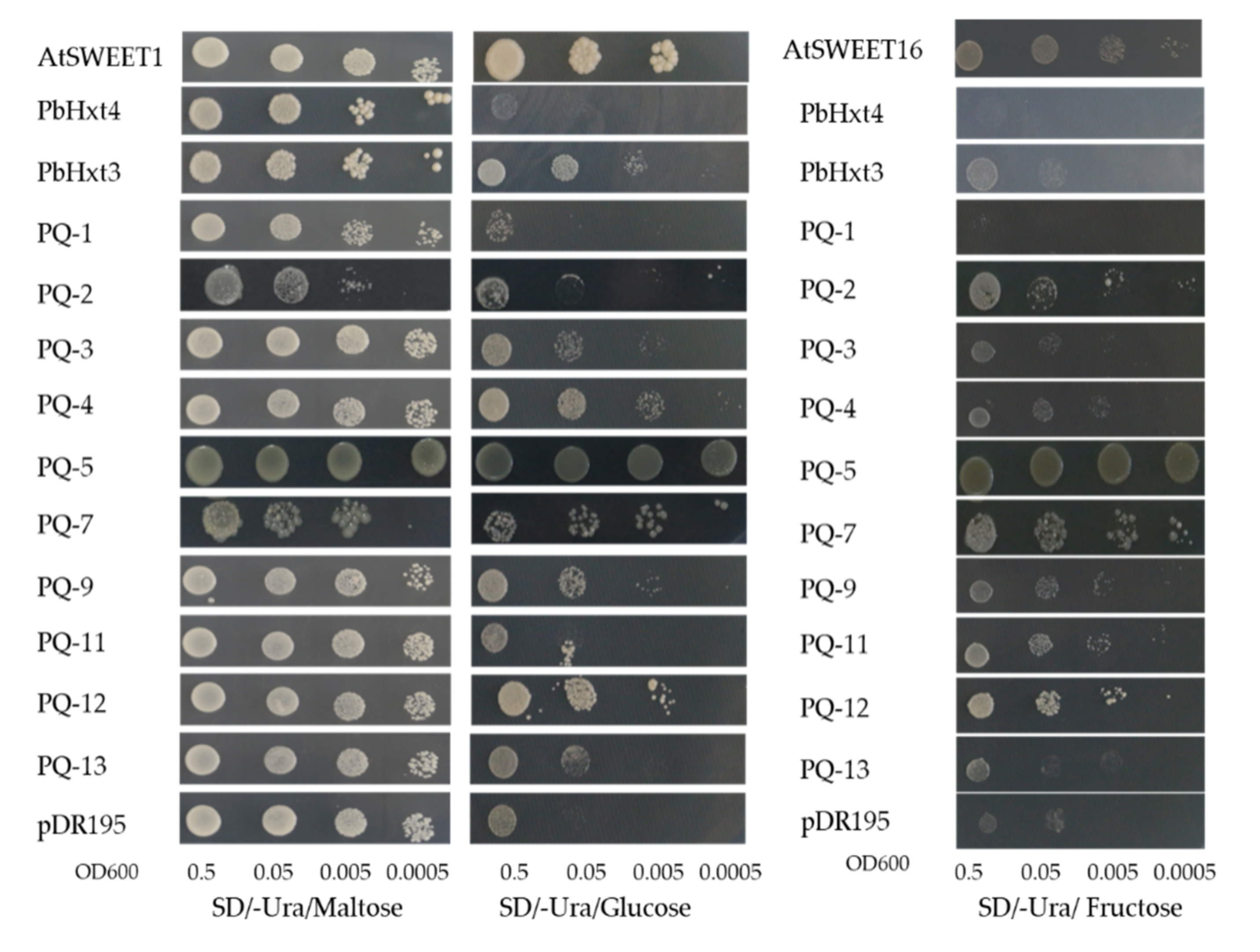
2.5. Hexose Transport Activity of P. brassicae Sugar Transporters in Yeast
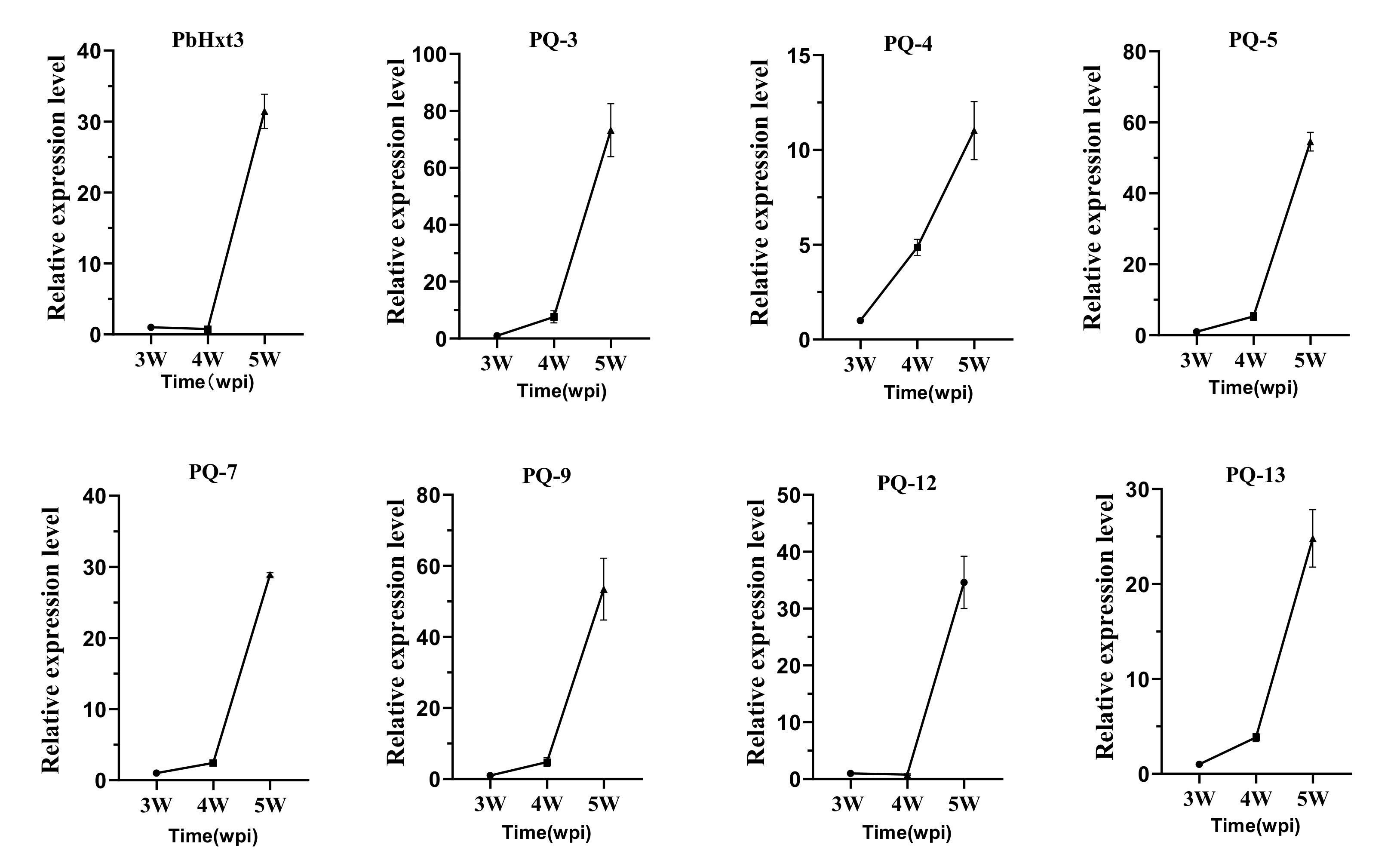
2.6. Expression Patterns of Glucose Transporters in P. brassicae
3. Discussion
4. Materials and Methods
4.1. Plant Material and P. brassicae Inoculation
4.2. Soluble Sugar Extraction and Content Determination
4.3. Total RNA Extraction and Quantitative Real-Time PCR
4.4. Screening Putative Sugar Transporters of P. brassicae
4.5. Transmembrane Helices and Motifs of P. brassicae Putative Sugar Transporters
4.6. Clustering Analysis of P. brassicae Glucose Transporters
4.7. Vector Construction and Functional Complementarity of Hexose-Deficient Yeast EBY.VW4000
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tao, Y.; Cheung, L.S.; Li, S.; Eom, J.S.; Chen, L.Q.; Xu, Y.; Perry, K.; Frommer, W.B.; Feng, L. Structure of a eukaryotic SWEET transporter in a homotrimeric complex. Nature 2015, 527, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q. SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytol. 2014, 201, 1150–1155. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; You, Y.; Fan, H.; Zhu, X.; Wang, Y.; Duan, Y.; Xuan, Y.; Chen, L. The Role of Sugar Transporter Genes during Early Infection by Root-Knot Nematodes. Int. J. Mol. Sci. 2018, 19, 302. [Google Scholar] [CrossRef] [PubMed]
- Walerowski, P.; Gundel, A.; Yahaya, N.; Truman, W.; Sobczak, M.; Olszak, M.; Rolfe, S.; Borisjuk, L.; Malinowski, R. Clubroot Disease Stimulates Early Steps of Phloem Differentiation and Recruits SWEET Sucrose Transporters within Developing Galls. Plant Cell 2018, 30, 3058–3073. [Google Scholar] [CrossRef]
- Rolfe, S.A.; Strelkov, S.E.; Links, M.G.; Clarke, W.E.; Robinson, S.J.; Djavaheri, M.; Malinowski, R.; Haddadi, P.; Kagale, S.; Parkin, I.A.; et al. The compact genome of the plant pathogen Plasmodiophora brassicae is adapted to intracellular interactions with host Brassica spp. BMC Genom. 2016, 17, 272. [Google Scholar] [CrossRef]
- Schwelm, A.; Fogelqvist, J.; Knaust, A.; Jülke, S.; Lilja, T.; Bonilla-Rosso, G.; Karlsson, M.; Shevchenko, A.; Dhandapani, V.; Choi, S.R.; et al. The Plasmodiophora brassicae genome reveals insights in its life cycle and ancestry of chitin synthases. Sci. Rep. 2015, 5, 11153. [Google Scholar] [CrossRef]
- Hwang, S.F.; Strelkov, S.E.; Feng, J.; Gossen, B.D.; Howard, R.J. Plasmodiophora brassicae: A review of an emerging pathogen of the Canadian canola (Brassica napus) crop. Mol. Plant Pathol. 2012, 13, 105–113. [Google Scholar] [CrossRef]
- Neuhauser, S.; Kirchmair, M.; Bulman, S.; Bass, D. Cross-kingdom host shifts of phytomyxid parasites. BMC Evol. Biol. 2014, 14, 33. [Google Scholar] [CrossRef]
- Burki, F.; Kudryavtsev, A.; Matz, M.V.; Aglyamova, G.V.; Bulman, S.; Fiers, M.; Keeling, P.J.; Pawlowski, J. Evolution of Rhizaria: New insights from phylogenomic analysis of uncultivated protists. BMC Evol. Biol. 2010, 10, 377. [Google Scholar] [CrossRef]
- Stjelja, S.; Fogelqvist, J.; Tellgren-Roth, C.; Dixelius, C. The architecture of the Plasmodiophora brassicae nuclear and mitochondrial genomes. Sci. Rep. 2019, 9, 15753. [Google Scholar] [CrossRef]
- Zhai, Z.; Keereetaweep, J.; Liu, H.; Xu, C.; Shanklin, J. The Role of Sugar Signaling in Regulating Plant Fatty Acid Synthesis. Front. Plant Sci. 2021, 12, 643843. [Google Scholar] [CrossRef] [PubMed]
- Bolouri-Moghaddam, M.R.; Le Roy, K.; Xiang, L.; Rolland, F.; Van den Ende, W. Sugar signalling and antioxidant network connections in plant cells. FEBS J. 2010, 277, 2022–2037. [Google Scholar] [CrossRef] [PubMed]
- Manck-Gotzenberger, J.; Requena, N. Arbuscular mycorrhiza Symbiosis Induces a Major Transcriptional Reprogramming of the Potato SWEET Sugar Transporter Family. Front. Plant Sci. 2016, 7, 487. [Google Scholar] [CrossRef] [PubMed]
- Roman-Reyna, V.; Rathjen, J.P. Apoplastic Sugar Extraction and Quantification from Wheat Leaves Infected with Biotrophic Fungi. Methods Mol. Biol. 2017, 1659, 125–134. [Google Scholar] [CrossRef]
- Kanwar, P.; Jha, G. Alterations in plant sugar metabolism: Signatory of pathogen attack. Planta 2019, 249, 305–318. [Google Scholar] [CrossRef]
- Kretschmer, M.; Croll, D.; Kronstad, J.W. Maize susceptibility to Ustilago maydis is influenced by genetic and chemical perturbation of carbohydrate allocation. Mol. Plant Pathol. 2017, 18, 1222–1237. [Google Scholar] [CrossRef]
- Buhtz, A.; Witzel, K.; Strehmel, N.; Ziegler, J.; Abel, S.; Grosch, R. Perturbations in the Primary Metabolism of Tomato and Arabidopsis thaliana Plants Infected with the Soil-Borne Fungus Verticillium dahliae. PLoS ONE 2015, 10, e0138242. [Google Scholar] [CrossRef]
- Kumar, Y.; Zhang, L.; Panigrahi, P.; Dholakia, B.B.; Dewangan, V.; Chavan, S.G.; Kunjir, S.M.; Wu, X.; Li, N.; Rajmohanan, P.R.; et al. Fusarium oxysporum mediates systems metabolic reprogramming of chickpea roots as revealed by a combination of proteomics and metabolomics. Plant Biotechnol. J. 2016, 14, 1589–1603. [Google Scholar] [CrossRef]
- Copley, T.R.; Aliferis, K.A.; Kliebenstein, D.J.; Jabaji, S.H. An integrated RNAseq-(1)H NMR metabolomics approach to understand soybean primary metabolism regulation in response to Rhizoctonia foliar blight disease. BMC Plant Biol. 2017, 17, 84. [Google Scholar] [CrossRef]
- Ghosh, S.; Kanwar, P.; Jha, G. Alterations in rice chloroplast integrity, photosynthesis and metabolome associated with pathogenesis of Rhizoctonia solani. Sci. Rep. 2017, 7, 41610. [Google Scholar] [CrossRef]
- Li, H.; Li, X.; Xuan, Y.; Jiang, J.; Wei, Y.; Piao, Z. Genome Wide Identification and Expression Profiling of SWEET Genes Family Reveals Its Role during Plasmodiophora brassicae-Induced Formation of Clubroot in Brassica rapa. Front. Plant Sci. 2018, 9, 207. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Sun, T.; Zhao, S.; Pan, L.; Liu, H.; Tian, X. Physiological change alters endophytic bacterial community in clubroot of tumorous stem mustard infected by Plasmodiophora brassicae. BMC Microbiol. 2020, 20, 244. [Google Scholar] [CrossRef] [PubMed]
- Bezrutczyk, M.; Yang, J.; Eom, J.S.; Prior, M.; Sosso, D.; Hartwig, T.; Szurek, B.; Oliva, R.; Vera-Cruz, C.; White, F.F.; et al. Sugar flux and signaling in plant-microbe interactions. Plant J. 2018, 93, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q.; Cheung, L.S.; Feng, L.; Tanner, W.; Frommer, W.B. Transport of sugars. Annu. Rev. Biochem. 2015, 84, 865–894. [Google Scholar] [CrossRef] [PubMed]
- Kühn, C.; Grof, C.P. Sucrose transporters of higher plants. Curr. Opin. Plant Biol. 2010, 13, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Barbi, F.; Vallon, L.; Guerrero-Galán, C.; Zimmermann, S.D.; Melayah, D.; Abrouk, D.; Doré, J.; Lemaire, M.; Fraissinet-Tachet, L.; Luis, P.; et al. Datamining and functional environmental genomics reassess the phylogenetics and functional diversity of fungal monosaccharide transporters. Appl. Microbiol. Biotechnol. 2021, 105, 647–660. [Google Scholar] [CrossRef] [PubMed]
- Yan, N. Structural Biology of the Major Facilitator Superfamily Transporters. Annu. Rev. Biophys. 2015, 44, 257–283. [Google Scholar] [CrossRef] [PubMed]
- Misra, V.A.; Wafula, E.K.; Wang, Y.; dePamphilis, C.W.; Timko, M.P. Genome-wide identification of MST, SUT and SWEET family sugar transporters in root parasitic angiosperms and analysis of their expression during host parasitism. BMC Plant Biol. 2019, 19, 196. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q.; Qu, X.Q.; Hou, B.H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef]
- Wright, E.M. Glucose transport families SLC5 and SLC50. Mol. Asp. Med. 2013, 34, 183–196. [Google Scholar] [CrossRef]
- Havukainen, S.; Pujol-Giménez, J.; Valkonen, M.; Westerholm-Parvinen, A.; Hediger, M.A.; Landowski, C.P. Electrophysiological characterization of a diverse group of sugar transporters from Trichoderma reesei. Sci. Rep. 2021, 11, 14678. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shu, Y.; Gu, C.; Fan, Y. The novel sugar transporter SLC50A1 as a potential serum-based diagnostic and prognostic biomarker for breast cancer. Cancer Manag. Res. 2019, 11, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q.; Hou, B.H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.Q.; Guo, W.J.; Kim, J.G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Bisson, L.F.; Fan, Q.; Walker, G.A. Sugar and Glycerol Transport in Saccharomyces cerevisiae. Adv. Exp. Med. Biol. 2016, 892, 125–168. [Google Scholar] [CrossRef]
- Nijland, J.G.; Vos, E.; Shin, H.Y.; de Waal, P.P.; Klaassen, P.; Driessen, A.J. Improving pentose fermentation by preventing ubiquitination of hexose transporters in Saccharomyces cerevisiae. Biotechnol. Biofuels 2016, 9, 158. [Google Scholar] [CrossRef] [PubMed]
- Scharff-Poulsen, P.; Moriya, H.; Johnston, M. Genetic Analysis of Signal Generation by the Rgt2 Glucose Sensor of Saccharomyces cerevisiae. G3 Genes Genomes Genet. 2018, 8, 2685–2696. [Google Scholar] [CrossRef]
- Cano, J.M.; Berrocal-Lobo, M.; Domínguez-Núñez, J.A. Growth of Amanita caesarea in the presence of Pseudomonas fluorescens and Bacillus cereus. Fungal Biol. 2017, 121, 825–833. [Google Scholar] [CrossRef]
- Ceccaroli, P.; Saltarelli, R.; Polidori, E.; Barbieri, E.; Guescini, M.; Ciacci, C.; Stocchi, V. Sugar transporters in the black truffle Tuber melanosporum: From gene prediction to functional characterization. Fungal Genet. Biol. 2015, 81, 52–61. [Google Scholar] [CrossRef]
- López, M.F.; Dietz, S.; Grunze, N.; Bloschies, J.; Weiß, M.; Nehls, U. The sugar porter gene family of Laccaria bicolor: Function in ectomycorrhizal symbiosis and soil-growing hyphae. New Phytol. 2010, 180, 365–378. [Google Scholar] [CrossRef]
- Sedaghatkish, A.; Gossen, B.D.; Yu, F.; Torkamaneh, D.; Mcdonald, M.R. Whole-genome DNA similarity and population structure of Plasmodiophora brassicae strains from Canada. BMC Genom. 2019, 20, 1–14. [Google Scholar] [CrossRef]
- Bi, K.; He, Z.; Gao, Z.; Zhao, Y.; Fu, Y.; Cheng, J.; Xie, J.; Jiang, D.; Chen, T. Integrated omics study of lipid droplets from Plasmodiophora brassicae. Sci. Rep. 2016, 6, 36965. [Google Scholar] [CrossRef] [PubMed]
- Fang, T.; Peng, Y.; Rao, Y.; Li, S.; Zeng, L. Genome-Wide Identification and Expression Analysis of Sugar Transporter (ST) Gene Family in Longan (Dimocarpus longan L.). Plants 2020, 9, 342. [Google Scholar] [CrossRef]
- Liu, L.; Qin, L.; Zhou, Z.; Hendriks, W.G.H.M.; Wei, Y.D. Refining the life cycle of Plasmodiophora brassicae. Phytopathology 2020, 110, 1704–1712. [Google Scholar] [CrossRef] [PubMed]
- Siemens, J.; González, M.C.; Wolf, S.; Hofmann, C.; Greiner, S.; Du, Y.; Rausch, T.; Roitsch, T.; Ludwig-Müller, J. Extracellular invertase is involved in the regulation of clubroot disease in Arabidopsis thaliana. Mol. Plant Pathol. 2011, 12, 247–262. [Google Scholar] [CrossRef] [PubMed]
- Proels, R.K.; Hückelhoven, R. Cell-wall invertases, key enzymes in the modulation of plant metabolism during defence responses. Mol. Plant Pathol. 2014, 15, 858–864. [Google Scholar] [CrossRef] [PubMed]
- Yahaya, N. Metabolic Interactions between Plasmodiophora brassicae and Arabidopsis Thaliana Plant. Ph.D. Thesis, University of Sheffield, Sheffield, UK, 2016. [Google Scholar]
- Vatsa-Portugal, P.; Walker, A.S.; Jacquens, L.; Clément, C.; Barka, E.A.; Vaillant-Gaveau, N. Inflorescences vs leaves: A distinct modulation of carbon metabolism process during Botrytis infection. Physiol. Plant. 2015, 154, 162–177. [Google Scholar] [CrossRef]
- Hayes, M.A.; Feechan, A.; Dry, I.B. Involvement of abscisic acid in the coordinated regulation of a stress-inducible hexose transporter (VvHT5) and a cell wall invertase in grapevine in response to biotrophic fungal infection. Plant Physiol. 2010, 153, 211–221. [Google Scholar] [CrossRef]
- Yuan, Q.; Yan, Y.; Sohail, M.A.; Liu, H.; Huang, J.; Hsiang, T.; Zheng, L. A Novel Hexose Transporter ChHxt6 Is Required for Hexose Uptake and Virulence in Colletotrichum higginsianum. Int. J. Mol. Sci. 2021, 22, 5963. [Google Scholar] [CrossRef]
- Ait Lahmidi, N.; Courty, P.E.; Brulé, D.; Chatagnier, O.; Arnould, C.; Doidy, J.; Berta, G.; Lingua, G.; Wipf, D.; Bonneau, L. Sugar exchanges in arbuscular mycorrhiza: RiMST5 and RiMST6, two novel Rhizophagus irregularis monosaccharide transporters, are involved in both sugar uptake from the soil and from the plant partner. Plant Physiol. Biochem. 2016, 107, 354–363. [Google Scholar] [CrossRef]
- Liu, J.; Liu, M.; Tan, L.; Huai, B.; Ma, X.; Pan, Q.; Zheng, P.; Wen, Y.; Zhang, Q.; Zhao, Q.; et al. AtSTP8, an endoplasmic reticulum-localised monosaccharide transporter from Arabidopsis, is recruited to the extrahaustorial membrane during powdery mildew infection. New Phytol. 2021, 230, 2404–2419. [Google Scholar] [CrossRef]
- Kayikci, Ö.; Nielsen, J. Glucose repression in Saccharomyces cerevisiae. FEMS Yeast Res. 2015, 15, fov068. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.S.; Shlykov, M.A.; Castillo, R.; Sun, E.I.; Saier, M.H., Jr. The major facilitator superfamily (MFS) revisited. FEBS J. 2012, 279, 2022–2035. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.C.; Zhao, Y.; Heng, J.; Jiang, D. Energy coupling mechanisms of MFS transporters. Protein Sci. 2015, 24, 1560–1579. [Google Scholar] [CrossRef]
- Chaudhary, N.; Kumari, I.; Sandhu, P.; Ahmed, M.; Akhter, Y. Proteome scale census of major facilitator superfamily transporters in Trichoderma reesei using protein sequence and structure based classification enhanced ranking. Gene 2016, 585, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Xuan, Y.H.; Hu, Y.B.; Chen, L.Q.; Sosso, D.; Ducat, D.C.; Hou, B.H.; Frommer, W.B. Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc. Natl. Acad. Sci. USA 2013, 110, E3685–E3694. [Google Scholar] [CrossRef]
- Patil, G.; Valliyodan, B.; Deshmukh, R.; Prince, S.; Nicander, B.; Zhao, M.; Sonah, H.; Song, L.; Lin, L.; Chaudhary, J.; et al. Soybean (Glycine max) SWEET gene family: Insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genom. 2015, 16, 520. [Google Scholar] [CrossRef]
- Jia, B.; Zhu, X.F.; Pu, Z.J.; Duan, Y.X.; Hao, L.J.; Zhang, J.; Chen, L.Q.; Jeon, C.O.; Xuan, Y.H. Integrative View of the Diversity and Evolution of SWEET and SemiSWEET Sugar Transporters. Front. Plant Sci. 2017, 8, 2178. [Google Scholar] [CrossRef]
- Wang, J.; Yan, C.; Li, Y.; Hirata, K.; Yamamoto, M.; Yan, N.; Hu, Q. Crystal structure of a bacterial homologue of SWEET transporters. Cell Res. 2014, 24, 1486–1489. [Google Scholar] [CrossRef]
- Hu, Y.B.; Sosso, D.; Qu, X.Q.; Chen, L.Q.; Ma, L.; Chermak, D.; Zhang, D.C.; Frommer, W.B. Phylogenetic evidence for a fusion of archaeal and bacterial SemiSWEETs to form eukaryotic SWEETs and identification of SWEET hexose transporters in the amphibian chytrid pathogen Batrachochytrium dendrobatidis. FASEB J. 2016, 30, 3644–3654. [Google Scholar] [CrossRef]
- Liu, N.; Wang, Q.; He, C.; An, B. CgMFS1, a Major Facilitator Superfamily Transporter, Is Required for Sugar Transport, Oxidative Stress Resistance, and Pathogenicity of Colletotrichum gloeosporioides from Hevea brasiliensis. Curr. Issues Mol. Biol. 2021, 43, 1548–1557. [Google Scholar] [CrossRef]
- Kryukov, A.A.; Gorbunova, A.O.; Kudriashova, T.R.; Yakhin, O.I.; Lubyanov, A.A.; Malikov, U.M.; Shishova, M.F.; Kozhemyakov, A.P.; Yurkov, A.P. Sugar transporters of the SWEET family and their role in arbuscular mycorrhiza. Vavilov J. Genet. Breed. 2021, 25, 754–760. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shang, L.; Zhan, Y.; Lin, M.; Liu, Z.; Yan, Y. Genome-Wide Analysis of Sugar Transporters Identifies the gtsA Gene for Glucose Transportation in Pseudomonas stutzeri A1501. Microorganisms 2020, 8, 592. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Piron, M.C.; Meyer, S.; Merdinoglu, D.; Bertsch, C.; Mestre, P. The SWEET family of sugar transporters in grapevine: VvSWEET4 is involved in the interaction with Botrytis cinerea. J. Exp. Bot. 2014, 65, 6589–6601. [Google Scholar] [CrossRef]
- Gravot, A.; Grillet, L.; Wagner, G.; Jubault, M.; Lariagon, C.; Baron, C.; Deleu, C.; Delourme, R.; Bouchereau, A.; Manzanares-Dauleux, M.J. Genetic and physiological analysis of the relationship between partial resistance to clubroot and tolerance to trehalose in Arabidopsis thaliana. New Phytol. 2011, 191, 1083–1094. [Google Scholar] [CrossRef]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2^(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013, 3, 71–85. [Google Scholar]
- Griffiths, A.G.; Barrett, B.A.; Simon, D.; Khan, A.K.; Bickerstaff, P.; Anderson, C.B.; Franzmayr, B.K.; Hancock, K.R.; Jones, C.S. An integrated genetic linkage map for white clover (Trifolium repens L.) with alignment to Medicago. BMC Genom. 2013, 14, 388. [Google Scholar] [CrossRef] [PubMed]
- Solis-Escalante, D.; van den Broek, M.; Kuijpers, N.G.; Pronk, J.T.; Boles, E.; Daran, J.M.; Daran-Lapujade, P. The genome sequence of the popular hexose-transport-deficient Saccharomyces cerevisiae strain EBY.VW4000 reveals LoxP/Cre-induced translocations and gene loss. FEMS Yeast Res. 2015, 15, fou004. [Google Scholar] [CrossRef][Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
| Gene Name | GenBank ID | Bp 1 | Aa 2 | MW 3 (kDa) | pI 4 | Domain 5 | Domain Position | TMH 6 | Subfamily |
|---|---|---|---|---|---|---|---|---|---|
| PbHxt1 | SPQ96265.1 | 1515 | 504 | 54.15 | 6.71 | Sugar_tr | 89–429 | 10 | MFS superfamily |
| PbHxt2 | SPQ93932.1 | 5379 | 1792 | 159.50 | 5.66 | Sugar_tr | 34–495 | 11 | |
| PbHxt3 | SPQ95410.1 | 1407 | 468 | 50.09 | 6.60 | Sugar_tr | 31–467 | 12 | |
| PbHxt4 | SPQ99469.1 | 1479 | 492 | 52.81 | 5.67 | Sugar_tr | 10–470 | 10 | |
| PQ-1 | SPQ94201.1 | 885 | 294 | 31.78 | 9.57 | PQ-loop | 35–93, 187–240 | 7 | SWEET superfamily |
| PQ-2 | SPQ94044.1 | 1041 | 346 | 38.05 | 9.23 | PQ-loop | 76–134, 252–308 | 7 | |
| PQ-3 | SPQ97464.1 | 879 | 292 | 31.43 | 8.36 | PQ-loop | 37–88, 184–237 | 8 | |
| PQ-4 | SPQ97733.1 | 864 | 287 | 31.97 | 8.50 | PQ-loop | 38–92, 182–238 | 6 | |
| PQ-5 | SPR00585.1 | 876 | 291 | 32.71 | 8.76 | PQ-loop | 37–93, 188–241 | 6 | |
| PQ-6 | CEO98791.1 | 729 | 242 | 26.62 | 8.60 | PQ-loop | 14–71, 144–200 | 7 | |
| PQ-7 | CEP03599.1 | 831 | 276 | 30.04 | 9.57 | PQ-loop | 12–63, 163–210 | 7 | |
| PQ-8 | SPQ93336.1 | 861 | 286 | 30.65 | 9.47 | PQ-loop | 17–73 | 5 | |
| PQ-9 | SPQ94026.1 | 690 | 229 | 25.31 | 9.62 | MtN3_slv | 19–100, 142–221 | 7 | |
| PQ-10 | SPQ96444.1 | 1824 | 607 | 69.30 | 6.8 | PQ-loop | 472–506 | 5 | |
| PQ-11 | SPQ97201.1 | 918 | 305 | 32.97 | 9.59 | PQ-loop | 24–80 | 6 | |
| PQ-12 | SPQ98447.1 | 736 | 245 | 26.01 | 8.81 | MtN3_slv | 139–219 | 6 | |
| PQ-13 | CEP03554.1 | 747 | 248 | 26.70 | 9.97 | PQ-loop | 42–99, 164–209 | 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, L.; Li, X.; Zhan, Z.; Piao, Z. Sugar Transporters in Plasmodiophora brassicae: Genome-Wide Identification and Functional Verification. Int. J. Mol. Sci. 2022, 23, 5264. https://doi.org/10.3390/ijms23095264
Kong L, Li X, Zhan Z, Piao Z. Sugar Transporters in Plasmodiophora brassicae: Genome-Wide Identification and Functional Verification. International Journal of Molecular Sciences. 2022; 23(9):5264. https://doi.org/10.3390/ijms23095264
Chicago/Turabian StyleKong, Liyan, Xiaonan Li, Zongxiang Zhan, and Zhongyun Piao. 2022. "Sugar Transporters in Plasmodiophora brassicae: Genome-Wide Identification and Functional Verification" International Journal of Molecular Sciences 23, no. 9: 5264. https://doi.org/10.3390/ijms23095264
APA StyleKong, L., Li, X., Zhan, Z., & Piao, Z. (2022). Sugar Transporters in Plasmodiophora brassicae: Genome-Wide Identification and Functional Verification. International Journal of Molecular Sciences, 23(9), 5264. https://doi.org/10.3390/ijms23095264






