Tissue Transglutaminase Knock-Out Preadipocytes and Beige Cells of Epididymal Fat Origin Possess Decreased Mitochondrial Functions Required for Thermogenesis
Abstract
1. Introduction
2. Results
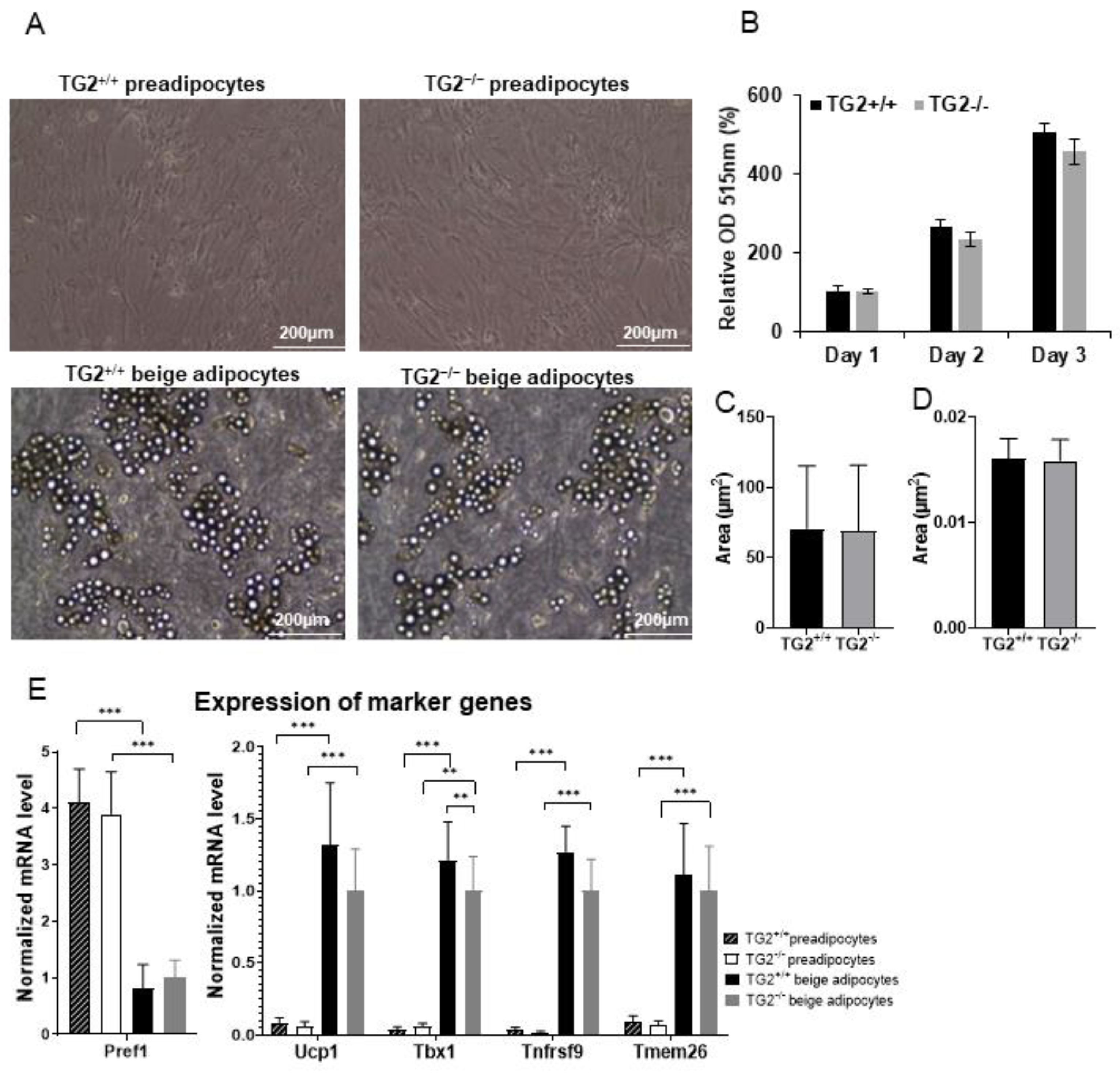
2.1. Differentiation of TG2−/− Preadipocytes Results in Altered Beige Adipocytes Phenotypically Similar to TG2+/+ Cells
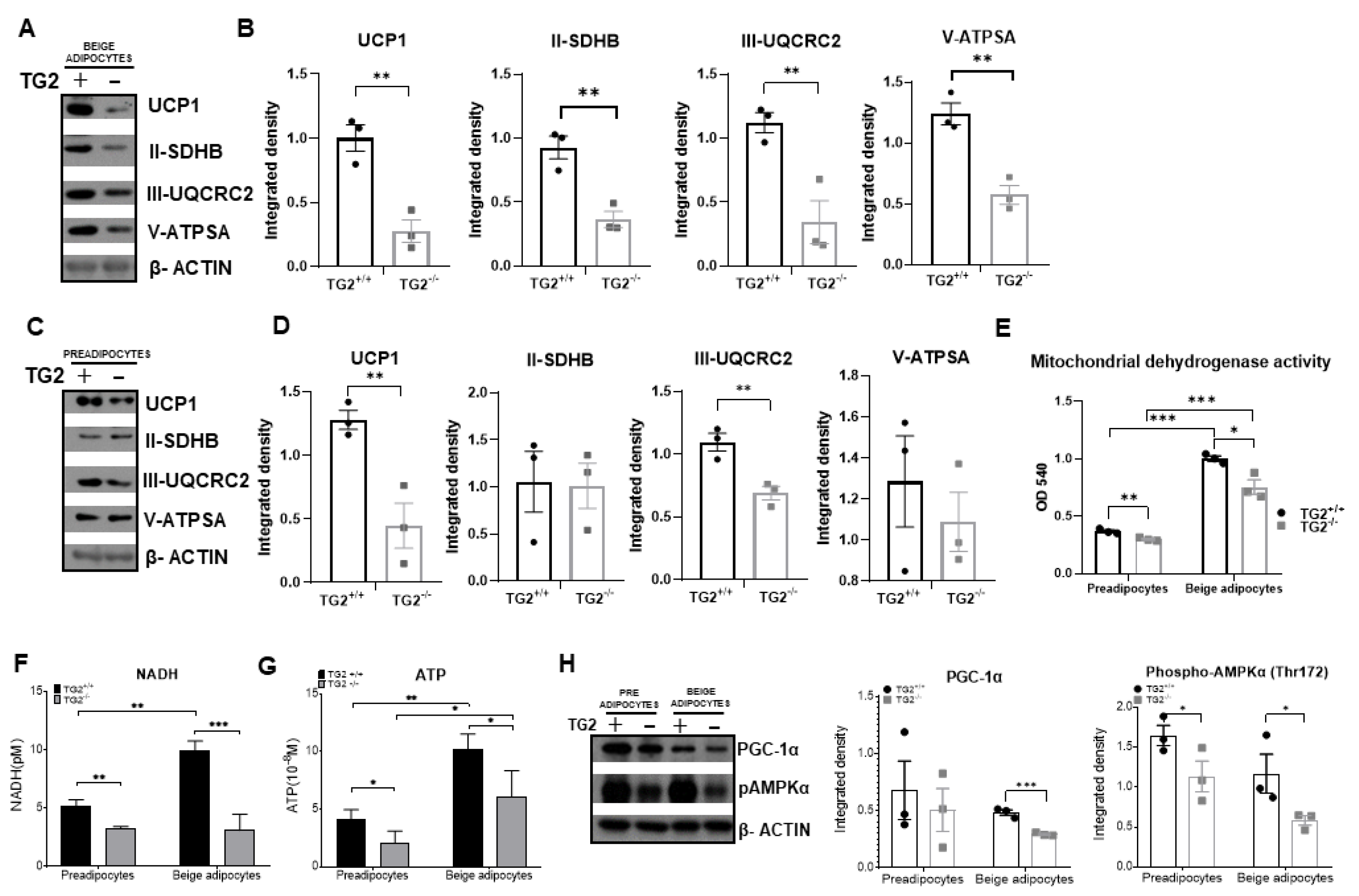
2.2. Lack of TG2 Alters the Level of Mitochondrial Membrane Proteins and Energy Production
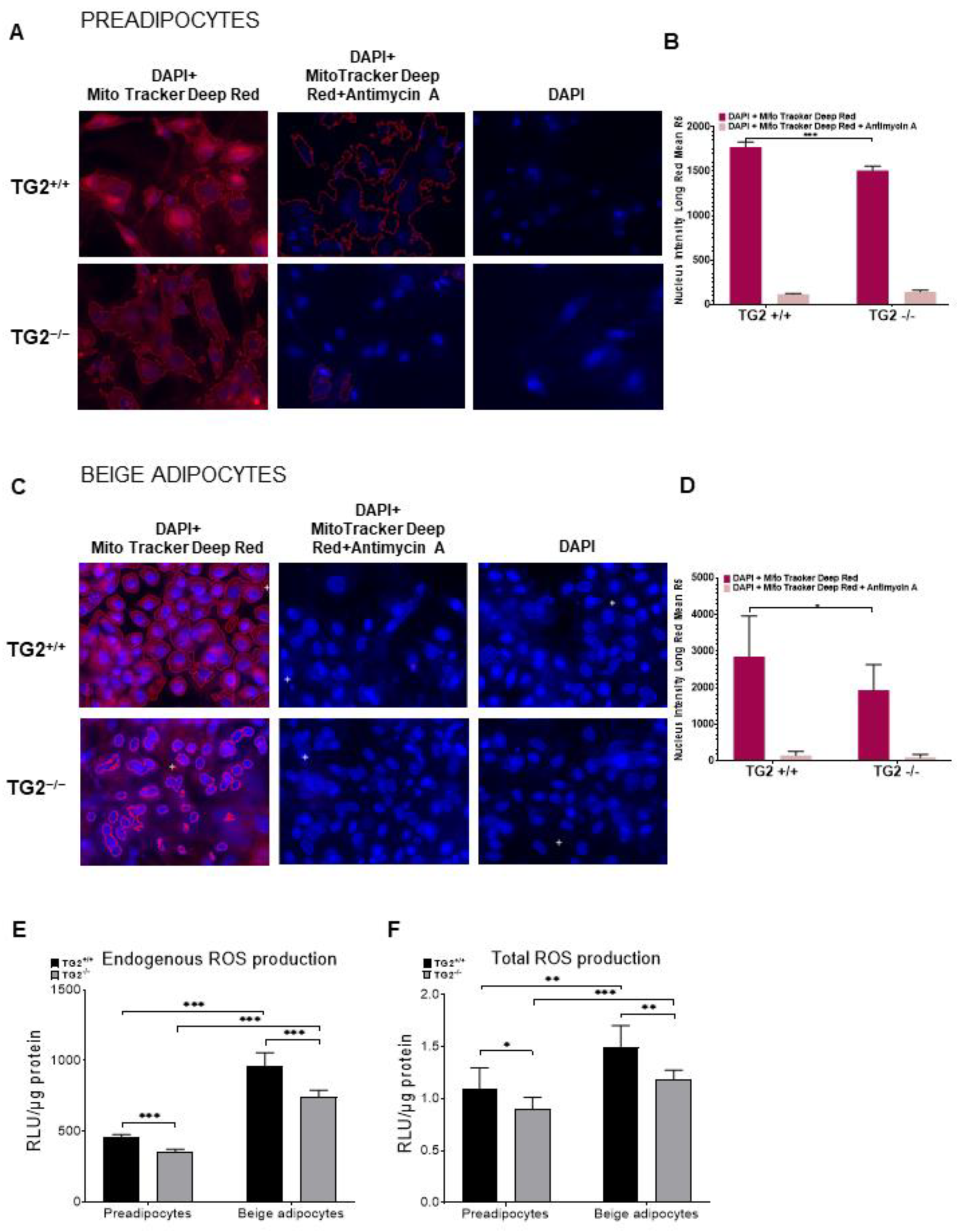
2.3. TG2−/− Preadipocytes and Beige Cells Generate Significantly Lower Membrane Potential in Mitochondria as Compared with TG2+/+ Cells
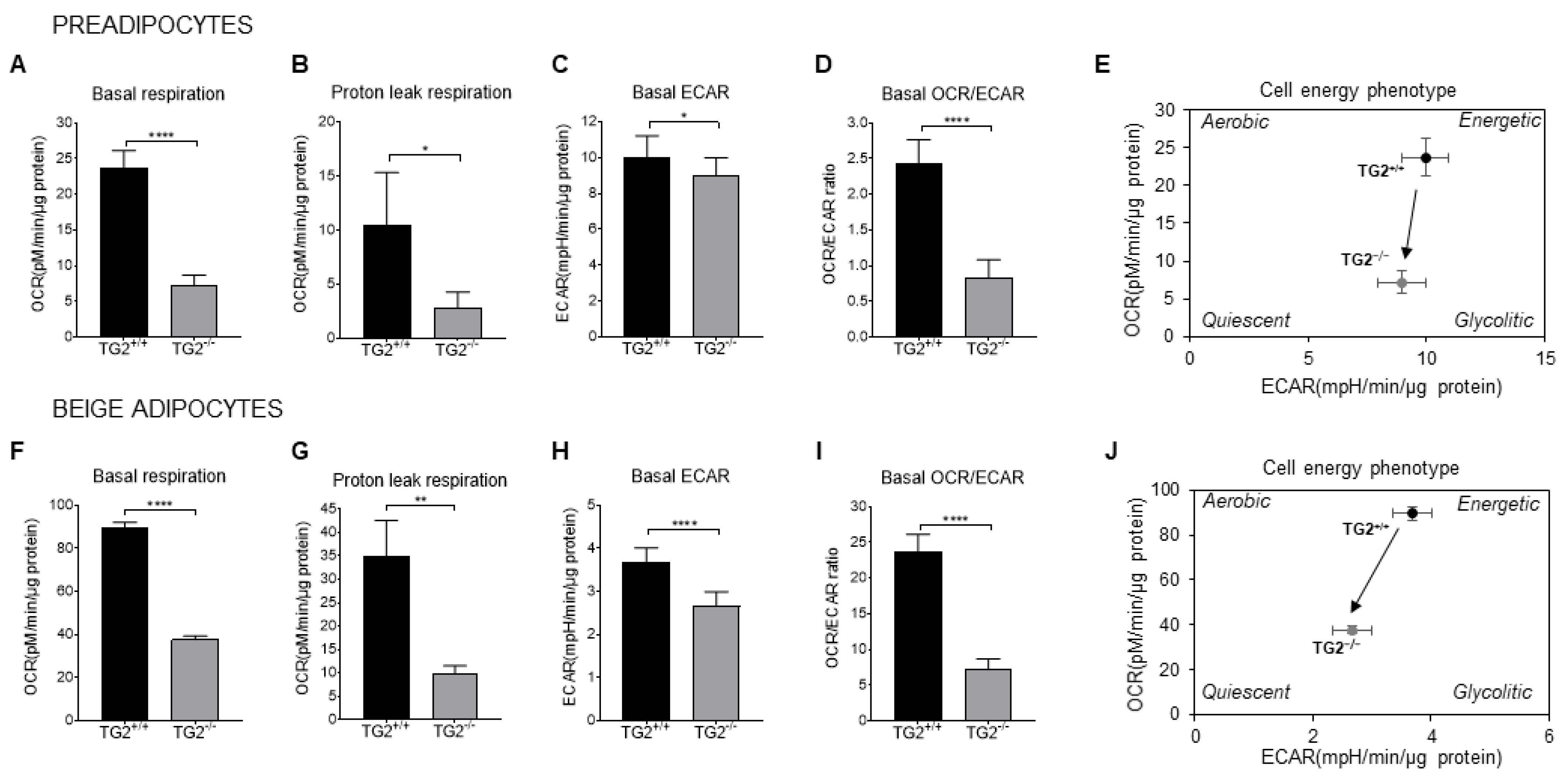
2.4. TG2−/− Preadipocytes and Beige Cells Are Metabolically Hypometabolic as Compared with TG2+/+
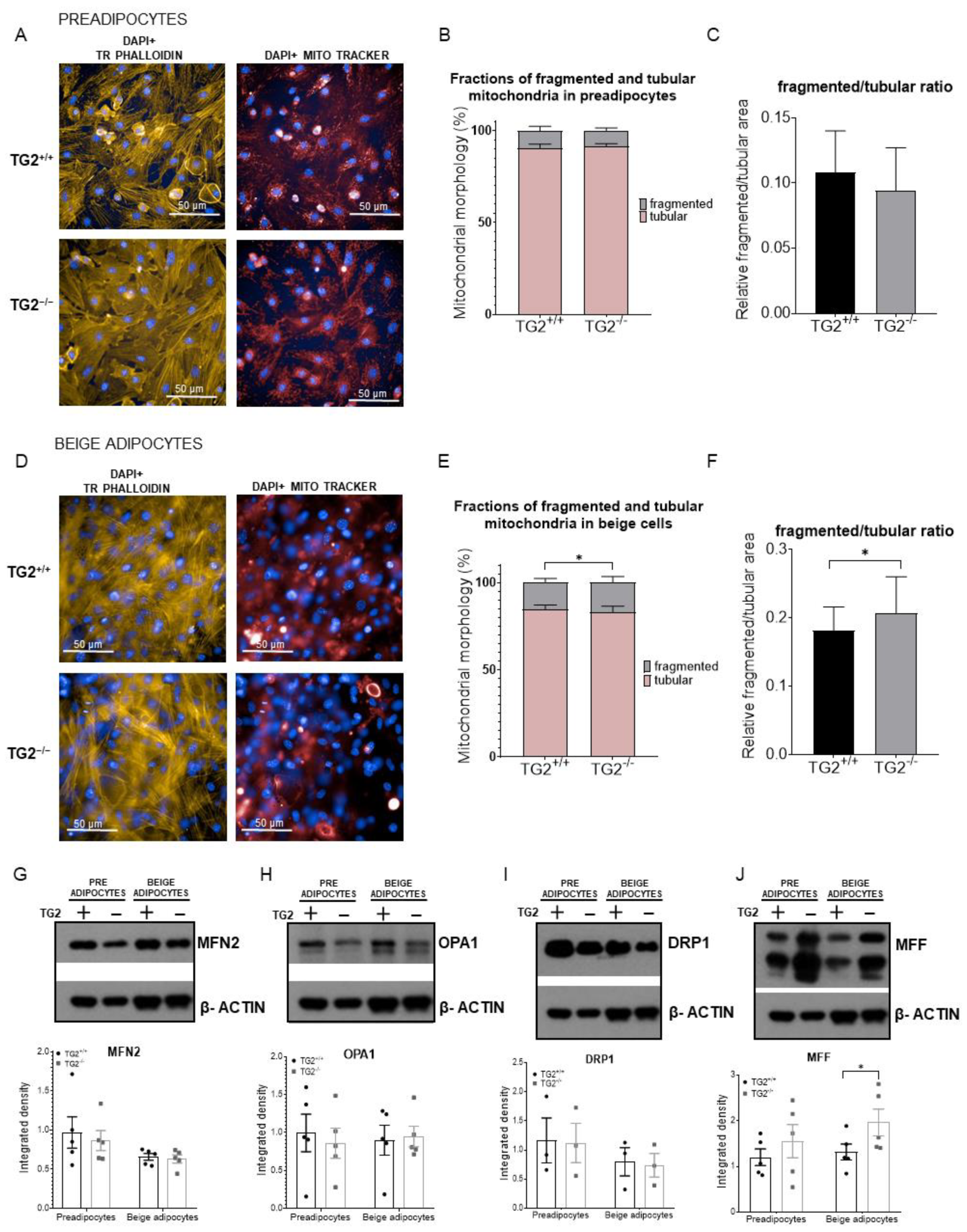
2.5. TG2−/− Beige Adipocytes Have More Fragmented Mitochondria as Compared with TG2+/+
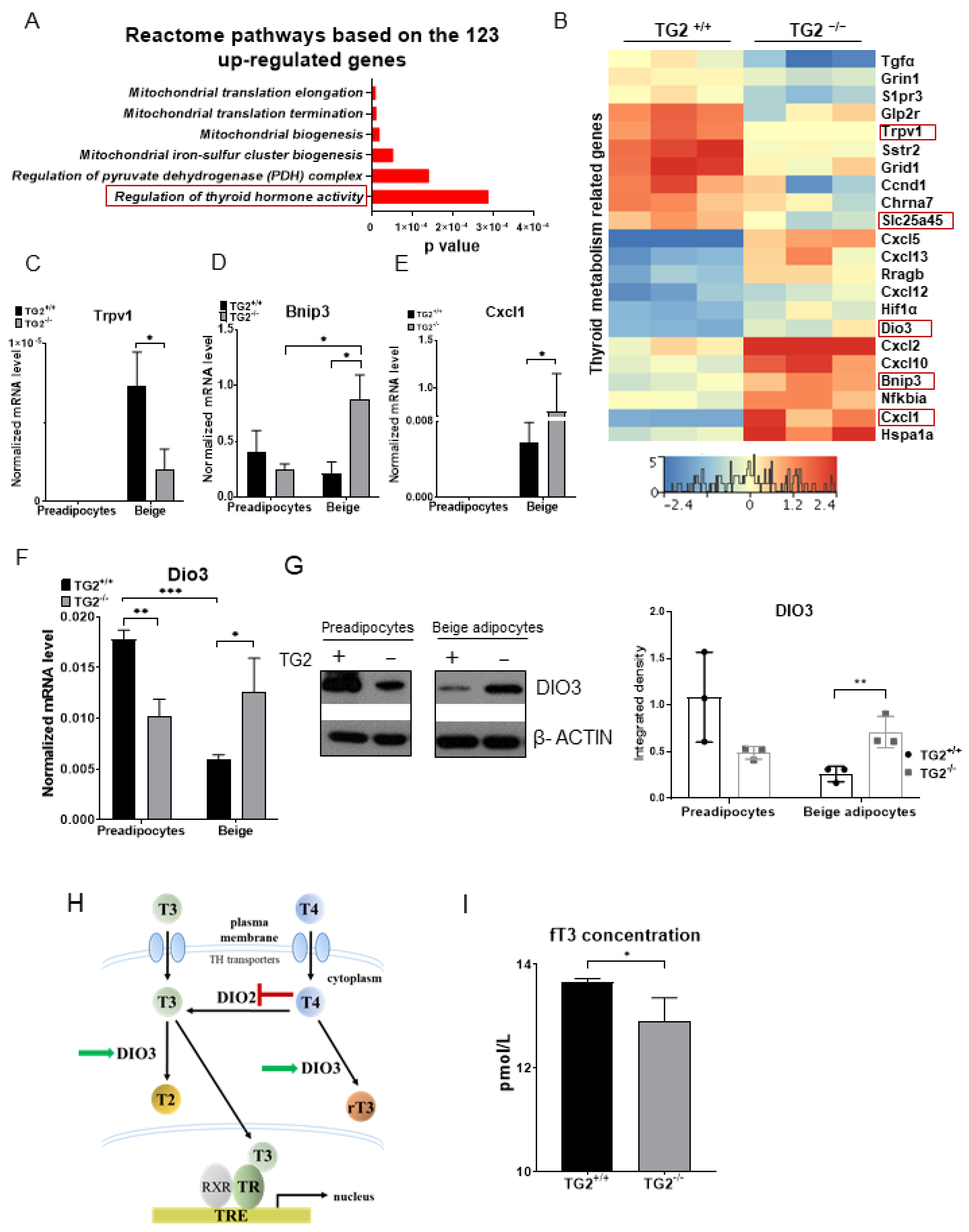
2.6. TG2−/− Beige Adipocytes Can Attenuate the Effect of Free Triiodothyronine
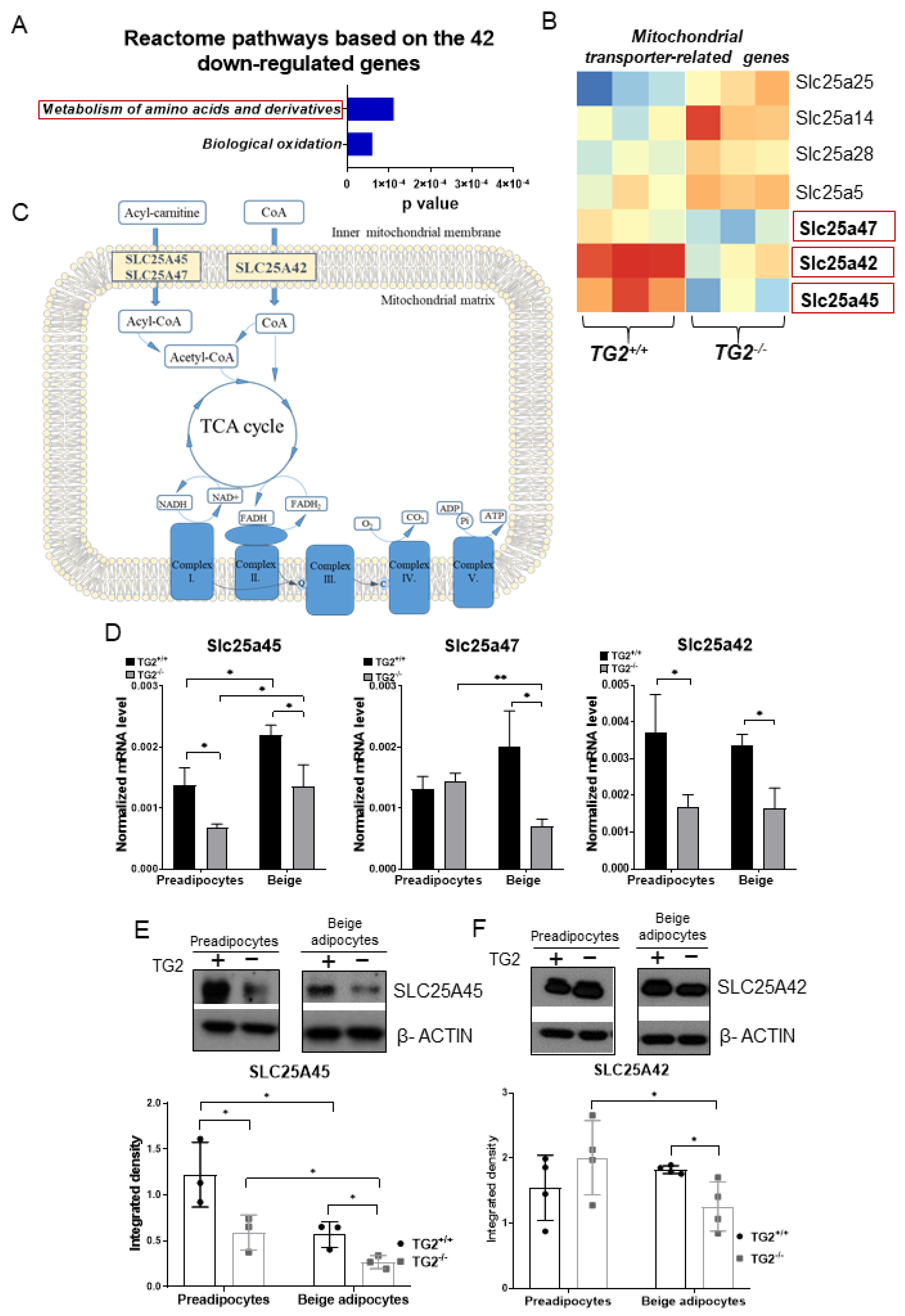
2.7. Expression of Transporters of Acylcarnitine, CoA, and Certain Amino Acids into the Mitochondria Is Lower in TG2−/− Cells as Compared with the TG2+/+ Controls
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Mice, Treatments, and Obtained Samples
4.3. Gene Expression Studies
4.4. Western Blots
4.5. Determination of Mitochondrial Dehydrogenase Activity
4.6. In Vitro Cell Proliferation Assays
4.7. Measurement of Oxygen Consumption and Extracellular Acidification Rate
4.8. Measurement of NADH and ATP Levels
4.9. Immunocytochemistry, Image Acquisition, and Analysis
4.10. Detection of Reactive Oxygen Species Production
4.11. Laser Scanning Cytometry
4.12. RNA-Sequencing
4.13. Measurement of Free Triiodothyronine
4.14. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eckert, R.L.; Kaartinen, M.T.; Nurminskaya, M.; Belkin, A.M.; Colak, G.; Johnson, G.V.; Mehta, K. Transglutaminase regulation of cell function. Physiol. Rev. 2014, 94, 383–417. [Google Scholar] [CrossRef] [PubMed]
- Fesus, L.; Piacentini, M. Transglutaminase 2: An enigmatic enzyme with diverse functions. Trends Biochem. Sci. 2002, 10, 534–539. [Google Scholar] [CrossRef]
- Katt, W.P.; Antonyak, M.A.; Cerione, R.A. Opening up about Tissue Transglutaminase: When Conformation Matters More than Enzymatic Activity. Med One 2018, 3, e180011. [Google Scholar] [PubMed]
- Folk, J.E.; Finlayson, J.S. The epsilon-(gamma-glutamyl) lysine crosslink and the catalytic role of transglutaminases. Adv. Protein Chem. 1977, 31, 1–133. [Google Scholar] [PubMed]
- Lee, K.N.; Birckbichler, P.J.; Patterson, M.K. GTP hydrolysis by guinea pig liver transglutaminase. Biochem. Biophys. Res. Commun. 1989, 162, 1370–1375. [Google Scholar] [CrossRef]
- Nakaoka, H.; Perez, D.M.; Baek, K.J.; Das, T.; Husain, A.; Misono, K.; Im, M.J.; Graham, R.M. Gh: A GTP binding protein with transglutaminase activity and receptor signaling function. Science 1994, 254, 1593–1596. [Google Scholar] [CrossRef]
- Feng, J.F.; Rhee, S.G.; Im, M.J. Evidence that phpospholipase delta-1 is the effector in the Gh (transglutaminase II)-mediated signalling. J. Biol. Chem. 1996, 271, 16451–16454. [Google Scholar] [CrossRef]
- Akimov, S.S.; Belkin, A.M. Cell-surface transglutaminase promotes fibronectin assembly via interaction with the gelatin-binding domain of fibronectin: A role in TGFβ-dependent matrix deposition. J. Cell Sci. 2001, 114, 2989–3000. [Google Scholar] [CrossRef]
- Fesus, L.; Thomazy, V.; Falus, A. Induction and activation of tissue transglutaminase during programmed cell death. FEBS Lett. 1987, 224, 104–108. [Google Scholar] [CrossRef]
- Iismaa, S.E.; Mearns, B.M.; Lorand, L.; Graham, R.M. Transglutaminases and disease: Lessons from genetically engineered mouse models and inherited disorders. Physiol. Rev. 2009, 89, 991–1023. [Google Scholar] [CrossRef]
- Szondy, Z.; Korponay-Szabó, I.; Király, R.; Sarang, Z.; Tsay, G.J. Transglutaminase 2 in human diseases. Biomedicine 2017, 7, 15. [Google Scholar] [CrossRef] [PubMed]
- Mangala, L.S.; Mehta, K. Tissue transglutaminase (TG2) in cancer biology. Prog. Exp. Tumor Res. 2015, 38, 125–138. [Google Scholar]
- Mehta, K.; Kumar, A.; Kim, H.I. Transglutaminase 2: A multi-tasking protein in the complex circuitry of inflammation and cancer. Biochem. Pharmacol. 2010, 80, 1921–1929. [Google Scholar] [CrossRef] [PubMed]
- Dudek, S.M.; Johnson, G.V.W. Transglutaminase facilitates the formation of polymers of the beta-amyloid peptide. Brain Res. 1994, 651, 129–133. [Google Scholar] [CrossRef]
- Norlund, M.A.; Lee, J.M.; Zainelli, G.M.; Muma, N.A. Elevated transglutaminase-induced bonds in PHF tau in Alzheimer’s disease. Brain Res. 1999, 851, 154–163. [Google Scholar] [CrossRef]
- Skill, N.J.; Griffin, M.; El Nahas, A.M.; Sanai, T.; Haylor, J.L.; Fisher, M.; Jamie, M.F.; Mould, N.N.; Johnson, T.S. Increases in renal ε-(γ-glutamyl)-lysine crosslinks result from compartment-specific changes in tissue transglutaminase in early experimental diabetic nephropathy: Pathologic implications. Lab. Investig. 2001, 81, 705–716. [Google Scholar] [CrossRef]
- Dieterich, W.; Ehnis, T.; Bauer, M.; Donner, P.; Volta, U.; Riecken, E.O.; Schuppan, D. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat. Med. 1997, 3, 797–801. [Google Scholar] [CrossRef]
- De Laurenzi, V.; Melino, G. Gene disruption of tissue transglutaminase. Mol. Cell. Biol. 2001, 21, 148–155. [Google Scholar] [CrossRef]
- Nanda, N.; Iismaa, S.E.; Owens, W.A.; Husain, A.; Mackay, F.; Graham, R.M. Targeted inactivation of Gh/tissue transglutaminase II. J. Biol. Chem. 2001, 276, 20673–20678. [Google Scholar] [CrossRef]
- Sarang, Z.; Madi, A.; Koy, C.; Varga, S.; Glocker, M.O.; Ucker, D.S.; Kuchay, S.; Chishti, A.H.; Melino, G.; Fesus, L. Tissue transglutaminase (TG2) facilitates phosphatidylserine exposure and calpain activity in calcium-induced death of erythrocytes. Cell Death Differ. 2007, 14, 1842–1844. [Google Scholar] [CrossRef][Green Version]
- Szondy, Z.; Sarang, Z.; Molnar, P.; Nemeth, T.; Piacentini, M.; Mastroberardino, P.G.; Falasca, L.; Aeschlimann, D.; Kovacs, J.; Kiss, I. Transglutaminase 2−/− mice reveals a phagocytosis-associated crosstalk between macrophages and apoptotic cells. Proc. Natl. Acad. Sci. USA 2003, 100, 7812–7817. [Google Scholar] [CrossRef]
- Sarang, Z.; Tóth, B.; Balajthy, Z.; Köröskényi, K.; Garabuczi, E.; Fésüs, L.; Szondy, Z. Some lessons from the tissue transglutaminase knockout mouse. Amino Acids 2009, 36, 625–631. [Google Scholar] [CrossRef]
- Sarang, Z.; Köröskényi, K.; Pallai, A.; Duro, E.; Melino, G.; Griffin, M.; Fesus, L.; Szondy, Z. Transglutaminase 2 null macrophages respond to lipopolysaccharide stimulation by elevated proinflammatory cytokine production due to an enhanced αvβ3 integrin-induced Src tyrosine kinase signaling. Immunol. Lett. 2011, 138, 71–78. [Google Scholar] [CrossRef]
- Bernassola, F.; Federici, M.; Corazzari, M.; Terrinoni, A.; Hribal, M.L.; De Laurenzi, V.; Ranalli, M.; Massa, O.; Sesti, G.; McLean, W.H. Role of transglutaminase 2 in glucose tolerance: Knockout mice studies and a putative mutation in a MODY patient. FASEB J. 2002, 16, 1371–1378. [Google Scholar] [CrossRef]
- Kajimura, S.; Spiegelman, B.M.; Seale, P. Brown and Beige Fat: Physiological Roles beyond Heat Generation. Cell Metab. 2015, 22, 546–559. [Google Scholar] [CrossRef]
- Lin, C.S.; Klingenberg, M. Isolation of the uncoupling protein from brown adipose tissue mitochondria. FEBS Lett. 1980, 113, 299–303. [Google Scholar] [CrossRef]
- Fedorenko, A.; Lishko, P.V.; Kirichok, Y. Mechanism of fatty-acid dependent UCP1 uncoupling in brown fat mitochondria. Cell 2012, 151, 400–413. [Google Scholar] [CrossRef]
- Mádi, A.; Cuaranta-Monroy, I.; Lénárt, K.; Pap, A.; Mezei, Z.A.; Kristóf, E.; Oláh, A.; Vámosi, G.; Bacsó, Z.; Bai, P. Browning deficiency and low mobilization of fatty acids in gonadal white adipose tissue lead to decreased cold-tolerance of transglutaminase 2 knock-out mice. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 1575–1586. [Google Scholar] [CrossRef]
- Myneni, V.D.; Melino, G.; Kaartinen, M.T. Transglutaminase 2–A novel inhibitor of adipogenesis. Cell Death Dis. 2015, 27, e1868. [Google Scholar] [CrossRef]
- Svensson, P.A.; Jernas, M.; Sjöholm, K.; Hoffmann, J.M.; Nilsson, B.E.; Hansson, M.; Carlsson, L.M.S. Gene expression in human brown adipose tissue. Int. J. Mol. Med. 2010, 27, 227–232. [Google Scholar] [CrossRef]
- Altshuler-Keylin, S.; Shinoda, K.; Hasegawa, Y.; Ikeda, K.; Hong, H.; Kang, Q.; Yang, Y.; Perera, R.M.; Debnath, J.; Kajimura, S. Beige Adipocyte Maintenance Is Regulated by Autophagy-Induced Mitochondrial Clearance. Cell Metab. 2016, 24, 402–419. [Google Scholar] [CrossRef] [PubMed]
- Cypess, A.M.; White, A.P.; Vernochet, C.; Schulz, T.J.; Xue, R.; Sass, C.A.; Huang, T.L.; Roberts-Toler, C.; Weiner, L.S.; Sze, C.; et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat. Med. 2013, 19, 635–639. [Google Scholar] [CrossRef] [PubMed]
- Gaidhu, M.P.; Fediuc, S.; Anthony, N.M.; So, M.; Mirpourian, M.; Perry, R.L.; Ceddia, R.B. Prolonged AICAR-induced AMP-kinase activation promotes energy dissipation in white adipocytes: Novel mechanisms integrating HSL and ATGL. J. Lipid Res. 2009, 50, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Wan, Z.; Root-McCaig, J.; Castellani, L.; Kemp, B.E.; Steinberg, G.R.; Wright, D.C. Evidence for the role of AMPK in regulating PGC-1 alpha expression and mitochondrial proteins in mouse epididymal adipose tissue. Obesity 2013, 22, 730–738. [Google Scholar] [CrossRef]
- Hallap, T.; Nagy, S.; Jaakma, U.; Johannisson, A.; Rodriguez-Martinez, H. Mitochondrial activity of frozen-thawed spermatozoa assessed by MitoTracker Deep Red 633. Theriogenology 2004, 63, 2311–2322. [Google Scholar] [CrossRef]
- Adam-Vizi, V. Production of reactive oxygen species in brain mitochondria: Contribution by electron transport chain and non-electron transport chain sources. Antioxid. Redox Signal. 2005, 7, 1140–1149. [Google Scholar] [CrossRef]
- Moro, M.A.; Almeida, A.; Bolaños, J.P.; Lizasoain, I. Mitochondrial respiratory chain and free radical generation in stroke. Free Radic. Biol. Med. 2005, 39, 1291–1304. [Google Scholar] [CrossRef]
- van der Bliek, A.M.; Shen, Q.; Kawajiri, S. Mechanisms of mitochondrial fission and fusion. Cold Spring Harb. Perspect. Biol. 2013, 5, a011072. [Google Scholar] [CrossRef]
- Peeters, R.P.; Hernandez, A.; Ng, L.; Ma, M.; Sharlin, D.S.; Pandey, M.; Simonds, W.F.; St Germain, D.L.; Forrest, D. Cerebellar abnormalities in mice lacking type 3 deiodinase and partial reversal of phenotype by deletion of thyroid hormone receptor α1. Endocrinology 2013, 154, 550–561. [Google Scholar] [CrossRef]
- Shi, L.Y.; Ma, Y.; Zhu, G.Y.; Liu, J.W.; Zhou, C.X.; Chen, L.J.; Wang, Y.; Li, R.C.; Yang, Z.X.; Zhang, D. Placenta-specific 1 regulates oocyte meiosis and fertilization through furin. FASEB J. 2018, 32, 5483–5494. [Google Scholar] [CrossRef]
- Sveinbjornsson, G.; Mikaelsdottir, E.; Palsson, R.; Indridason, O.S.; Holm, H.; Jonasdottir, A.; Helgason, A.; Sigurdsson, S.; Jonasdottir, A.; Sigurdsson, A. Rare mutations associating with serum creatinine and chronic kidney disease. Hum. Mol. Genet. 2014, 23, 6935–6943. [Google Scholar] [CrossRef] [PubMed]
- Porcelli, V.; Fiermonte, G.; Longo, A.; Palmieri, F. The human gene SLC25A29, of solute carrier family 25, encodes a mitochondrial transporter of basic amino acids. J. Biol. Chem. 2014, 289, 13374–13384. [Google Scholar] [CrossRef] [PubMed]
- Haitina, T.; Lindblom, J.; Renström, T.; Fredriksson, R. Fourteen novel human members of mitochondrial solute carrier family 25 (SLC25) widely expressed in the central nervous system. Genomics 2006, 88, 779–790. [Google Scholar] [CrossRef] [PubMed]
- Fiermonte, G.; Paradies, E.; Todisco, S.; Marobbio, C.M.; Palmieri, F. A novel member of solute carrier family 25 (SLC25A42) is a transporter of coenzyme A and adenosine 3’,5’-diphosphate in human mitochondria. J. Biol. Chem. 2009, 284, 18152–18159. [Google Scholar] [CrossRef]
- Almannai, M.; Alasmari, A.; Alqasmi, A.; Faqeih, E.; Al Mutairi, F.; Alotaibi, M.; Samman, M.M.; Eyaid, W.; Aljadhai, Y.I.; Shamseldin, H.E. Expanding the phenotype of SLC25A42-associated mitochondrial encephalomyopathy. Clin. Genet. 2018, 93, 1097–1102. [Google Scholar] [CrossRef] [PubMed]
- Chou, F.S.; Wang, P.S. The SLC25A42 Transcript Is a Biomarker for Fetal Reprogramming in Response to Placental Insufficiency in Preterm Newborns Under 32 Weeks Gestation-A Pilot Study. Front. Pediatr. 2020, 8, 459. [Google Scholar] [CrossRef]
- Bird, M.J.; Adant, I.; Windmolders, P.; Vander Elst, I.; Felgueira, C.; Altassan, R.; Gruenert, S.C.; Ghesquière, B.; Witters, P.; Cassiman, D. Oxygraphy Versus Enzymology for the Biochemical Diagnosis of Primary Mitochondrial Disease. Metabolites 2019, 9, 220. [Google Scholar] [CrossRef]
- Arianti, R.; Vinnai, B.Á.; Tóth, B.B.; Shaw, A.; Csősz, É.; Vámos, A.; Győry, F.; Fischer-Posovszky, P.; Wabitsch, M.; Kristóf, E.; et al. ASC-1 transporter-dependent amino acid uptake is required for the efficient thermogenic response of human adipocytes to adrenergic stimulation. FEBS Lett. 2021, 595, 2085–2098. [Google Scholar] [CrossRef]
- Snell, K.; Duff, D.A. Alanine release by rat adipose tissue in vitro. Biochem. Biophys. Res. Commun. 1977, 77, 925–931. [Google Scholar] [CrossRef]
- Fisher, F.M.; Kleiner, S.; Douris, N.; Fox, E.C.; Mepani, R.J.; Verdeguer, F.; Wu, J.; Kharitonenkov, A.; Flier, J.S.; Maratos-Flier, E. FGF21 regulates PGC-1α and browning of white adipose tissues in adaptive thermogenesis. Genes Dev. 2012, 26, 271–281. [Google Scholar] [CrossRef]
- Ye, L.; Wu, J.; Cohen, P.; Kazak, L.; Khandekar, M.J.; Jedrychowski, M.P.; Zeng, X.; Gygi, S.P.; Spiegelman, B.M. Fat cells directly sense temperature to activate thermogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 12480–12485. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Petkova, A.P.; Granneman, J.G. Identification of an Adipogenic Niche for Adipose Tissue Remodeling and Restoration. Cell Metab. 2013, 18, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Markan, K.R.; Boland, L.K.; King-McAlpin, A.Q.; Claflin, K.E.; Leaman, M.P.; Kemerling, M.K.; Stonewall, M.M.; Amendt, B.A.; Ankrum, J.A.; Potthoff, M.J. Adipose TBX1 regulates β-adrenergic sensitivity in subcutaneous adipose tissue and thermogenic capacity in vivo. Mol. Metab. 2020, 36, 100965. [Google Scholar] [CrossRef] [PubMed]
- Westphal, S.; Gantert, T.; Kless, C.; Hüttinger, K.; Klingenspor, M.; Fromme, T. Fibroblast growth factor 8b induces uncoupling protein 1 expression in epididymal white preadipocytes. Sci. Rep. 2019, 9, 44878. [Google Scholar] [CrossRef]
- Rossin, F.; D’Eletto, M.; Falasca, L.; Sepe, S.; Cocco, S.; Fimia, G.M.; Campanella, M.; Mastroberardino, P.G.; Farrace, M.G.; Piacentini, M. Transglutaminase 2 ablation leads to mitophagy impairment associated with a metabolic shift towards aerobic glycolysis. Cell Death Differ. 2014, 22, 408–418. [Google Scholar] [CrossRef]
- Piacentini, M.; Farrace, M.G.; Piredda, L.; Matarrese, P.; Ciccosanti, F.; Falasca, L.; Rodolfo, C.; Giammarioli, A.M.; Verderio, E.; Griffin, M. Transglutaminase overexpression sensitizes neuronal cell lines to apoptosis by increasing mitochondrial membrane potential and cellular oxidative stress. J. Neurochem. 2002, 81, 1061–1072. [Google Scholar] [CrossRef]
- Battaglia, G.; Farrace, M.G.; Mastroberardino, P.G.; Viti, I.; Fimia, G.M.; Van Beeumen, J.; Devreese, B.; Melino, G.; Molinaro, G.; Busceti, C.L. Transglutaminase 2 ablation leads to defective function of mitochondrial respiratory complex I affecting neuronal vulnerability in experimental models of extrapyramidal disorders. J. Neurochem. 2007, 100, 36–49. [Google Scholar] [CrossRef]
- Szondy, Z.; Mastroberardino, P.G.; Varadi, J.; Farrace, M.G.; Nagy, N.; Bak, I.; Viti, I.; Wieckowski, M.R.; Melino, G.; Rizzuto, R. Tissue transglutaminase (TG2) protects cardiomyocytes against ischemia/reperfusion injury by regulating ATP synthesis. Cell Death Differ. 2006, 13, 1827–1829. [Google Scholar] [CrossRef]
- Liu, R.; Chan, D.C. The mitochondrial fission receptor Mff selectively recruits oligomerized Drp1. Mol. Biol. Cell 2015, 26, 4466–4477. [Google Scholar] [CrossRef]
- Jheng, H.F.; Tsai, P.J.; Guo, S.M.; Kuo, L.H.; Chang, C.S.; Su, I.J.; Chang, C.R.; Tsai, Y.S. Mitochondrial fission contributes to mitochondrial dysfunction and insulin resistance in skeletal muscle. Mol. Cell. Biol. 2012, 32, 309–319. [Google Scholar] [CrossRef]
- Harmuth, T.; Prell-Schicker, C.; Weber, J.J.; Gellerich, F.; Funke, C.; Drießen, S.; Magg, J.C.D.; Krebiehl, G.; Wolburg, H.; Hayer, S.N. Mitochondrial Morphology, Function and Homeostasis Are Impaired by Expression of an N-terminal Calpain Cleavage Fragment of Ataxin-3. Front. Mol. Neurosci. 2018, 11, 368. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Marcos, P.J.; Auwerx, J. Regulation of PGC-1α, a nodal regulator of mitochondrial biogenesis. Am. J. Clin. Nutr. 2011, 93, 884S–890S. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Rhee, J.; St-Pierre, J.; Handschin, C.; Puigserver, P.; Lin, J.; Jäeger, S.; Erdjument-Bromage, H.; Tempst, P.; Spiegelman, B.M. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1α: Modulation by p38 MAPK. Genes Dev. 2004, 18, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Jäger, S.; Handschin, C.; St-Pierre, J.; Spiegelman, B.M. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1α. Proc. Natl. Acad. Sci. USA 2007, 104, 12017–12022. [Google Scholar] [CrossRef]
- Knutti, D.; Kressler, D.; Kralli, A. Regulation of the transcriptional coactivator PGC-1 via MAPK-sensitive interaction with a repressor. Proc. Natl. Acad. Sci. USA 2001, 98, 9713–9718. [Google Scholar] [CrossRef]
- Rossin, F.; Costa, R.; Bordi, M.; D’Eletto, M.; Occhigrossi, L.; Farrace, M.G.; Barlev, N.; Ciccosanti, F.; Muccioli, S.; Chieregato, L. Transglutaminase Type 2 regulates the Wnt/beta-catenin pathway in vertebrates. Cell Death Dis. 2021, 12, 249. [Google Scholar] [CrossRef]
- Yau, W.W.; Yen, P.M. Thermogenesis in Adipose Tissue Activated by Thyroid Hormone. Int. J. Mol. Sci. 2020, 21, 3020. [Google Scholar] [CrossRef]
- Guilherme, A.; Yenilmez, B.; Bedard, A.H.; Henriques, F.; Liu, D.; Lee, A.; Goldstein, L.; Kelly, M.; Nicoloro, S.M.; Chen, M. Control of Adipocyte Thermogenesis and Lipogenesis through beta3-Adrenergic and Thyroid Hormone Signal Integration. Cell Rep. 2020, 31, 107598. [Google Scholar] [CrossRef]
- Lindsey, R.C.; Mohan, S. Thyroid hormone acting via TRbeta induces expression of browning genes in mouse bone marrow adipose tissue. Endocrine 2017, 56, 109–120. [Google Scholar] [CrossRef]
- Paquette, M.A.; Dong, H.; Gagné, R.; Williams, A.; Malowany, M.; Wade, M.G.; Yauk, C.L. Thyroid hormone-regulated gene expression in juvenile mouse liver: Identification of thyroid response elements using microarray profiling and in silico analyses. BMC Genom. 2011, 12, 634. [Google Scholar] [CrossRef]
- Xue, H.; Wang, Z.; Hua, Y.; Ke, S.; Wang, Y.; Zhang, J.; Pan, Y.H.; Huang, W.; Irwin, D.M.; Zhang, S. Molecular signatures and functional analysis of beige adipocytes induced from in vivo intra-abdominal adipocytes. Sci. Adv. 2018, 4, eaar5319. [Google Scholar] [CrossRef] [PubMed]
- Vistica, D.T.; Skehan, P.; Scudiero, D.; Monks, A.; Pittman, A.; Boyd, M.R. Tetrazolium-based assays for cellular viability: A critical examination of selected parameters affecting formazan production. Cancer Res. 1991, 51, 2515–2520. [Google Scholar] [PubMed]
- Fodor, T.; Szanto, M.; Abdul-Rahman, O.; Nagy, L.; Der, A.; Kiss, B.; Bai, P. Combined Treatment of MCF-7 Cells with AICAR and Methotrexate, Arrests Cell Cycle and Reverses Warburg Metabolism through AMP-Activated Protein Kinase (AMPK) and FOXO1. PLoS ONE 2016, 11, e0150232. [Google Scholar] [CrossRef]
- Berg, S.; Kutra, D.; Kroeger, T.; Straehle, C.N.; Kausler, B.X.; Haubold, C.; Schiegg, M.; Ales, J.; Beier, T.; Rudy, M. Ilastik: Interactive machine learning for (bio)image analysis. Nat. Methods 2019, 16, 1226–1232. [Google Scholar] [CrossRef]
- Lamprecht, M.R.; Sabatini, D.M.; Carpenter, A.E. CellProfiler™: Free, versatile software for automated biological image analysis. BioTechniques 2007, 42, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Jambrovics, K.; Uray, I.P.; Keillor, J.W.; Fésüs, L.; Balajthy, Z. Benefits of Combined All-Trans Retinoic Acid and Arsenic Trioxide Treatment of Acute Promyelocytic Leukemia Cells and Further Enhancement by Inhibition of Atypically Expressed Transglutaminase 2. Cancers 2020, 12, 648. [Google Scholar] [CrossRef]
- Zielonka, J.; Lambeth, J.D.; Kalyanaraman, B. On the use of L-012, a luminol-based chemiluminescent probe, for detecting superoxide and identifying inhibitors of NADPH oxidase: A reevaluation. Free Radic. Biol. Med. 2013, 65, 1310–1314. [Google Scholar] [CrossRef]
- Rastogi, R.P.; Richa, K.A.; Tyagi, M.B.; Sinha, R.P. Molecular Mechanisms of Ultraviolet Radiation-Induced DNA Damage and Repair. J. Nucleic Acids 2010, 56, 1–32. [Google Scholar] [CrossRef]
- Budai, Z.; Ujlaky-Nagy, L.; Kis, G.N.; Antal, M.; Bankó, C.; Bacsó, Z.; Szondy, Z.; Sarang, Z. Macrophages engulf apoptotic and primary necrotic thymocytes through similar phosphatidylserine-dependent mechanisms. FEBS Open Biol. 2019, 13, 446–456. [Google Scholar] [CrossRef]
- Tóth, B.B.; Arianti, R.; Shaw, A.; Vámos, A.; Veréb, Z.; Póliska, S.; Győry, F.; Bacso, Z.; Fésüs, L.; Kristóf, E. FTO Intronic SNP Strongly Influences Human Neck Adipocyte Browning Determined by Tissue and PPARγ Specific Regulation: A Transcriptome Analysis. Cells 2020, 9, 987. [Google Scholar] [CrossRef]
- Kazerouni, F.; Amirrasouli, H. Performance characteristics of three automated immunoassays for thyroid hormones. Casp. J. Intern. Med. Spring 2012, 3, 400–404. [Google Scholar]
- Semlitsch, G.; Buchinger, W.; Reiterer, E.; Binter, G.; Rainer, F. Free triiodothyronine (FT3) measurement using an electrochemiluminescence immunoassay in patients with autoantibodies to triiodothyronine. Acta Med. Aust. 2000, 27, 54–55. [Google Scholar] [CrossRef]
- Nagy, L.; Márton, J.; Vida, A.; Kis, G.; Bokor, É.; Kun, S.; Gönczi, M.; Docsa, T.; Tóth, A.; Antal, M.; et al. Glycogen phosphorylase inhibition improves beta-cell function. Br. J. Pharmacol. 2018, 175, 301–319. [Google Scholar] [CrossRef] [PubMed]
- Armenta, J.M.; Cortes, D.F.; Pisciotta, J.M.; Shuman, J.L.; Blakeslee, K.; Rasoloson, D.; Ogunbiyi, O.; Sullivan, D.J.; Shulaev, V. A sensitive and rapid method for amino acid quantitation in malaria biological samples using AccQ•Tag UPLC-ESI-MS/MS with multiple reaction monitoring. Anal. Chem. 2011, 82, 548–558. [Google Scholar] [CrossRef] [PubMed]
| Antibody | Dilution | Suppliers |
|---|---|---|
| β-ACTIN | 1:5000 | Sigma Aldrich (#A2066) |
| UCP1 | 1:1000 | Sigma Aldrich (#SAB1404511) |
| TOMM20 | 1:1000 | Abcam (#ab56783) |
| PGC1α | 1:1000 | Santa Cruz Biotechnology (#D1112) |
| Phospho-AMPKα (Thr172) | 1:1000 | Cell Signaling (#2535) |
| OXPHOS | 1:1000 | Abcam (#ab110411) |
| OPA1 | 1:1500 | Novus Biologicals (#NB110-55290) |
| MFN2 | 1:1000 | Sigma Aldrich (#WH0009927M3) |
| MFF | 1:1000 | Proteintech (#17090-1-AP) |
| DRP1 | 1:1000 | BD Biosciences (#611112) |
| Anti-rabbit IgG, HRP conjugated secondary antibody | 1:10,000 | Advansta (R-05072-500) |
| Anti-mouse IgG, HRP conjugated secondary antibody | 1:10,000 | Advansta (R-05071-500) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lénárt, K.; Bankó, C.; Ujlaki, G.; Póliska, S.; Kis, G.; Csősz, É.; Antal, M.; Bacso, Z.; Bai, P.; Fésüs, L.; et al. Tissue Transglutaminase Knock-Out Preadipocytes and Beige Cells of Epididymal Fat Origin Possess Decreased Mitochondrial Functions Required for Thermogenesis. Int. J. Mol. Sci. 2022, 23, 5175. https://doi.org/10.3390/ijms23095175
Lénárt K, Bankó C, Ujlaki G, Póliska S, Kis G, Csősz É, Antal M, Bacso Z, Bai P, Fésüs L, et al. Tissue Transglutaminase Knock-Out Preadipocytes and Beige Cells of Epididymal Fat Origin Possess Decreased Mitochondrial Functions Required for Thermogenesis. International Journal of Molecular Sciences. 2022; 23(9):5175. https://doi.org/10.3390/ijms23095175
Chicago/Turabian StyleLénárt, Kinga, Csaba Bankó, Gyula Ujlaki, Szilárd Póliska, Gréta Kis, Éva Csősz, Miklós Antal, Zsolt Bacso, Péter Bai, László Fésüs, and et al. 2022. "Tissue Transglutaminase Knock-Out Preadipocytes and Beige Cells of Epididymal Fat Origin Possess Decreased Mitochondrial Functions Required for Thermogenesis" International Journal of Molecular Sciences 23, no. 9: 5175. https://doi.org/10.3390/ijms23095175
APA StyleLénárt, K., Bankó, C., Ujlaki, G., Póliska, S., Kis, G., Csősz, É., Antal, M., Bacso, Z., Bai, P., Fésüs, L., & Mádi, A. (2022). Tissue Transglutaminase Knock-Out Preadipocytes and Beige Cells of Epididymal Fat Origin Possess Decreased Mitochondrial Functions Required for Thermogenesis. International Journal of Molecular Sciences, 23(9), 5175. https://doi.org/10.3390/ijms23095175







