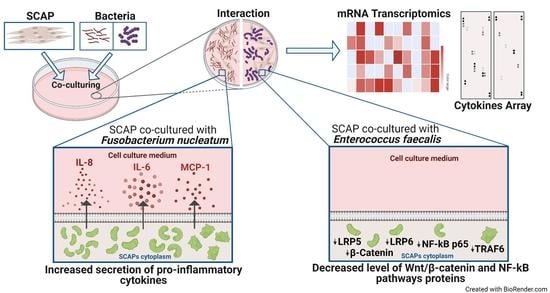
Combined Transcriptomic and Protein Array Cytokine Profiling of Human Stem Cells from Dental Apical Papilla Modulated by Oral Bacteria
Abstract
:1. Introduction
2. Results
2.1. Oral Bacteria Isolated from Root Canals Modulate SCAP Cytokine Profiling and Their Osteogenic and Immunomodulatory Potentials in a Species-Dependent Manner
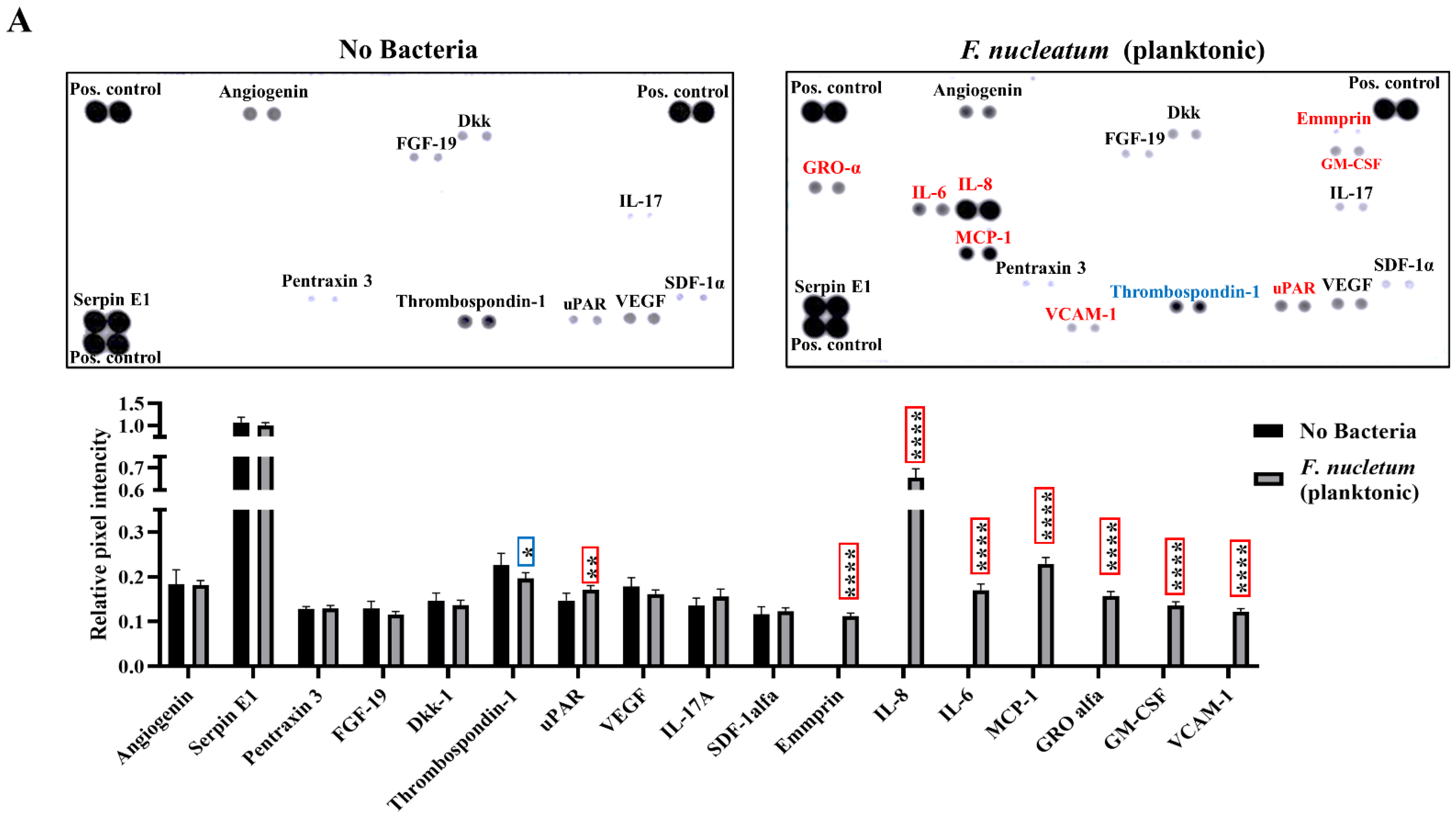
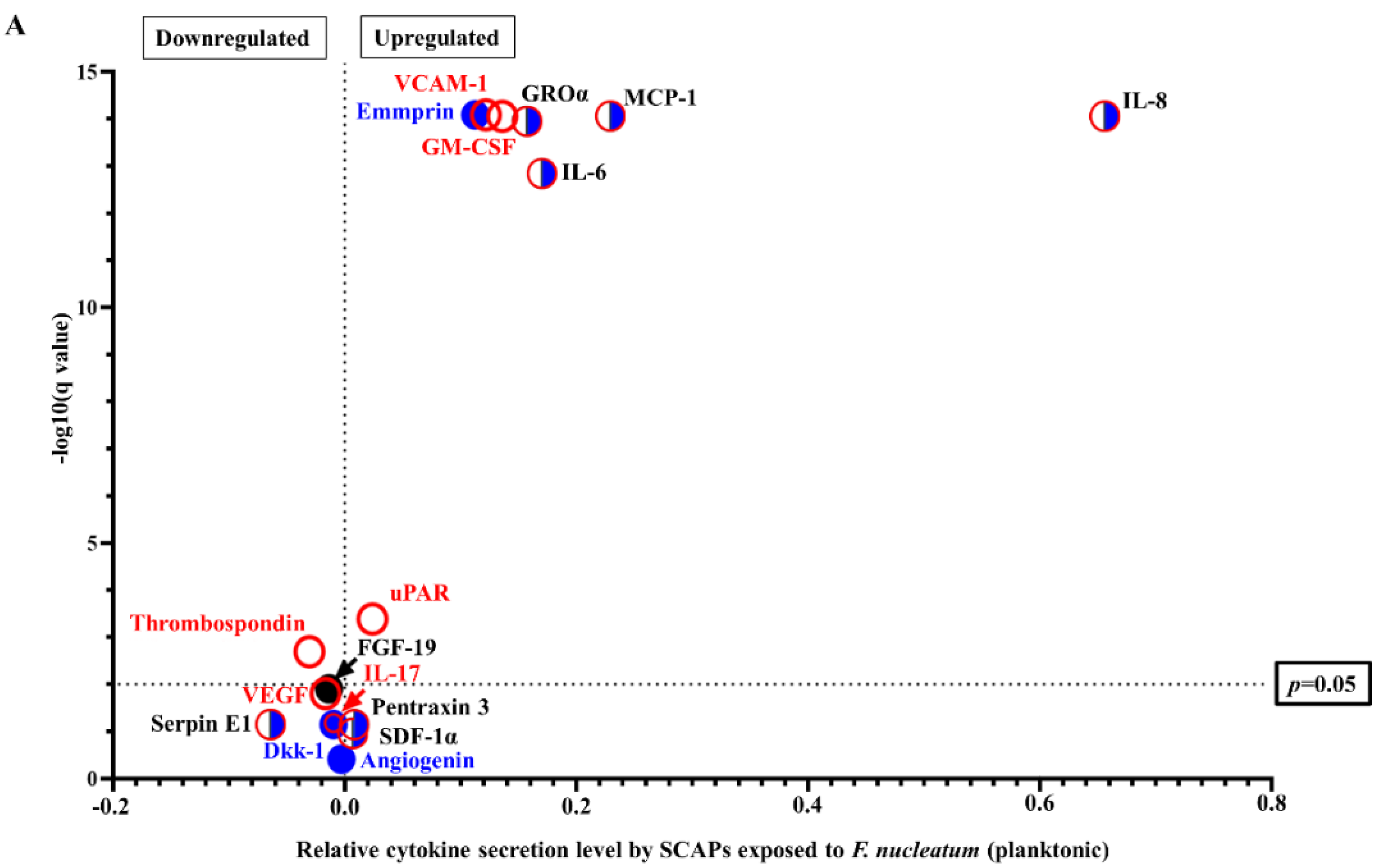
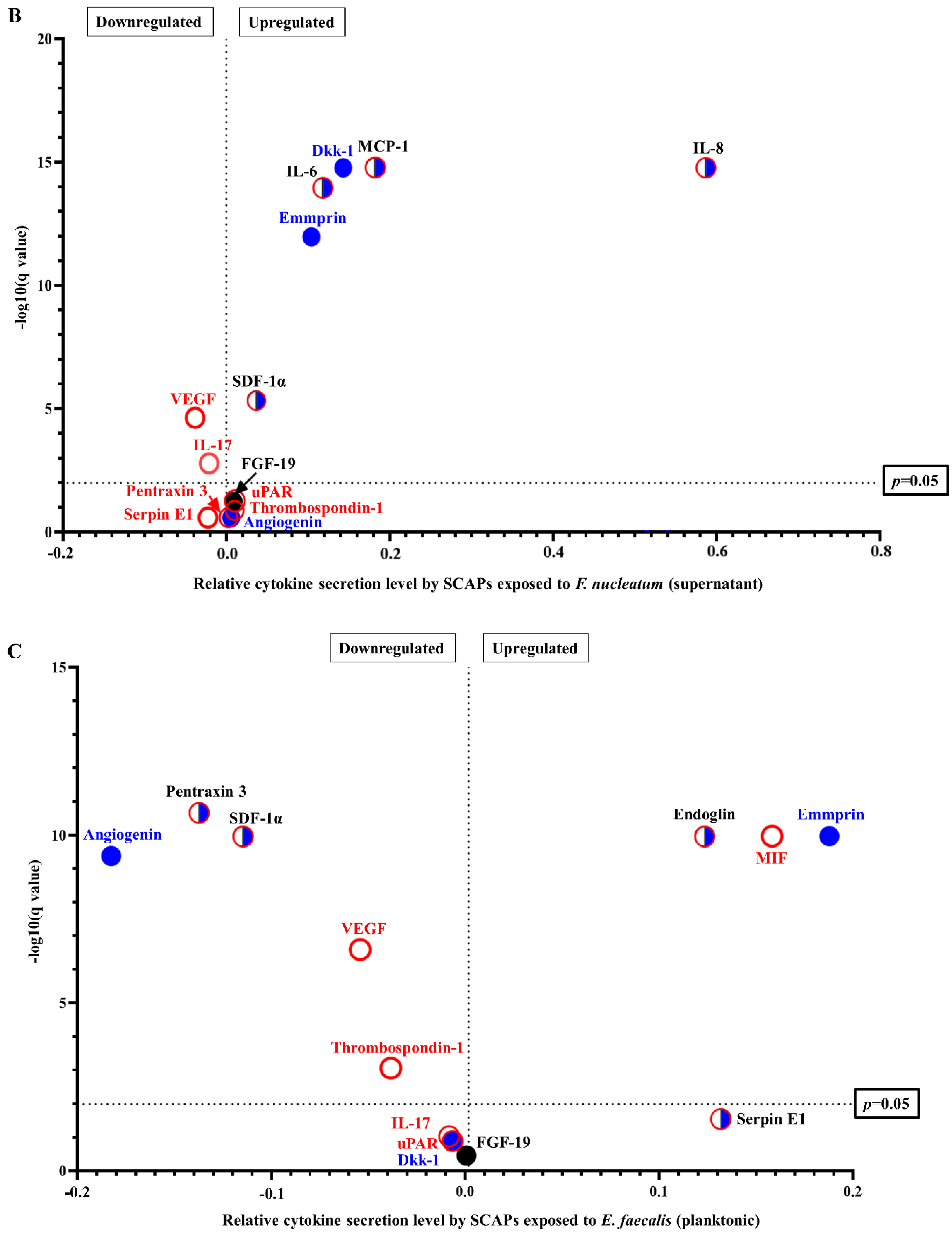
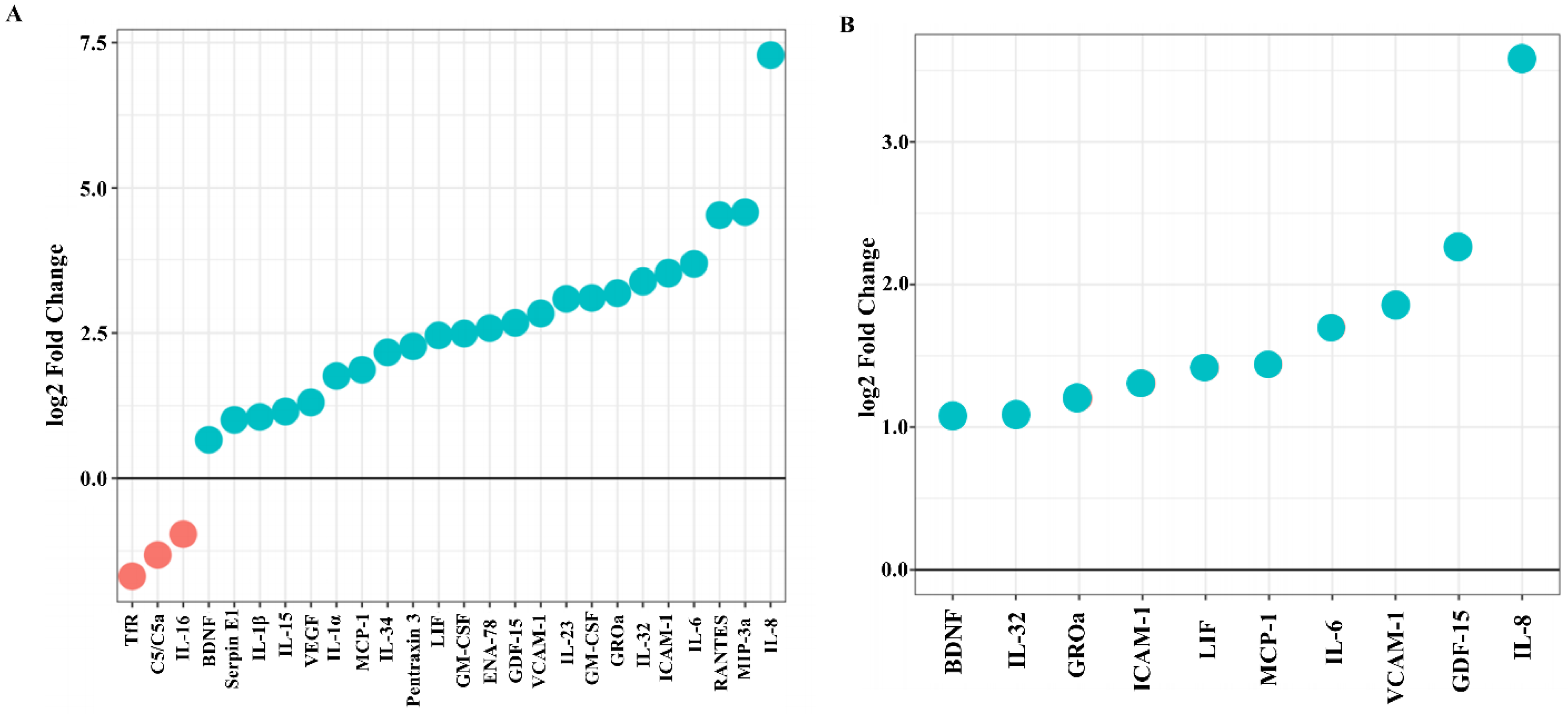
2.2. F. nucleatum Triggers While E. faecalis Inhibits Pro-Inflammatory Chemokine and Cytokine Generation, Such as IL-6, IL-8, and MCP-1, at the mRNA and Protein Levels
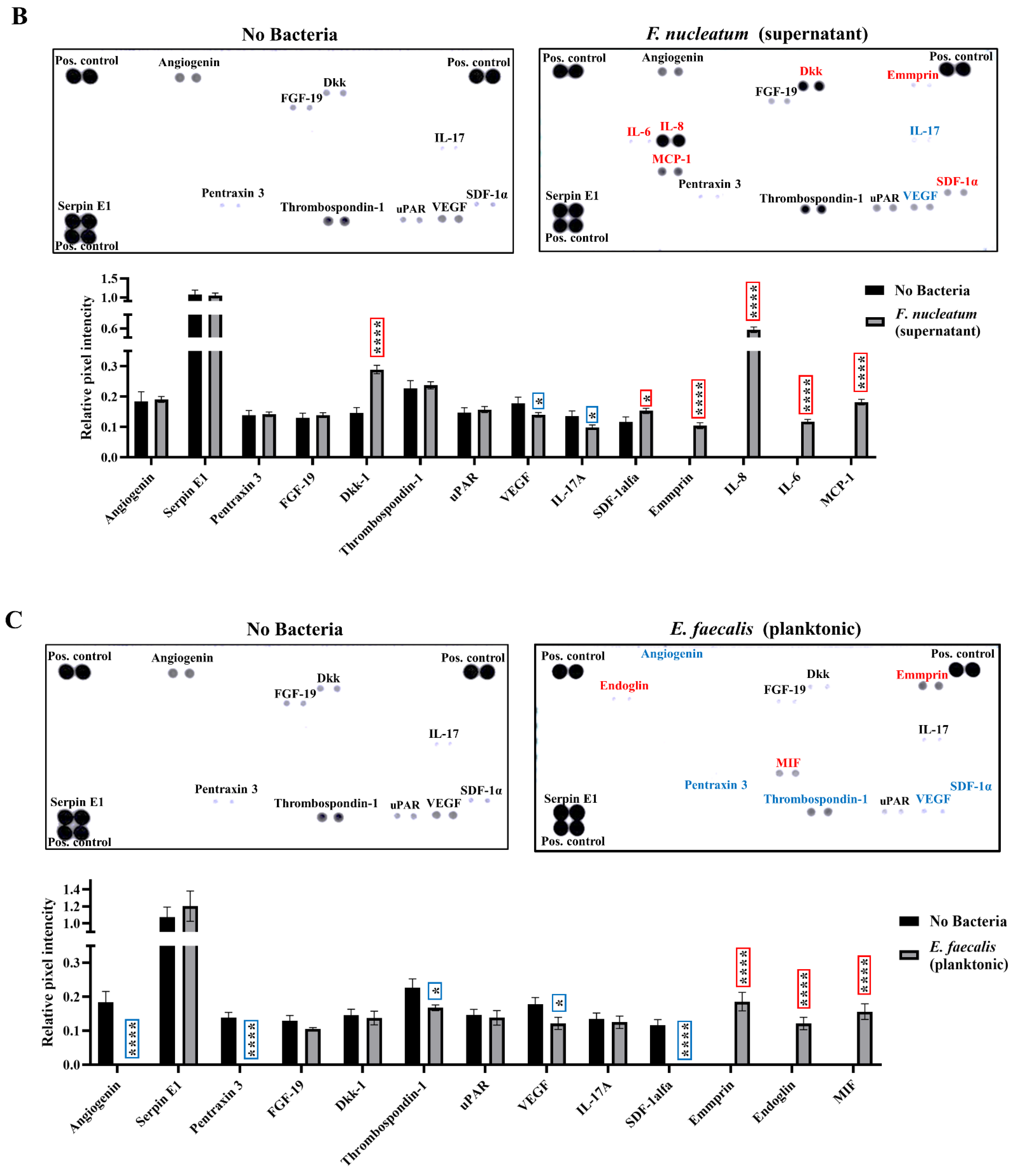
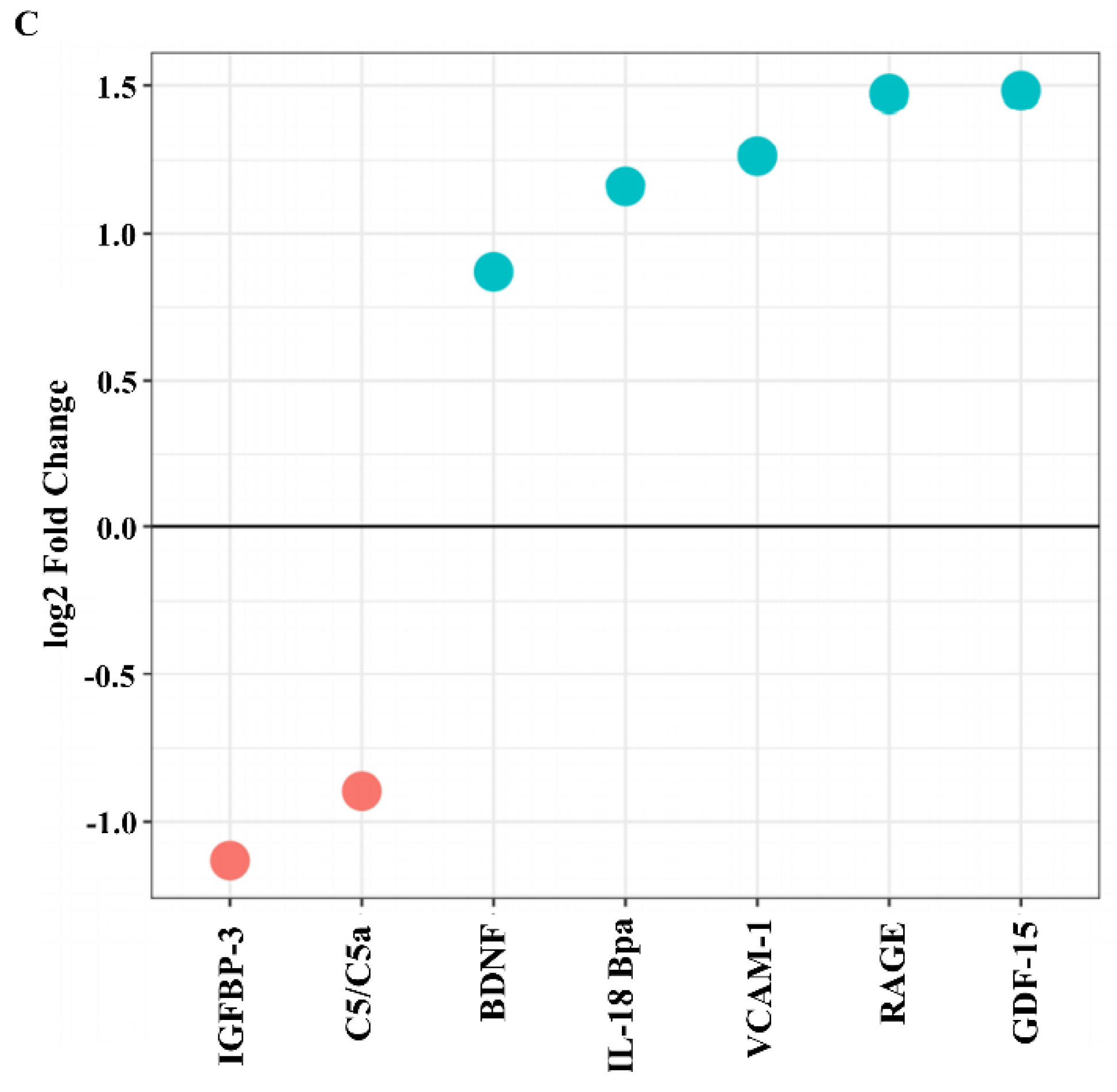
2.3. SCAP Modulated by F. nucleatum Express Cytokine mRNA and Translate It into Protein, as Shown for IL-6, IL-8, GM-CSF, and MCP-1. However, the Gene and Protein Expressions Diverged When SCAP Were Modulated by E. faecalis
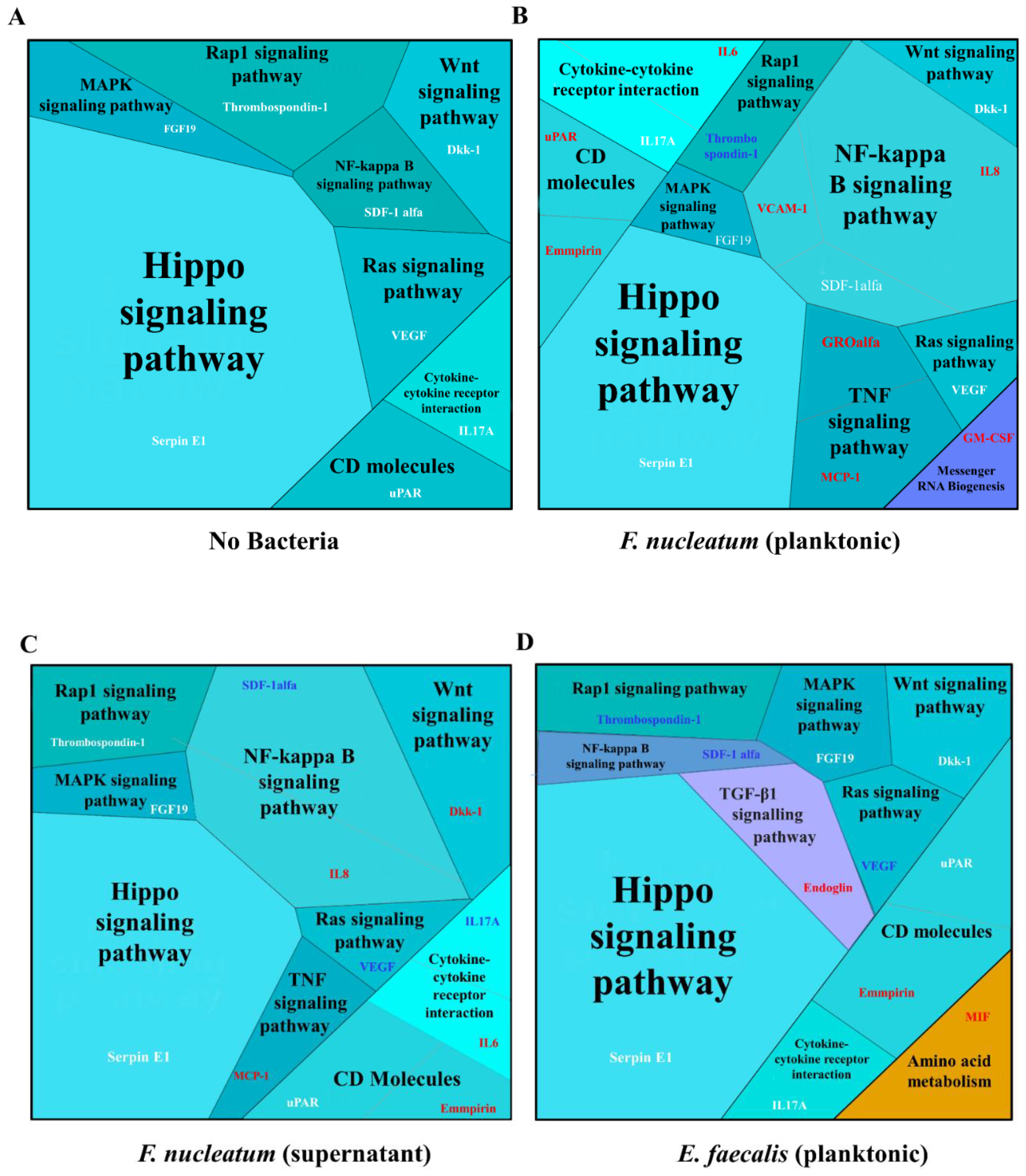
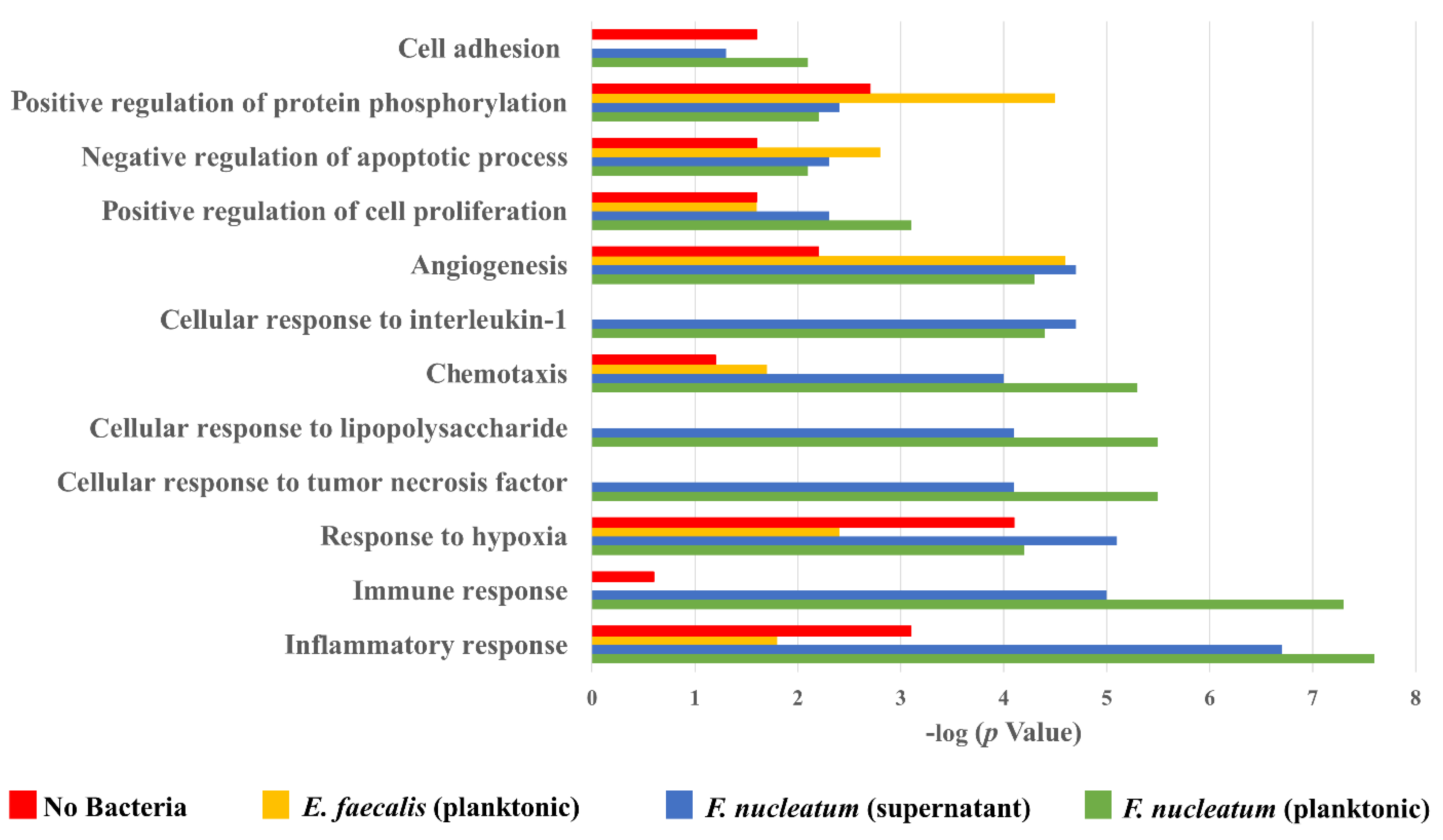
2.4. Biological Processes Such as Immune Response, Inflammatory Response, and Response to Hypoxia Processes Were Decreased When SCAP Were Modulated by E. faecalis but Enhanced in Cases of SCAP Co-Cultured with F. nucleatum
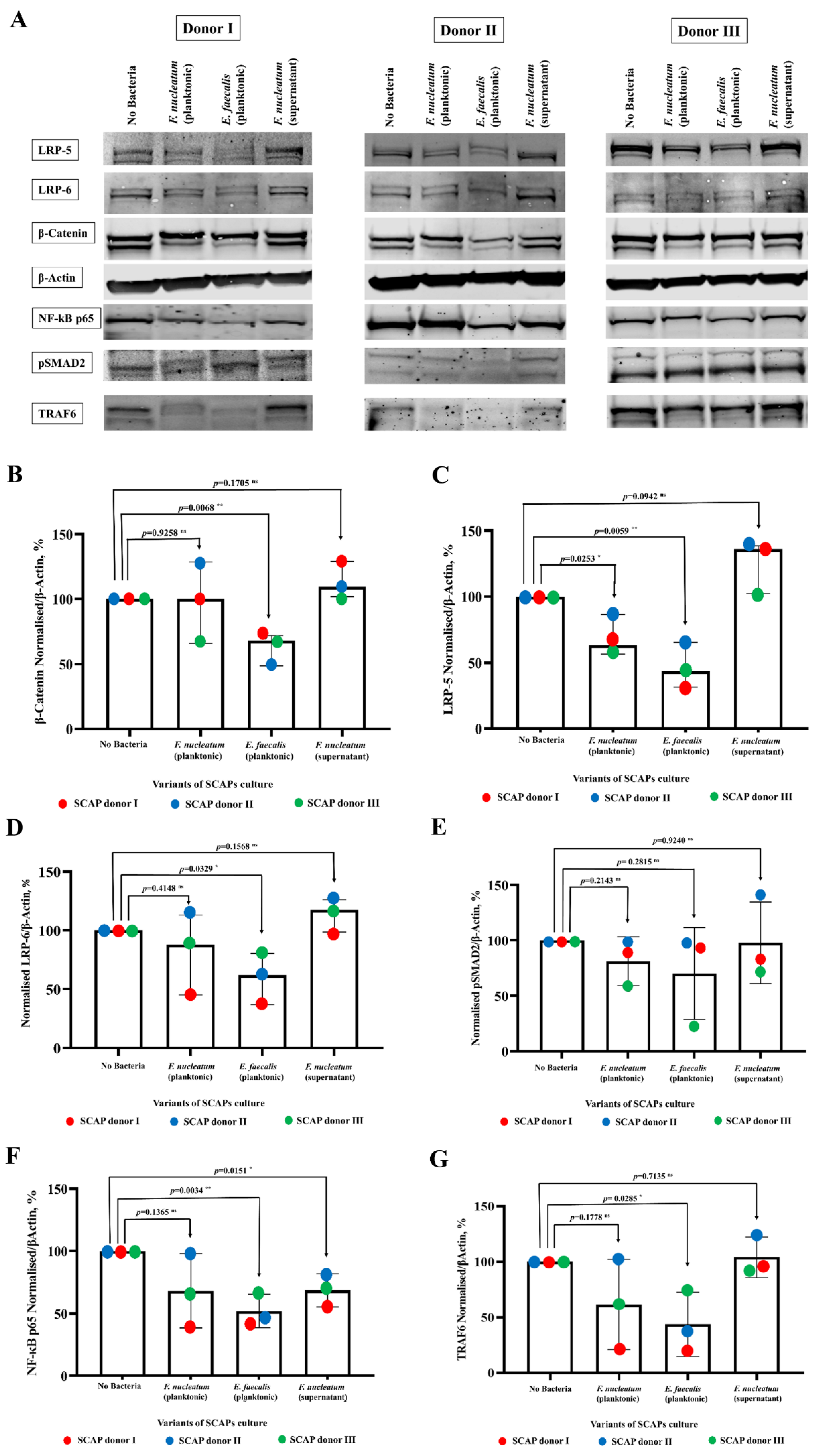
2.5. E. faecalis Led to the Decrease of Key Protein Levels of Wnt/β-Catenin and NF-κB Signaling Pathways Playing Crucial Role in Bone Formation
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Supernatants, and Growth Conditions
4.2. Isolation, Culture, and Characterization of Human SCAP
4.3. Variant of Treatments Included in Co-Culture Experiments
4.4. Detection of Secreted Cytokines by Protein Array
4.5. mRNA Preparation and Transcriptome Sequencing Analysis
4.6. Protein Interaction Mapping
4.7. Preparation of Total Cell Lysate for SDS-PAGE Electrophoresis
4.8. SDS-PAGE and Immunoblot Analysis
4.9. Antibodies and Reagents
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aas, J.A.; Paster, B.J.; Stokes, L.N.; Olsen, I.; Dewhirst, F.E. Defining the Normal Bacterial Flora of the Oral Cavity. J. Clin. Microbiol. 2005, 43, 5721–5732. [Google Scholar] [CrossRef] [Green Version]
- Nair, P.N.R. Pathogenesis of Apical Periodontitis and the Causes of Endodontic Failures. Crit. Rev. Oral Biol. Med. 2004, 15, 348–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cvek, M. Prognosis of luxated non-vital maxillary incisors treated with calcium hydroxide and filled with gutta-percha: A retrospective clinical study. Dent. Traumatol. 1992, 8, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.E.; Garcia-Godoy, F.; Hargreaves, K.M. Regenerative Endodontics: A Review of Current Status and a Call for Action. J. Endod. 2007, 33, 377–390. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, F.; Kempf, V.A.J. Interaction of bacteria and stem cells in health and disease. FEMS Microbiol. Rev. 2019, 43, 162–180. [Google Scholar] [CrossRef] [PubMed]
- Chatzivasileiou, K.; Kriebel, K.; Steinhoff, G.; Kreikemeyer, B.; Lang, H. Do oral bacteria alter the regenerative potential of stem cells? A concise review. J. Cell. Mol. Med. 2015, 19, 2067–2074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nada, O.A.; El Backly, R.M. Stem Cells from the Apical Papilla (SCAP) as a Tool for Endogenous Tissue Regeneration. Front. Bioeng. Biotechnol. 2018, 6, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yilmaz, Ö. The chronicles of Porphyromonas gingivalis: The microbium, the human oral epithelium and their interplay. Microbiology 2008, 154, 2897–2903. [Google Scholar] [CrossRef]
- Chen, Q.; Hou, T.; Luo, F.; Wu, X.; Xie, Z.; Xu, J. Involvement of Toll-Like Receptor 2 and Pro-Apoptotic Signaling Pathways in Bone Remodeling in Osteomyelitis. Cell. Physiol. Biochem. 2014, 34, 1890–1900. [Google Scholar] [CrossRef]
- Baldwin, A.S. Series Introduction: The transcription factor NF-κB and human disease. J. Clin. Investig. 2001, 107, 3–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.W. Fusobacterium nucleatum: A commensal-turned pathogen. Curr. Opin. Microbiol. 2015, 23, 141–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujii, R.; Saito, Y.; Tokura, Y.; Nakagawa, K.-I.; Okuda, K.; Ishihara, K. Characterization of bacterial flora in persistent apical periodontitis lesions. Oral Microbiol. Immunol. 2009, 24, 502–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manoharan, L.; Brundin, M.; Rakhimova, O.; de Paz, L.C.; Romani Vestman, N. New Insights into the Microbial Profiles of Infected Root Canals in Traumatized Teeth. J. Clin. Med. 2020, 9, 3877. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.A.; Garrett, W.S. Fusobacterium nucleatum—Symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 2019, 17, 156–166. [Google Scholar] [CrossRef]
- Bolstad, A.I.; Jensen, H.B.; Bakken, V. Taxonomy, biology, and periodontal aspects of Fusobacterium nucleatum. Clin. Microbiol. Rev. 1996, 9, 55–71. [Google Scholar] [CrossRef]
- Fisher, K.; Phillips, C. The ecology, epidemiology and virulence of Enterococcus. Microbiology 2009, 155, 1749–1757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.-Q.; Zhang, C.-F.; Chu, C.-H.; Zhu, X.-F. Prevalence of Enterococcus faecalis in saliva and filled root canals of teeth associated with apical periodontitis. Int. J. Oral Sci. 2012, 4, 19–23. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Huang, Z.; Tang, Z.; Huang, Y.; Huang, M.; Liu, H.; Ziebolz, D.; Schmalz, G.; Jia, B.; Zhao, J. More than Just a Periodontal Pathogen—The Research Progress on Fusobacterium nucleatum. Front. Cell. Infect. Microbiol. 2022, 12, 815318. [Google Scholar] [CrossRef]
- Van Dyke, T.E.; Bartold, P.M.; Reynolds, E.C. The Nexus between Periodontal Inflammation and Dysbiosis. Front. Immunol. 2020, 11, 511. [Google Scholar] [CrossRef]
- Papapanou, P.N.; Sanz, M.; Buduneli, N.; Dietrich, T.; Feres, M.; Fine, D.H.; Flemmig, T.F.; Garcia, R.; Giannobile, W.V.; Graziani, F.; et al. Periodontitis: Consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Periodontol. 2018, 89, S173–S182. [Google Scholar] [CrossRef] [Green Version]
- Kolenbrander, P.E. Oral Microbial Communities: Biofilms, Interactions, and Genetic Systems. Annu. Rev. Microbiol. 2000, 54, 413–437. [Google Scholar] [CrossRef] [PubMed]
- Rakhimova, O.; Schmidt, A.; Landström, M.; Johansson, A.; Kelk, P.; Romani Vestman, N. Cytokine Secretion, Viability, and Real-Time Proliferation of Apical-Papilla Stem Cells Upon Exposure to Oral Bacteria. Front. Cell. Infect. Microbiol. 2021, 10, 620801. [Google Scholar] [CrossRef] [PubMed]
- Qiang, Y.-W.; Barlogie, B.; Rudikoff, S.; Shaughnessy, J.D. Dkk1-induced inhibition of Wnt signaling in osteoblast differentiation is an underlying mechanism of bone loss in multiple myeloma. Bone 2008, 42, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Liebermeister, W.; Noor, E.; Flamholz, A.; Davidi, D.; Bernhardt, J.; Milo, R. Visual account of protein investment in cellular functions. Proc. Natl. Acad. Sci. USA 2014, 111, 8488–8493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, J.; Wang, Z.; Tang, E.; Fan, Z.; McCauley, L.; Franceschi, R.; Guan, K.; Krebsbach, P.H.; Wang, C.-Y. Inhibition of osteoblastic bone formation by nuclear factor-κB. Nat. Med. 2009, 15, 682–689. [Google Scholar] [CrossRef]
- Rahman, M.S.; Akhtar, N.; Jamil, H.M.; Banik, R.S.; Asaduzzaman, S.M. TGF-β/BMP signaling and other molecular events: Regulation of osteoblastogenesis and bone formation. Bone Res. 2015, 3, 15005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lomaga, M.A.; Yeh, W.-C.; Sarosi, I.; Duncan, G.S.; Furlonger, C.; Ho, A.; Morony, S.; Capparelli, C.; Van, G.; Kaufman, S.; et al. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 1999, 13, 1015–1024. [Google Scholar] [CrossRef] [Green Version]
- Paganelli, A.; Trubiani, O.; Diomede, F.; Pisciotta, A.; Paganelli, R. Immunomodulating Profile of Dental Mesenchymal Stromal Cells: A Comprehensive Overview. Front. Oral Heal. 2021, 2, 635055. [Google Scholar] [CrossRef]
- Wang, Q.; Zhao, L.; Xu, C.; Zhou, J.; Wu, Y. Fusobacterium nucleatum stimulates monocyte adhesion to and transmigration through endothelial cells. Arch. Oral Biol. 2019, 100, 86–92. [Google Scholar] [CrossRef]
- Johnson, L.; Almeida-da-Silva, C.L.C.; Takiya, C.M.; Figliuolo, V.; Rocha, G.M.; Weissmüller, G.; Scharfstein, J.; Coutinho-Silva, R.; Ojcius, D.M. Oral infection of mice with Fusobacterium nucleatum results in macrophage recruitment to the dental pulp and bone resorption. Biomed. J. 2018, 41, 184–193. [Google Scholar] [CrossRef]
- Kang, W.; Ji, X.; Zhang, X.; Tang, D.; Feng, Q. Persistent Exposure to Fusobacterium nucleatum Triggers Chemokine/Cytokine Release and Inhibits the Proliferation and Osteogenic Differentiation Capabilities of Human Gingiva-Derived Mesenchymal Stem Cells. Front. Cell. Infect. Microbiol. 2019, 9, 429. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Hu, C.-H.; Zhou, C.-H.; Cui, X.-X.; Yang, K.; Deng, C.; Xia, J.-J.; Wu, Y.; Liu, L.-C.; Jin, Y. DKK1 rescues osteogenic differentiation of mesenchymal stem cells isolated from periodontal ligaments of patients with diabetes mellitus induced periodontitis. Sci. Rep. 2015, 5, 13142. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, J.F.; Rôças, I.N. Diversity of Endodontic Microbiota Revisited. J. Dent. Res. 2009, 88, 969–981. [Google Scholar] [CrossRef] [PubMed]
- Darveau, R.P. Periodontitis: A polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 2010, 8, 481–490. [Google Scholar] [CrossRef]
- Butera, A.; Gallo, S.; Maiorani, C.; Molino, D.; Chiesa, A.; Preda, C.; Esposito, F.; Scribante, A. Probiotic Alternative to Chlorhexidine in Periodontal Therapy: Evaluation of Clinical and Microbiological Parameters. Microorganisms 2020, 9, 69. [Google Scholar] [CrossRef]
- Ghazalpour, A.; Bennett, B.; Petyuk, V.A.; Orozco, L.; Hagopian, R.; Mungrue, I.N.; Farber, C.R.; Sinsheimer, J.; Kang, H.M.; Furlotte, N.; et al. Comparative Analysis of Proteome and Transcriptome Variation in Mouse. PLoS Genet. 2011, 7, e1001393. [Google Scholar] [CrossRef] [Green Version]
- Gao, B.; Roux, P.P. Translational control by oncogenic signaling pathways. Biochim. Biophys. Acta-Gene Regul. Mech. 2015, 1849, 753–765. [Google Scholar] [CrossRef]
- Sauro, H.M. Control and regulation of pathways via negative feedback. J. R. Soc. Interface 2017, 14, 20160848. [Google Scholar] [CrossRef]
- Xin, X.; Zeng, X.; Gu, H.; Li, M.; Tan, H.; Jin, Z.; Hua, T.; Shi, R.; Wang, H. CD147/EMMPRIN overexpression and prognosis in cancer: A systematic review and meta-analysis. Sci. Rep. 2016, 6, 32804. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Chen, L.; Wang, C.; Chen, J.; Zhang, X.; Hu, Y.; Niu, X.; Pei, D.; He, Z.; Bi, Y. Extracellular matrix metalloproteinase inducer enhances host resistance against pseudomonas aeruginosa infection through MAPK signaling pathway. Am. J. Transl. Res. 2016, 8, 5619–5627. [Google Scholar]
- Jacquin, C.; Koczon-Jaremko, B.; Aguila, H.L.; Leng, L.; Bucala, R.; Kuchel, G.A.; Lee, S.-K. Macrophage migration inhibitory factor inhibits osteoclastogenesis. Bone 2009, 45, 640–649. [Google Scholar] [CrossRef] [PubMed]
- Onuora, S. MIF drives inflammation and bone formation in AS. Nat. Rev. Rheumatol. 2017, 13, 451. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Henen, M.A.; Hinck, A.P. Structural biology of betaglycan and endoglin, membrane-bound co-receptors of the TGF-beta family. Exp. Biol. Med. 2019, 244, 1547–1558. [Google Scholar] [CrossRef] [PubMed]
- David, C.J.; Massagué, J. Contextual determinants of TGFβ action in development, immunity and cancer. Nat. Rev. Mol. Cell Biol. 2018, 19, 419–435. [Google Scholar] [CrossRef] [PubMed]
- Morandini, A.C.F.; Sipert, C.R.; Gasparoto, T.H.; Greghi, S.L.A.; Passanezi, E.; Rezende, M.L.R.; Sant’ana, A.P.; Campanelli, A.P.; Garlet, G.P.; Santos, C.F. Differential Production of Macrophage Inflammatory Protein-1α, Stromal-Derived Factor-1, and IL-6 by Human Cultured Periodontal Ligament and Gingival Fibroblasts Challenged With Lipopolysaccharide From P. gingivalis. J. Periodontol. 2010, 81, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Finlay, B.B.; McFadden, G. Anti-Immunology: Evasion of the Host Immune System by Bacterial and Viral Pathogens. Cell 2006, 124, 767–782. [Google Scholar] [CrossRef] [Green Version]
- Elashiry, M.M.; Elashiry, M.; Zeitoun, R.; Elsayed, R.; Tian, F.; Saber, S.E.; Elashry, S.H.; Tay, F.R.; Cutler, C.W. Enterococcus faecalis Induces Differentiation of Immune-Aberrant Dendritic Cells from Murine Bone Marrow-Derived Stem Cells. Infect. Immun. 2020, 88, 11. [Google Scholar] [CrossRef]
- Chong, K.K.L.; Tay, W.H.; Janela, B.; Yong, A.M.H.; Liew, T.H.; Madden, L.; Keogh, D.; Barkham, T.M.S.; Ginhoux, F.; Becker, D.L.; et al. Enterococcus faecalis Modulates Immune Activation and Slows Healing During Wound Infection. J. Infect. Dis. 2017, 216, 1644–1654. [Google Scholar] [CrossRef] [Green Version]
- Tien, B.Y.Q.; Goh, H.M.S.; Chong, K.K.L.; Bhaduri-Tagore, S.; Holec, S.; Dress, R.; Ginhoux, F.; Ingersoll, M.A.; Williams, R.B.H.; Kline, K.A. Enterococcus faecalis Promotes Innate Immune Suppression and Polymicrobial Catheter-Associated Urinary Tract Infection. Infect. Immun. 2017, 85, e00378-17. [Google Scholar] [CrossRef] [Green Version]
- Molina, M.A.; Díaz, A.M.; Hesse, C.; Ginter, W.; Gentilini, M.V.; Nuñez, G.G.; Canellada, A.M.; Sparwasser, T.; Berod, L.; Castro, M.S.; et al. Immunostimulatory Effects Triggered by Enterococcus faecalis CECT7121 Probiotic Strain Involve Activation of Dendritic Cells and Interferon-Gamma Production. PLoS ONE 2015, 10, e0127262. [Google Scholar] [CrossRef] [Green Version]
- Clevers, H.; Loh, K.M.; Nusse, R. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 2014, 346, 1248012. [Google Scholar] [CrossRef] [PubMed]
- Thesleff, I. From understanding tooth development to bioengineering of teeth. Eur. J. Oral Sci. 2018, 126, 67–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norwitz, N.G.; Mota, A.S.; Misra, M.; Ackerman, K.E. LRP5, Bone Density, and Mechanical Stress: A Case Report and Literature Review. Front. Endocrinol. 2019, 10, 184. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Slee, R.B.; Fukai, N.; Rawadi, G.; Roman-Roman, S.; Reginato, A.M.; Wang, H.; Cundy, T.; Glorieux, F.H.; Lev, D.; et al. LDL Receptor-Related Protein 5 (LRP5) Affects Bone Accrual and Eye Development. Cell 2001, 107, 513–523. [Google Scholar] [CrossRef] [Green Version]
- Walsh, M.C.; Kim, G.K.; Maurizio, P.L.; Molnar, E.E.; Choi, Y. TRAF6 autoubiquitination-independent activation of the NFkappaB and MAPK pathways in response to IL-1 and RANKL. PLoS ONE 2008, 3, e4064. [Google Scholar] [CrossRef] [Green Version]
- Silva-García, O.; Valdez-Alarcón, J.J.; Baizabal-Aguirre, V.M. Wnt/β-Catenin Signaling as a Molecular Target by Pathogenic Bacteria. Front. Immunol. 2019, 10, 2135. [Google Scholar] [CrossRef]
- Yang, H.; Li, G.; Han, N.; Zhang, X.; Cao, Y.; Cao, Y.; Fan, Z. Secreted frizzled-related protein 2 promotes the osteo/odontogenic differentiation and paracrine potentials of stem cells from apical papilla under inflammation and hypoxia conditions. Cell Prolif. 2020, 53, e12694. [Google Scholar] [CrossRef]
- Iida, K.; Takeda-Kawaguchi, T.; Tezuka, Y.; Kunisada, T.; Shibata, T.; Tezuka, K. Hypoxia enhances colony formation and proliferation but inhibits differentiation of human dental pulp cells. Arch. Oral Biol. 2010, 55, 648–654. [Google Scholar] [CrossRef]
- Utting, J.C.; Robins, S.P.; Brandao-Burch, A.; Orriss, I.R.; Behar, J.; Arnett, T.R. Hypoxia inhibits the growth, differentiation and bone-forming capacity of rat osteoblasts. Exp. Cell Res. 2006, 312, 1693–1702. [Google Scholar] [CrossRef]
- Fiedler, T.; Salamon, A.; Adam, S.; Herzmann, N.; Taubenheim, J.; Peters, K. Impact of bacteria and bacterial components on osteogenic and adipogenic differentiation of adipose-derived mesenchymal stem cells. Exp. Cell Res. 2013, 319, 2883–2892. [Google Scholar] [CrossRef]
- Vanacker, J.; Viswanath, A.; De Berdt, P.; Everard, A.; Cani, P.D.; Bouzin, C.; Feron, O.; Diogenes, A.; Leprince, J.G.; des Rieux, A. Hypoxia Modulates the Differentiation Potential of Stem Cells of the Apical Papilla. J. Endod. 2014, 40, 1410–1418. [Google Scholar] [CrossRef]
- Pettersson, L.F.; Kingham, P.J.; Wiberg, M.; Kelk, P. In Vitro Osteogenic Differentiation of Human Mesenchymal Stem Cells from Jawbone Compared with Dental Tissue. Tissue Eng. Regen. Med. 2017, 14, 763–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schisterman, E.F.; Vexler, A. To pool or not to pool, from whether to when: Applications of pooling to biospecimens subject to a limit of detection. Paediatr. Perinat. Epidemiol. 2008, 22, 486–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Zhang, Y.; Park, C.; Bennett, C.; Thornton, M.; Kim, D. Rapid and accurate alignment of nucleotide conversion sequencing reads with HISAT-3N. Genome Res. 2021, 31, 1290–1295. [Google Scholar] [CrossRef]
- Wang, L.; Wang, S.; Li, W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics 2012, 28, 2184–2185. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zymovets, V.; Razghonova, Y.; Rakhimova, O.; Aripaka, K.; Manoharan, L.; Kelk, P.; Landström, M.; Romani Vestman, N. Combined Transcriptomic and Protein Array Cytokine Profiling of Human Stem Cells from Dental Apical Papilla Modulated by Oral Bacteria. Int. J. Mol. Sci. 2022, 23, 5098. https://doi.org/10.3390/ijms23095098
Zymovets V, Razghonova Y, Rakhimova O, Aripaka K, Manoharan L, Kelk P, Landström M, Romani Vestman N. Combined Transcriptomic and Protein Array Cytokine Profiling of Human Stem Cells from Dental Apical Papilla Modulated by Oral Bacteria. International Journal of Molecular Sciences. 2022; 23(9):5098. https://doi.org/10.3390/ijms23095098
Chicago/Turabian StyleZymovets, Valeriia, Yelyzaveta Razghonova, Olena Rakhimova, Karthik Aripaka, Lokeshwaran Manoharan, Peyman Kelk, Maréne Landström, and Nelly Romani Vestman. 2022. "Combined Transcriptomic and Protein Array Cytokine Profiling of Human Stem Cells from Dental Apical Papilla Modulated by Oral Bacteria" International Journal of Molecular Sciences 23, no. 9: 5098. https://doi.org/10.3390/ijms23095098
APA StyleZymovets, V., Razghonova, Y., Rakhimova, O., Aripaka, K., Manoharan, L., Kelk, P., Landström, M., & Romani Vestman, N. (2022). Combined Transcriptomic and Protein Array Cytokine Profiling of Human Stem Cells from Dental Apical Papilla Modulated by Oral Bacteria. International Journal of Molecular Sciences, 23(9), 5098. https://doi.org/10.3390/ijms23095098