Open the Technical Black Box of Tumor Mutational Burden (TMB): Factors Affecting Harmonization and Standardization of Panel-Based TMB
Abstract
:1. Introduction
2. Harmonization and Standardization of Panel-Based TMB
2.1. Preanalytic Factors (Sample and DNA Issues)
2.2. Sequencing Factors
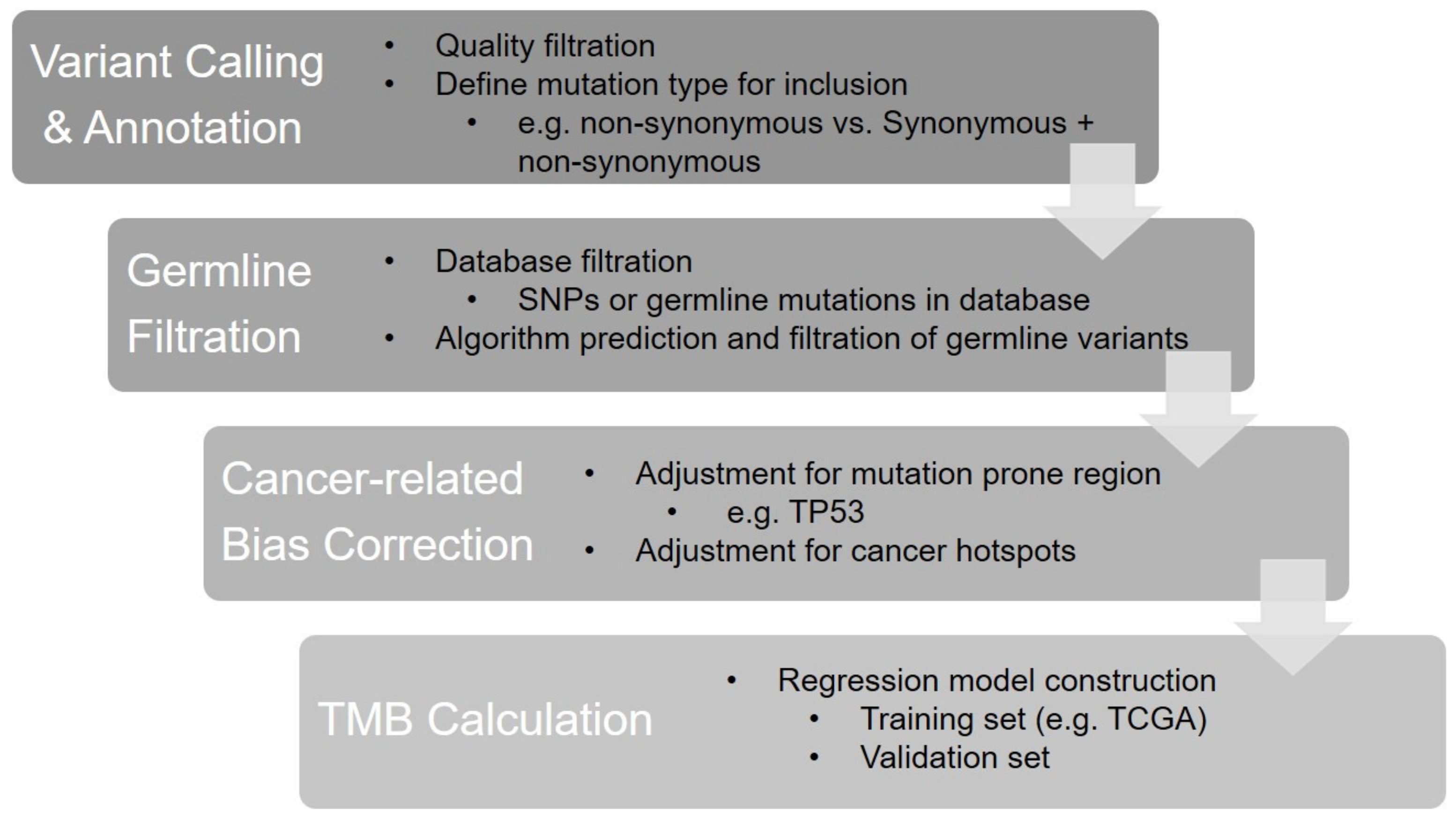
2.3. Bioinformatics Factors
2.4. Interpretation and Reporting
| Cancer | Trials/Types | Treatment | Method | TMB Cutoff | RR | PFS | OS |
|---|---|---|---|---|---|---|---|
| Various cancer types, previously treated | KEYNOTE-158 [9] Single-arm phase II | Pembrolizumab | F1 CDx assay | ≧10 mut/Mb | 29% | 2.1 months | 11.7 months |
| NSCLC | CheckMate227 [7,55,60] Phase III | Nivolumab plus ipilimumab vs. platinum-doublet chemotherapy | F1 CDx assay | ≧10 mut/Mb | 45.3% vs. 26.9% | 7.2 vs. 5.5 months (p < 0.001) | NA |
| NSCLC | Checkmate9LA [61,62] Phase III | Nivolumab plus ipilimumab plus platinum-doublet chemotherapy x 2 cycles vs. platinum-doublet chemotherapy | F1 CDx assay | ≧10 mut/Mb | 46 vs. 28% | 8.9 vs. 4.7 months | mOS:15.0 vs. 10.8 months |
| NSCLC | Checkmate026 [8] Phase III | Nivolumab vs. platinum-doublet chemotherapy | CGP by research lab | ≥243 somatic missense mutations per sample | 47 vs. 28% | 9.7 vs. 5.8 months | OS: no difference |
| NSCLC | Checkmate568 [56] Phase II | Nivolumab plus low-dose ipilimumab | F1 CDx assay | ≧10 mut/Mb | 43.8% | 7.1 months | NA |
| NSCLC | BIRCH [63] Phase II | Atezolizumab | F1 CDx assay | ≧10 mut/Mb | 25% versus 14% | NA | NA |
| NSCLC | POPLAR [63] Randomized phase II | atezolizumab versus docetaxel | F1 CDx assay | ≧10 mut/Mb | 20% versus 4% | 7.3 versus 2.8 months | 16.2 versus 8.3 months |
| NSCLC | MYSTIC [64] | Durvalumab versus Durvalumab plus tremelimumab vs. chemotherapy | F1 CDx assay | ≧10 mut/Mb | NA | NA | 18.6 versus 16.6 versus 11.9 months |
| UC | IMvigor211 [65] | Atezolizumab versus chemotherapy | F1 CDx assay | >9.65 mut/Mb | NA | NA | 11.3 versus 8.3 months HR:0.68 (0.51–0.90) |
| Melanoma | IMspire170 [66] | Cobimetinib plus atezolizumab versuspembrolizumab | F1 CDx assay | >10 mut/Mb | NA | NR versus 3.7 months in cobimetinib plus atezolizumab arm (p = 0.0004) NR versus 3.6 months in pembrolizumab arm (p = 0.002) | NA |
| Melanoma | Checkmate-067 [67] | Nivolumab versus nivolumab plus ipilimumab versus ipilimumab | WES | >median | Nivolumab 62.1% versus 31.5% Nivolumab plus ipilimumab 64.8% versus 51.0% Ipilimumab 25.5% versus 14.3% | HR 0.45 in nivolumab arm; HR 0.55 in nivolumab plus ipilimumab arm; HR 0.60 in ipilimumab arm | HR 0.46 in nivolumab arm; HR 0.53 in nivolumab plus ipilimumab arm; HR 0.52 in ipilimumab arm |
3. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Tran, E.; Ahmadzadeh, M.; Lu, Y.C.; Gros, A.; Turcotte, S.; Robbins, P.F.; Gartner, J.J.; Zheng, Z.; Li, Y.F.; Ray, S.; et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 2015, 350, 1387–1390. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Yelensky, R.; Jooss, K.; Chan, T.A. Update on Tumor Neoantigens and Their Utility: Why It Is Good to Be Different. Trends Immunol. 2018, 39, 536–548. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Cancer immunology. Mutational landscape determines sensitivity to PD–1 blockade in non–small cell lung cancer. Science 2015, 348, 124–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodman, A.M.; Kato, S.; Bazhenova, L.; Patel, S.P.; Frampton, G.M.; Miller, V.; Stephens, P.J.; Daniels, G.A.; Kurzrock, R. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol. Cancer Ther. 2017, 16, 2598–2608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snyder, A.; Makarov, V.; Merghoub, T.; Yuan, J.; Zaretsky, J.M.; Desrichard, A.; Walsh, L.A.; Postow, M.A.; Wong, P.; Ho, T.S.; et al. Genetic basis for clinical response to CTLA–4 blockade in melanoma. N. Engl. J. Med. 2014, 371, 2189–2199. [Google Scholar] [CrossRef] [Green Version]
- Van Allen, E.M.; Miao, D.; Schilling, B.; Shukla, S.A.; Blank, C.; Zimmer, L.; Sucker, A.; Hillen, U.; Foppen, M.H.G.; Goldinger, S.M.; et al. Genomic correlates of response to CTLA–4 blockade in metastatic melanoma. Science 2015, 350, 207–211. [Google Scholar] [CrossRef] [Green Version]
- Hellmann, M.D.; Ciuleanu, T.E.; Pluzanski, A.; Lee, J.S.; Otterson, G.A.; Audigier–Valette, C.; Minenza, E.; Linardou, H.; Burgers, S.; Salman, P.; et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N. Engl. J. Med. 2018, 378, 2093–2104. [Google Scholar] [CrossRef]
- Carbone, D.P.; Reck, M.; Paz–Ares, L.; Creelan, B.; Horn, L.; Steins, M.; Felip, E.; van den Heuvel, M.M.; Ciuleanu, T.E.; Badin, F.; et al. First–Line Nivolumab in Stage IV or Recurrent Non–Small–Cell Lung Cancer. N. Engl. J. Med. 2017, 376, 2415–2426. [Google Scholar] [CrossRef]
- Marabelle, A.; Fakih, M.; Lopez, J.; Shah, M.; Shapira–Frommer, R.; Nakagawa, K.; Chung, H.C.; Kindler, H.L.; Lopez–Martin, J.A.; Miller, W.H., Jr.; et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: Prospective biomarker analysis of the multicohort, open–label, phase 2 KEYNOTE–158 study. Lancet Oncol. 2020, 21, 1353–1365. [Google Scholar] [CrossRef]
- Powles, T.; Duran, I.; van der Heijden, M.S.; Loriot, Y.; Vogelzang, N.J.; De Giorgi, U.; Oudard, S.; Retz, M.M.; Castellano, D.; Bamias, A.; et al. Atezolizumab versus chemotherapy in patients with platinum–treated locally advanced or metastatic urothelial carcinoma (IMvigor211): A multicentre, open–label, phase 3 randomised controlled trial. Lancet 2018, 391, 748–757. [Google Scholar] [CrossRef]
- Rosenberg, J.E.; Hoffman–Censits, J.; Powles, T.; van der Heijden, M.S.; Balar, A.V.; Necchi, A.; Dawson, N.; O’Donnell, P.H.; Balmanoukian, A.; Loriot, Y.; et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum–based chemotherapy: A single–arm, multicentre, phase 2 trial. Lancet 2016, 387, 1909–1920. [Google Scholar] [CrossRef] [Green Version]
- Balar, A.V.; Galsky, M.D.; Rosenberg, J.E.; Powles, T.; Petrylak, D.P.; Bellmunt, J.; Loriot, Y.; Necchi, A.; Hoffman–Censits, J.; Perez–Gracia, J.L.; et al. Atezolizumab as first–line treatment in cisplatin–ineligible patients with locally advanced and metastatic urothelial carcinoma: A single–arm, multicentre, phase 2 trial. Lancet 2017, 389, 67–76. [Google Scholar] [CrossRef] [Green Version]
- Hellmann, M.D.; Callahan, M.K.; Awad, M.M.; Calvo, E.; Ascierto, P.A.; Atmaca, A.; Rizvi, N.A.; Hirsch, F.R.; Selvaggi, G.; Szustakowski, J.D.; et al. Tumor Mutational Burden and Efficacy of Nivolumab Monotherapy and in Combination with Ipilimumab in Small–Cell Lung Cancer. Cancer Cell 2018, 33, 853–861.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turajlic, S.; Litchfield, K.; Xu, H.; Rosenthal, R.; McGranahan, N.; Reading, J.L.; Wong, Y.N.S.; Rowan, A.; Kanu, N.; Al Bakir, M.; et al. Insertion–and–deletion–derived tumour–specific neoantigens and the immunogenic phenotype: A pan–cancer analysis. Lancet Oncol. 2017, 18, 1009–1021. [Google Scholar] [CrossRef] [Green Version]
- Marcus, L.; Fashoyin–Aje, L.A.; Donoghue, M.; Yuan, M.; Rodriguez, L.; Gallagher, P.S.; Philip, R.; Ghosh, S.; Theoret, M.R.; Beaver, J.A.; et al. FDA Approval Summary: Pembrolizumab for the Treatment of Tumor Mutational Burden–High Solid Tumors. Clin. Can. Res. Off. J. Am. Assoc. Cancer Res. 2021, 27, 4685–4689. [Google Scholar] [CrossRef]
- Cheng, D.T.; Mitchell, T.N.; Zehir, A.; Shah, R.H.; Benayed, R.; Syed, A.; Chandramohan, R.; Liu, Z.Y.; Won, H.H.; Scott, S.N.; et al. Memorial Sloan Kettering–Integrated Mutation Profiling of Actionable Cancer Targets (MSK–IMPACT): A Hybridization Capture–Based Next–Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J. Mol. Diagn. JMD 2015, 17, 251–264. [Google Scholar] [CrossRef]
- Jibiki, T.; Nishimura, H.; Sengoku, S.; Kodama, K. Regulations, Open Data and Healthcare Innovation: A Case of MSK–IMPACT and Its Implications for Better Cancer Care. Cancers 2021, 13, 3448. [Google Scholar] [CrossRef]
- Stenzinger, A.; Allen, J.D.; Maas, J.; Stewart, M.D.; Merino, D.M.; Wempe, M.M.; Dietel, M. Tumor mutational burden standardization initiatives: Recommendations for consistent tumor mutational burden assessment in clinical samples to guide immunotherapy treatment decisions. Genes Chromosomes Cancer 2019, 58, 578–588. [Google Scholar] [CrossRef] [Green Version]
- Merino, D.M.; McShane, L.M.; Fabrizio, D.; Funari, V.; Chen, S.J.; White, J.R.; Wenz, P.; Baden, J.; Barrett, J.C.; Chaudhary, R.; et al. Establishing guidelines to harmonize tumor mutational burden (TMB): In silico assessment of variation in TMB quantification across diagnostic platforms: Phase I of the Friends of Cancer Research TMB Harmonization Project. J. Immunother. Cancer 2020, 8, e000147. [Google Scholar] [CrossRef] [Green Version]
- Vega, D.M.; Yee, L.M.; McShane, L.M.; Williams, P.M.; Chen, L.; Vilimas, T.; Fabrizio, D.; Funari, V.; Newberg, J.; Bruce, L.K.; et al. Aligning tumor mutational burden (TMB) quantification across diagnostic platforms: Phase II of the Friends of Cancer Research TMB Harmonization Project. Ann. Oncol Off. J. Eur. Soc. Med. Oncol. ESMO 2021, 32, 1626–1636. [Google Scholar] [CrossRef]
- Chen, H.; Luthra, R.; Goswami, R.S.; Singh, R.R.; Roy–Chowdhuri, S. Analysis of Pre–Analytic Factors Affecting the Success of Clinical Next–Generation Sequencing of Solid Organ Malignancies. Cancers 2015, 7, 1699–1715. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, M.; Sedmak, D.; Jewell, S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am. J. Pathol. 2002, 161, 1961–1971. [Google Scholar] [CrossRef] [Green Version]
- Quy, P.N.; Kanai, M.; Fukuyama, K.; Kou, T.; Kondo, T.; Yamamoto, Y.; Matsubara, J.; Hiroshima, A.; Mochizuki, H.; Sakuma, T.; et al. Association Between Preanalytical Factors and Tumor Mutational Burden Estimated by Next–Generation Sequencing–Based Multiplex Gene Panel Assay. Oncologist 2019, 24, e1401–e1408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchhalter, I.; Rempel, E.; Endris, V.; Allgauer, M.; Neumann, O.; Volckmar, A.L.; Kirchner, M.; Leichsenring, J.; Lier, A.; von Winterfeld, M.; et al. Size matters: Dissecting key parameters for panel–based tumor mutational burden analysis. Int. J. Cancer. 2019, 144, 848–858. [Google Scholar] [CrossRef] [Green Version]
- Qiu, P.; Pang, L.; Arreaza, G.; Maguire, M.; Chang, K.C.; Marton, M.J.; Levitan, D. Data Interoperability of Whole Exome Sequencing (WES) Based Mutational Burden Estimates from Different Laboratories. Int. J. Mol. Sci. 2016, 17, 651. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.; Sasson, A.; Srinivasan, S.; Golhar, R.; Greenawalt, D.M.; Geese, W.J.; Green, G.; Zerba, K.; Kirov, S.; Szustakowski, J. Bioinformatic Methods and Bridging of Assay Results for Reliable Tumor Mutational Burden Assessment in Non–Small–Cell Lung Cancer. Mol. Diagn. Ther. 2019, 23, 507–520. [Google Scholar] [CrossRef] [Green Version]
- Yarchoan, M.; Hopkins, A.; Jaffee, E.M. Tumor Mutational Burden and Response Rate to PD–1 Inhibition. N. Engl. J. Med. 2017, 377, 2500–2501. [Google Scholar] [CrossRef]
- Samstein, R.M.; Lee, C.H.; Shoushtari, A.N.; Hellmann, M.D.; Shen, R.; Janjigian, Y.Y.; Barron, D.A.; Zehir, A.; Jordan, E.J.; Omuro, A.; et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019, 51, 202–206. [Google Scholar] [CrossRef]
- Campbell, B.B.; Light, N.; Fabrizio, D.; Zatzman, M.; Fuligni, F.; de Borja, R.; Davidson, S.; Edwards, M.; Elvin, J.A.; Hodel, K.P.; et al. Comprehensive Analysis of Hypermutation in Human Cancer. Cell 2017, 171, 1042–1056.e10. [Google Scholar] [CrossRef] [Green Version]
- Bettoni, F.; Koyama, F.C.; de Avelar Carpinetti, P.; Galante, P.A.F.; Camargo, A.A.; Asprino, P.F. A straightforward assay to evaluate DNA integrity and optimize next–generation sequencing for clinical diagnosis in oncology. Exp. Mol. Pathol. 2017, 103, 294–299. [Google Scholar] [CrossRef]
- Wang, Z.; Duan, J.; Cai, S.; Han, M.; Dong, H.; Zhao, J.; Zhu, B.; Wang, S.; Zhuo, M.; Sun, J.; et al. Assessment of Blood Tumor Mutational Burden as a Potential Biomarker for Immunotherapy in Patients With Non–Small Cell Lung Cancer With Use of a Next–Generation Sequencing Cancer Gene Panel. JAMA Oncol. 2019, 5, 696–702. [Google Scholar] [CrossRef] [PubMed]
- Schuurbiers, M.; Huang, Z.; Saelee, S.; Javey, M.; de Visser, L.; van den Broek, D.; Monkhorst, K.; Heuvel, M.V.D.; Lovejoy, A.F.; Klass, D. Biological and technical factors in the assessment of blood–based tumor mutational burden (bTMB) in patients with NSCLC. J. Immunother. Cancer 2022, 10, e004064. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Duan, J.; Wang, G.; Zhao, J.; Xu, J.; Han, J.; Zhao, Z.; Zhao, J.; Zhu, B.; Zhuo, M.; et al. Allele Frequency–Adjusted Blood–Based Tumor Mutational Burden as a Predictor of Overall Survival for Patients With NSCLC Treated With PD–(L)1 Inhibitors. J. Thorac. Oncol. 2020, 15, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Hideaki, B.; Yoshiaki, N.; Hiroya, T.; Manabu, S.; Hisateru, Y.; Taito, E.; Takashi, O.; Tadamichi, D.; Taroh, S.; Kentaro, Y.; et al. Impact of a metastatic site on circulating tumor DNA (ctDNA) analysis in patients (pts) with metastatic colorectal cancer (mCRC). J. Clin. Oncol. 2021, 39, 3554. Available online: https://ascopubs.org/doi/abs/10.1200/JCO.2021.39.15_suppl.3554 (accessed on 27 February 2022). [CrossRef]
- De Mattos–Arruda, L.; Siravegna, G. How to use liquid biopsies to treat patients with cancer. ESMO Open 2021, 6, 100060. [Google Scholar] [CrossRef]
- Chan, H.T.; Nagayama, S.; Chin, Y.M.; Otaki, M.; Hayashi, R.; Kiyotani, K.; Fukunaga, Y.; Ueno, M.; Nakamura, Y.; Low, S.K. Clinical significance of clonal hematopoiesis in the interpretation of blood liquid biopsy. Mol. Oncol. 2020, 14, 1719–1730. [Google Scholar] [CrossRef]
- Khagi, Y.; Goodman, A.M.; Daniels, G.A.; Patel, S.P.; Sacco, A.G.; Randall, J.M.; Bazhenova, L.A.; Kurzrock, R. Hypermutated Circulating Tumor DNA: Correlation with Response to Checkpoint Inhibitor–Based Immunotherapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 5729–5736. [Google Scholar] [CrossRef] [Green Version]
- Gandara, D.R.; Paul, S.M.; Kowanetz, M.; Schleifman, E.; Zou, W.; Li, Y.; Rittmeyer, A.; Fehrenbacher, L.; Otto, G.; Malboeuf, C.; et al. Blood–based tumor mutational burden as a predictor of clinical benefit in non–small–cell lung cancer patients treated with atezolizumab. Nat. Med. 2018, 24, 1441–1448. [Google Scholar] [CrossRef]
- Kim, E.S.; Velcheti, V.; Mekhail, T.; Yun, C.; Shagan, S.M.; Hu, S.; Chae, Y.K.; Leal, T.A.; Dowell, J.E.; Tsai, M.L.; et al. Blood–based tumor mutational burden as a biomarker for atezolizumab in non–small cell lung cancer: The phase 2 B–F1RST trial. Nat. Med. 2022. [Google Scholar] [CrossRef]
- Zehir, A.; Benayed, R.; Shah, R.H.; Syed, A.; Middha, S.; Kim, H.R.; Srinivasan, P.; Gao, J.; Chakravarty, D.; Devlin, S.M.; et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017, 23, 703–713. [Google Scholar] [CrossRef]
- Budczies, J.; Allgauer, M.; Litchfield, K.; Rempel, E.; Christopoulos, P.; Kazdal, D.; Endris, V.; Thomas, M.; Frohling, S.; Peters, S.; et al. Optimizing panel–based tumor mutational burden (TMB) measurement. Ann. Oncol Off. J. Eur. Soc. Med. Oncol. ESMO 2019, 30, 1496–1506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allgauer, M.; Budczies, J.; Christopoulos, P.; Endris, V.; Lier, A.; Rempel, E.; Volckmar, A.L.; Kirchner, M.; von Winterfeld, M.; Leichsenring, J.; et al. Implementing tumor mutational burden (TMB) analysis in routine diagnostics–a primer for molecular pathologists and clinicians. Transl. Lung Cancer Res. 2018, 7, 703–715. [Google Scholar] [CrossRef] [PubMed]
- Sha, D.; Jin, Z.; Budczies, J.; Kluck, K.; Stenzinger, A.; Sinicrope, F.A. Tumor Mutational Burden as a Predictive Biomarker in Solid Tumors. Cancer Discov. 2020, 10, 1808–1825. [Google Scholar] [CrossRef] [PubMed]
- Svensson, F.; Lang, T.; Johansson, M.E.V.; Hansson, G.C. The central exons of the human MUC2 and MUC6 mucins are highly repetitive and variable in sequence between individuals. Sci. Rep. 2018, 8, 17503. [Google Scholar] [CrossRef] [Green Version]
- Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Germano, G.; Lamba, S.; Rospo, G.; Barault, L.; Magri, A.; Maione, F.; Russo, M.; Crisafulli, G.; Bartolini, A.; Lerda, G.; et al. Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature 2017, 552, 116–120. [Google Scholar] [CrossRef]
- Rospo, G.; Lorenzato, A.; Amirouchene–Angelozzi, N.; Magri, A.; Cancelliere, C.; Corti, G.; Negrino, C.; Amodio, V.; Montone, M.; Bartolini, A.; et al. Evolving neoantigen profiles in colorectal cancers with DNA repair defects. Genome Med. 2019, 11, 42. [Google Scholar] [CrossRef] [Green Version]
- Siravegna, G.; Lazzari, L.; Crisafulli, G.; Sartore–Bianchi, A.; Mussolin, B.; Cassingena, A.; Martino, C.; Lanman, R.B.; Nagy, R.J.; Fairclough, S.; et al. Radiologic and Genomic Evolution of Individual Metastases during HER2 Blockade in Colorectal Cancer. Cancer Cell 2018, 34, 148–162.e7. [Google Scholar] [CrossRef] [Green Version]
- Parikh, K.; Huether, R.; White, K.; Hoskinson, D.; Beaubier, N.; Dong, H.; Adjei, A.A.; Mansfield, A.S. Tumor Mutational Burden From Tumor–Only Sequencing Compared With Germline Subtraction From Paired Tumor and Normal Specimens. JAMA Netw. Open 2020, 3, e200202. [Google Scholar] [CrossRef] [Green Version]
- Stenzinger, A.; Endris, V.; Budczies, J.; Merkelbach–Bruse, S.; Kazdal, D.; Dietmaier, W.; Pfarr, N.; Siebolts, U.; Hummel, M.; Herold, S.; et al. Harmonization and Standardization of Panel–Based Tumor Mutational Burden Measurement: Real–World Results and Recommendations of the Quality in Pathology Study. J. Thorac. Oncol. 2020, 15, 1177–1189. [Google Scholar] [CrossRef]
- Hellmann, M.D.; Nathanson, T.; Rizvi, H.; Creelan, B.C.; Sanchez–Vega, F.; Ahuja, A.; Ni, A.; Novik, J.B.; Mangarin, L.M.B.; Abu–Akeel, M.; et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non–Small–Cell Lung Cancer. Cancer Cell 2018, 33, 843–852.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riaz, N.; Havel, J.J.; Makarov, V.; Desrichard, A.; Urba, W.J.; Sims, J.S.; Hodi, F.S.; Martin–Algarra, S.; Mandal, R.; Sharfman, W.H.; et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell 2017, 171, 934–949.e16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, T.A.; Yarchoan, M.; Jaffee, E.; Swanton, C.; Quezada, S.A.; Stenzinger, A.; Peters, S. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann. Oncol Off. J. Eur. Soc. Med. Oncol. ESMO 2019, 30, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, A.; Sholl, L.; Reardon, B.; Taylor–Weiner, A.; Amin–Mansour, A.; Miao, D.; Liu, D.; Oliver, N.; MacConaill, L.; Ducar, M.; et al. The impact of tumor profiling approaches and genomic data strategies for cancer precision medicine. Genome Med. 2016, 8, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hellmann, M.D.; Paz–Ares, L.; Bernabe Caro, R.; Zurawski, B.; Kim, S.W.; Carcereny Costa, E.; Park, K.; Alexandru, A.; Lupinacci, L.; de la Mora Jimenez, E.; et al. Nivolumab plus Ipilimumab in Advanced Non–Small–Cell Lung Cancer. N. Engl. J. Med. 2019, 381, 2020–2031. [Google Scholar] [CrossRef]
- Ready, N.; Hellmann, M.D.; Awad, M.M.; Otterson, G.A.; Gutierrez, M.; Gainor, J.F.; Borghaei, H.; Jolivet, J.; Horn, L.; Mates, M.; et al. First–Line Nivolumab Plus Ipilimumab in Advanced Non–Small–Cell Lung Cancer (CheckMate 568): Outcomes by Programmed Death Ligand 1 and Tumor Mutational Burden as Biomarkers. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2019, 37, 992–1000. [Google Scholar] [CrossRef]
- Foote, M.B.; Maron, S.B.; Cercek, A.; Argiles, G.; Rousseau, B.; Diaz, L.A., Jr. TMB cut–offs fail to predict benefit of PD–1 blockade in gastroesophageal adenocarcinoma in KEYNOTE–061. Ann. Oncol Off. J. Eur. Soc. Med. Oncol. ESMO 2021, 32, 1188–1189. [Google Scholar] [CrossRef]
- Shitara, K.; Lunceford, J. Response to the letter to the Editor: TMB cut–offs fail to predict benefit of PD–1 blockade in gastroesophageal adenocarcinoma in KEYNOTE–061. Ann. Oncol Off. J. Eur. Soc. Med. Oncol. ESMO 2021, 32, 1303–1304. [Google Scholar] [CrossRef]
- Cristescu, R.; Mogg, R.; Ayers, M.; Albright, A.; Murphy, E.; Yearley, J.; Sher, X.; Liu, X.Q.; Lu, H.; Nebozhyn, M.; et al. Pan–tumor genomic biomarkers for PD–1 checkpoint blockade–based immunotherapy. Science 2018, 362, eaar3593. [Google Scholar] [CrossRef] [Green Version]
- Paz–Ares, L.G.; Ramalingam, S.S.; Ciuleanu, T.E.; Lee, J.S.; Urban, L.; Caro, R.B.; Park, K.; Sakai, H.; Ohe, Y.; Nishio, M.; et al. First–Line Nivolumab Plus Ipilimumab in Advanced NSCLC: 4–Year Outcomes From the Randomized, Open–Label, Phase 3 CheckMate 227 Part 1 Trial. J. Thorac. Oncol. 2022, 17, 289–308. [Google Scholar] [CrossRef]
- Paz–Ares, L.; Ciuleanu, T.E.; Cobo, M.; Schenker, M.; Zurawski, B.; Menezes, J.; Richardet, E.; Bennouna, J.; Felip, E.; Juan–Vidal, O.; et al. First–line nivolumab plus ipilimumab combined with two cycles of chemotherapy in patients with non–small–cell lung cancer (CheckMate 9LA): An international, randomised, open–label, phase 3 trial. Lancet Oncol. 2021, 22, 198–211. [Google Scholar] [CrossRef]
- Paz–Ares, L.; Ciuleanu, T.E.; Cobo, M.; Schenker, M.; Zurawski, B.; Menezes, J.; Richardet, E.; Bennouna, J.; Felip, E.; Juan–Vidal, O.; et al. 98O First–line nivolumab (NIVO) + ipilimumab (IPI) + 2 cycles chemotherapy (chemo) vs 4 cycles chemo in advanced non–small cell lung cancer (aNSCLC): Association of blood and tissue tumor mutational burden (TMB) with efficacy in CheckMate 9LA. J. Thorac. Oncol. 2021, 16, S750–S751. Available online: https://www.jto.org/article/S1556-0864(21)01940-7/fulltext (accessed on 27 February 2022). [CrossRef]
- Kowanetz, M.; Zou, W.; Shames, D.S.; Cummings, C.; Rizvi, N.; Spira, A.I.; Frampton, G.M.; Leveque, V.; Flynn, S.; Mocci, S.; et al. Tumor mutation load assessed by FoundationOne (FM1) is associated with improved efficacy of atezolizumab (atezo) in patients with advanced NSCLC. Ann. Oncol. 2016, 27, vi23. Available online: https://www.annalsofoncology.org/article/S0923-7534(19)43709-5/fulltext (accessed on 27 February 2022). [CrossRef]
- Rizvi, N.A.; Cho, B.C.; Reinmuth, N.; Lee, K.H.; Luft, A.; Ahn, M.J.; van den Heuvel, M.M.; Cobo, M.; Vicente, D.; Smolin, A.; et al. Durvalumab With or Without Tremelimumab vs Standard Chemotherapy in First–line Treatment of Metastatic Non–Small Cell Lung Cancer: The MYSTIC Phase 3 Randomized Clinical Trial. JAMA Oncol. 2020, 6, 661–674. [Google Scholar] [CrossRef] [Green Version]
- Powles, T.; Loriot, Y.; Ravaud, A.; Vogelzang, N.J.; Duran, I.; Retz, M.; De Giorgi, U.; Oudard, S.; Bamias, A.; Koeppen, H.; et al. Atezolizumab (atezo) vs. chemotherapy (chemo) in platinum–treated locally advanced or metastatic urothelial carcinoma (mUC): Immune biomarkers, tumor mutational burden (TMB), and clinical outcomes from the phase III IMvigor211 study. J. Clin. Oncol. 2018, 36, 409. Available online: https://ascopubs.org/doi/abs/10.1200/JCO.2018.36.6_suppl.409 (accessed on 27 February 2022). [CrossRef]
- Gogas, H.; Dreno, B.; Larkin, J.; Demidov, L.; Stroyakovskiy, D.; Eroglu, Z.; Francesco Ferrucci, P.; Pigozzo, J.; Rutkowski, P.; Mackiewicz, J.; et al. Cobimetinib plus atezolizumab in BRAF(V600) wild–type melanoma: Primary results from the randomized phase III IMspire170 study. Ann. Oncol Off. J. Eur. Soc. Med. Oncol. ESMO 2021, 32, 384–394. [Google Scholar] [CrossRef]
- Hodi, F.S.; Wolchok, J.D.; Schadendorf, D.; Larkin, J.; Long, G.V.; Qian, X.; Saci, A.; Young, T.C.; Srinivasan, S.; Chang, H.; et al. TMB and Inflammatory Gene Expression Associated with Clinical Outcomes following Immunotherapy in Advanced Melanoma. Cancer Immunol. Res. 2021, 9, 1202–1213. [Google Scholar] [CrossRef]
| FoCR Study | Design | Purpose |
|---|---|---|
| Phase I [19] | In silico analysis using TCGA data | Validate bioinformatics algorithms. Standardize panel-based TMB estimates by comparing reference WES TMB value. |
| Phase II [20] | Analysis using clinical samples (FFPE tissue) | Evaluate variation between TMB panels. |
| Phase III * | Retrospective analysis of clinical samples with ICI treatment response | Validate cutoffs of TMB for clinical application. |
| Testing Process | Factors Affecting TMB Results | Effects on TMB Estimation |
|---|---|---|
| Sample collection and DNA extraction |
| |
| Sequencing |
| |
| Bioinformatics algorithm |
|
|
| Interpretation and reporting |
|
| Laboratories/Panels | Mutation Type Included | Known Pathogenic Variant Removal | Germline Variant Removal Approach |
|---|---|---|---|
| ACTOnco+ | Non-synonymous + synonymous | Yes | Algorithm-based |
| AZ650 | Non-synonymous + synonymous | No | Matching normal tissue |
| OncoPanel v3.1 | Non-synonymous only | No | Algorithm-based |
| SureSelectXT | Non-synonymous only | No | Algorithm-based |
| FoundationOne CDx | Non-synonymous + synonymous | Yes | Algorithm-based |
| TruSight Oncology (TSO500) | Non-synonymous + synonymous | Yes | Algorithm-based |
| JHOP2 | Non-synonymous + synonymous | Yes | Algorithm-based |
| MSK-IMPACT | Non-synonymous only | No | Matching normal tissue |
| NeoTYPE Discovery Profile for Solid Tumors | Non-synonymous + synonymous | No | Algorithm-based |
| Ion AmpliSeq Comprehensive Cancer Panel | Non-synonymous only | No | Algorithm-based |
| PGDx elio tissue complete | Non-synonymous + synonymous | Yes | Algorithm-based |
| QIAseq TMB panel | Non-synonymous only | No | Algorithm-based |
| Oncomine Comprehensive Assay Plus (OCA Plus) | Non-synonymous only | No | Algorithm-based |
| Oncomine Tumor Mutation Load Assay (OTMLA) | Non-synonymous only | No | Algorithm-based |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sung, M.-T.; Wang, Y.-H.; Li, C.-F. Open the Technical Black Box of Tumor Mutational Burden (TMB): Factors Affecting Harmonization and Standardization of Panel-Based TMB. Int. J. Mol. Sci. 2022, 23, 5097. https://doi.org/10.3390/ijms23095097
Sung M-T, Wang Y-H, Li C-F. Open the Technical Black Box of Tumor Mutational Burden (TMB): Factors Affecting Harmonization and Standardization of Panel-Based TMB. International Journal of Molecular Sciences. 2022; 23(9):5097. https://doi.org/10.3390/ijms23095097
Chicago/Turabian StyleSung, Meng-Ta, Yeh-Han Wang, and Chien-Feng Li. 2022. "Open the Technical Black Box of Tumor Mutational Burden (TMB): Factors Affecting Harmonization and Standardization of Panel-Based TMB" International Journal of Molecular Sciences 23, no. 9: 5097. https://doi.org/10.3390/ijms23095097
APA StyleSung, M.-T., Wang, Y.-H., & Li, C.-F. (2022). Open the Technical Black Box of Tumor Mutational Burden (TMB): Factors Affecting Harmonization and Standardization of Panel-Based TMB. International Journal of Molecular Sciences, 23(9), 5097. https://doi.org/10.3390/ijms23095097







