Profile of Innate Immunity in Gilthead Seabream Larvae Reflects Mortality upon Betanodavirus Reassortant Infection and Replication
Abstract
:1. Introduction
2. Results
2.1. Both NNV Reassortants Produce Mortality in Seabream Larvae
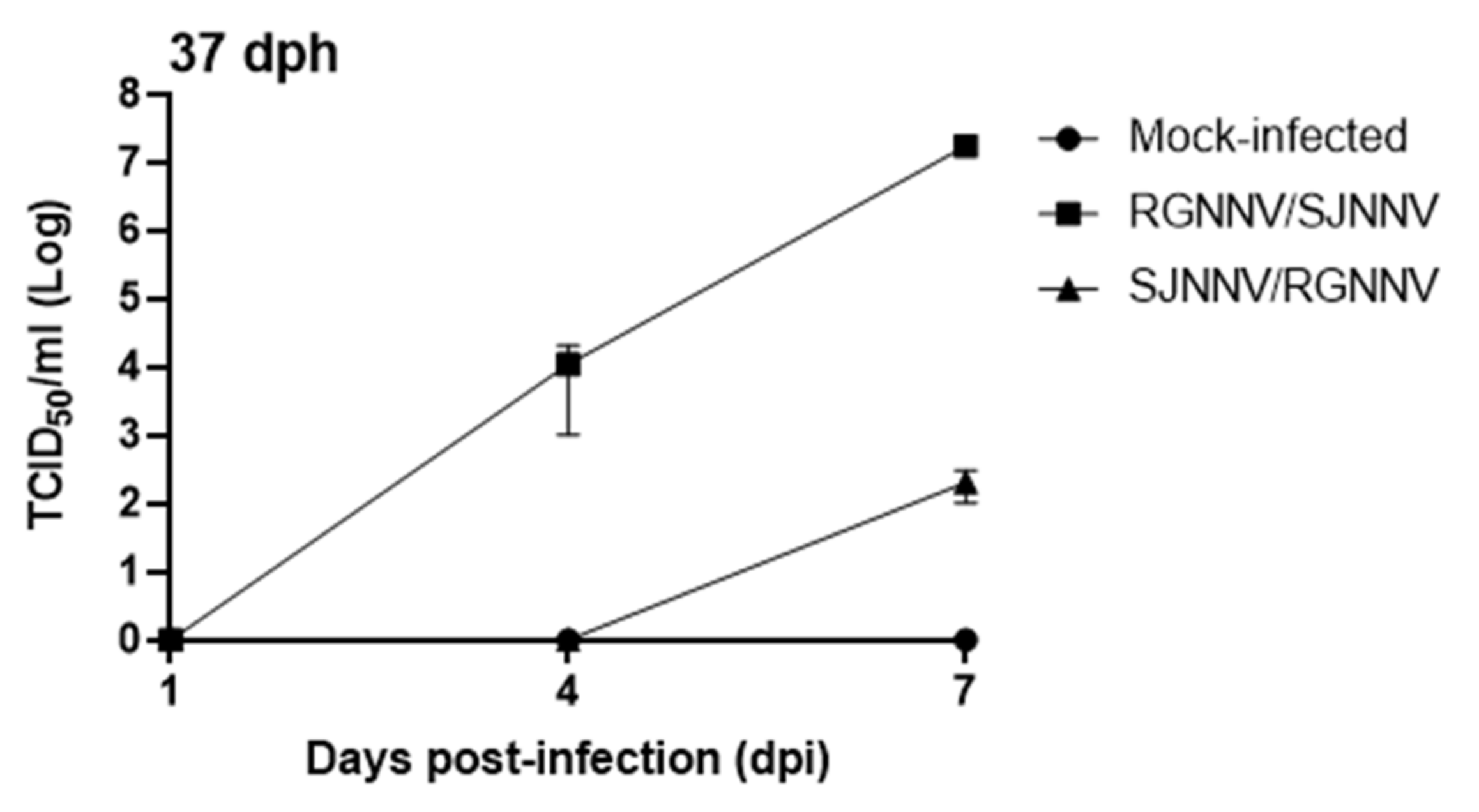
2.2. NNV Reassortants Replicate and Produce Infective Particles in Seabream Larvae
2.3. Nkl Protein Is Increased in 37 dph Larvae Infected by SJNNV/RGNNV
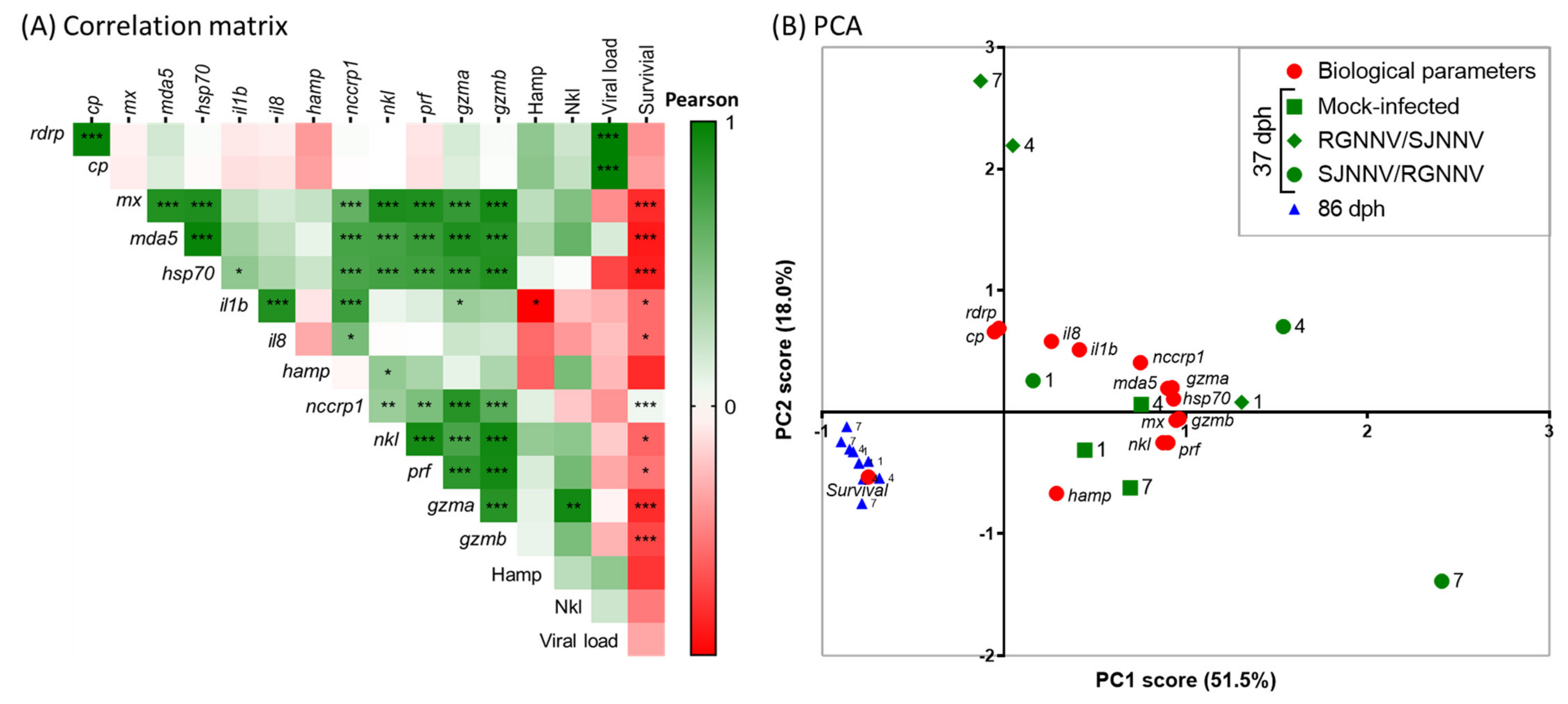
2.4. Larval Age Is More Important than NNV Reassortant or Infection Time Factors
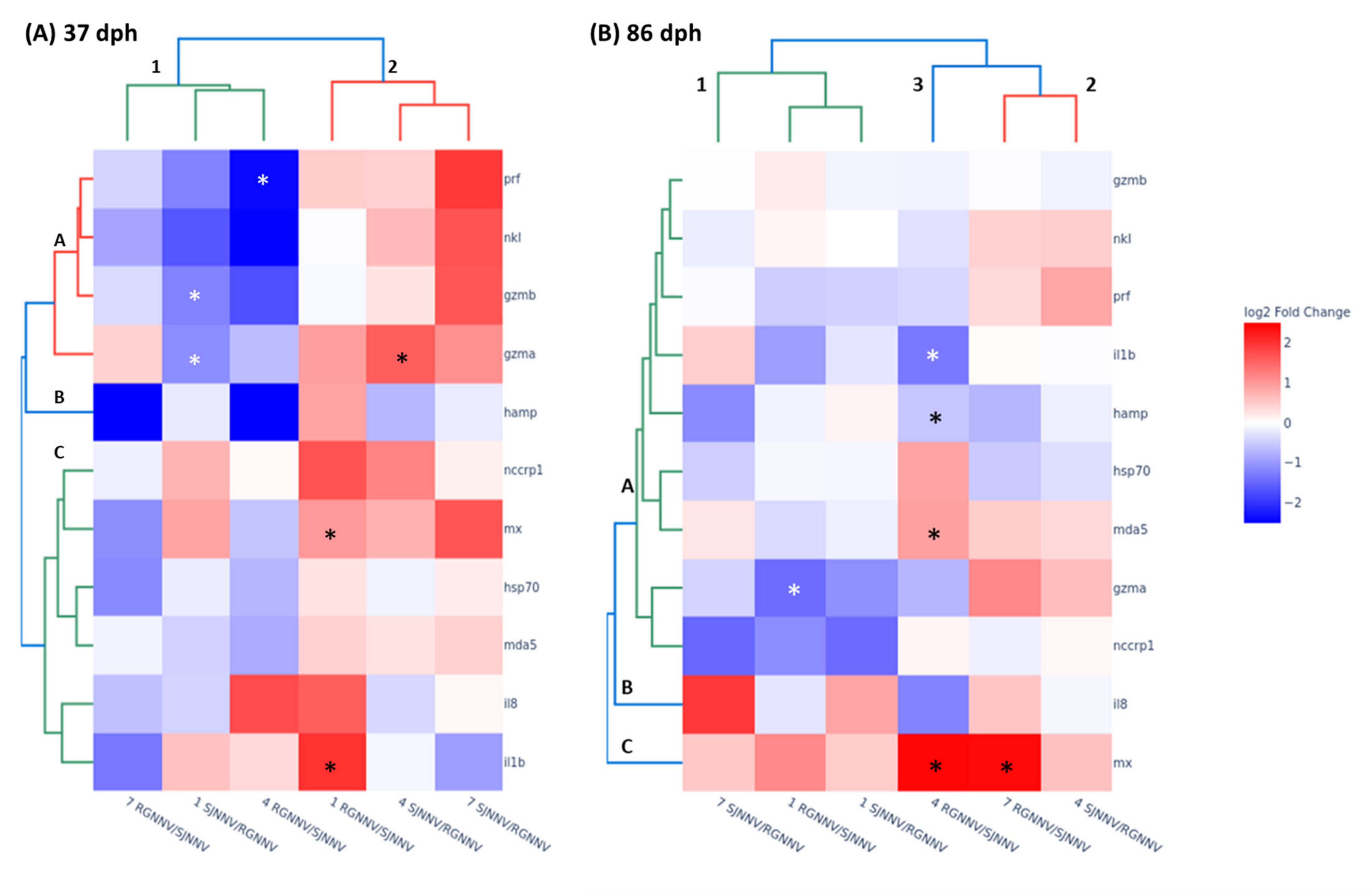
2.5. Immune Genes Are Differently Modulated by NNV Reassortants
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Challenge with Nodavirus and Sampling
4.3. Gene Expression by Real-Time PCR (qPCR)
4.4. Virus Isolation and Titration
4.5. Detection of Hamp and Nkl Proteins by ELISA
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mori, K.-I.; Nakai, T.; Muroga, K.; Arimoto, M.; Mushiake, K.; Furusawa, I. Properties of a new virus belonging to nodaviridae found in larval striped jack (Pseudocaranx dentex) with nervous necrosis. Virology 1992, 187, 368–371. [Google Scholar] [CrossRef]
- Iwamoto, T.; Mise, K.; Takeda, A.; Okinaka, Y.; Mori, K.I.; Arimoto, M.; Okuno, T.; Nakai, T. Characterization of striped jack nervous necrosis virus subgenomic RNA3 and biological activities of its encoded protein B2. J. Gen. Virol. 2005, 86, 2807–2816. [Google Scholar] [CrossRef]
- Nagai, T.; Nishizawa, T. Sequence of the non-structural protein gene encoded by RNA1 of striped jack nervous necrosis virus. J. Gen. Virol. 1999, 80, 3019–3022. [Google Scholar] [CrossRef] [PubMed]
- Bandín, I.; Souto, S. Betanodavirus and VER disease: A 30-year research review. Pathogens 2020, 9, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toffolo, V.; Negrisolo, E.; Maltese, C.; Bovo, G.; Belvedere, P.; Colombo, L.; Valle, L.D. Phylogeny of betanodaviruses and molecular evolution of their RNA polymerase and coat proteins. Mol. Phylogenet. Evol. 2007, 43, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Olveira, J.G.; Souto, S.; Dopazo, C.P.; Thiéry, R.; Barja, J.L.; Bandín, I. Comparative analysis of both genomic segments of betanodaviruses isolated from epizootic outbreaks in farmed fish species provides evidence for genetic reassortment. J. Gen. Virol. 2009, 90, 2940–2951. [Google Scholar] [CrossRef]
- Iwamoto, T.; Okinaka, Y.; Mise, K.; Mori, K.-I.; Arimoto, M.; Okuno, T.; Nakai, T. Identification of host-specificity determinants in betanodaviruses by using reassortants between striped jack nervous necrosis virus and sevenband grouper nervous necrosis virus. J. Virol. 2004, 78, 1256–1262. [Google Scholar] [CrossRef] [Green Version]
- Panzarin, V.; Cappellozza, E.; Mancin, M.; Milani, A.; Toffan, A.; Terregino, C.; Cattoli, G. In vitro study of the replication capacity of the RGNNV and the SJNNV betanodavirus genotypes and their natural reassortants in response to temperature. Vet. Res. 2014, 45, 56. [Google Scholar] [CrossRef] [Green Version]
- Castric, J.; Thiéry, R.; Jeffroy, J.; de Kinkelin, P.; Raymond, J. Sea bream Sparus aurata, an asymptomatic contagious fish host for nodavirus. Dis. Aquat. Organ. 2001, 47, 33–38. [Google Scholar] [CrossRef] [Green Version]
- Chaves-Pozo, E.; Guardiola, F.A.; Meseguer, J.; Esteban, M.A.; Cuesta, A. Nodavirus infection induces a great innate cell-mediated cytotoxic activity in resistant, Gilthead seabream, and susceptible, European sea bass, teleost fish. Fish Shellfish Immunol. 2012, 33, 1159–1166. [Google Scholar] [CrossRef]
- Moreno, P.; Lopez-Jimena, B.; Randelli, E.; Scapigliati, G.; Buonocore, F.; Garcia-Rosado, E.; Borrego, J.J.; Alonso, M.C. Immuno-related gene transcription and antibody response in nodavirus (RGNNV and SJNNV)-infected European sea bass (Dicentrarchus labrax L.). Fish Shellfish Immunol. 2018, 78, 270–278. [Google Scholar] [CrossRef]
- NaveenKumar, S.; Hassan, M.A.; Mahmoud, M.A.; Al-Ansari, A.; Al-Shwared, W.K. Betanodavirus infection in reared marine fishes along the arabian gulf. Aquac. Int. 2017, 25, 1543–1554. [Google Scholar] [CrossRef]
- Toffan, A.; Pascoli, F.; Pretto, T.; Panzarin, V.; Abbadi, M.; Buratin, A.; Quartesan, R.; Gijón, D.; Padrós, F. Viral nervous necrosis in Gilthead sea bream (Sparus aurata) caused by reassortant betanodavirus RGNNV/SJNNV: An emerging threat for Mediterranean aquaculture. Sci. Rep. 2017, 7, 46755. [Google Scholar] [CrossRef]
- Volpe, E.; Gustinelli, A.; Caffara, M.; Errani, F.; Quaglio, F.; Fioravanti, M.L.; Ciulli, S. Viral nervous necrosis outbreaks caused by the RGNNV/SJNNV reassortant betanodavirus in Gilthead sea bream (Sparus aurata) and European sea bass (Dicentrarchus labrax). Aquaculture 2020, 523, 735155. [Google Scholar] [CrossRef]
- Toffan, A.; Biasini, L.; Pretto, T.; Abbadi, M.; Buratin, A.; Franch, R.; Dalla Rovere, G.; Panzarin, V.M.; Marsella, A.; Bargelloni, L.; et al. Age dependency of RGNNV/SJNNV viral encephalo-retinopathy in Gilthead sea bream (Sparus aurata). Aquaculture 2021, 539, 736605. [Google Scholar] [CrossRef]
- Peruzza, L.; Pascoli, F.; Dalla Rovere, G.; Franch, R.; Ferraresso, S.; Babbucci, M.; Biasini, L.; Abbadi, M.; Panzarin, V.; Toffan, A.; et al. Transcriptome analysis reveals a complex response to the RGNNV/SJNNV reassortant nervous necrosis virus strain in sea bream larvae. Fish Shellfish Immunol. 2021, 114, 282–292. [Google Scholar] [CrossRef]
- Chérif, N.; Thiéry, R.; Castric, J.; Biacchesi, S.; Brémont, M.; Thabti, F.; Limem, L.; Hammami, S. Viral encephalopathy and retinopathy of Dicentrarchus labrax and Sparus aurata farmed in tunisia. Vet. Res. Commun. 2009, 33, 345–353. [Google Scholar] [CrossRef]
- Moretti, A.; Pedini Fernández-Criado, M.; Cittolin, G.; Guidastri, R. Manual on Hatchery Production of Seabass and Gilthead Seabream: Volume 1. Food and Agriculture Organization of the United Nations (FAO): Rome, Italy, 1999. [Google Scholar]
- Guo, Y.X.; Chan, S.-W.; Kwang, J. Membrane association of greasy grouper nervous necrosis virus protein A and characterization of its mitochondrial localization targeting signal. J. Virol. 2004, 78, 6498–6508. [Google Scholar] [CrossRef] [Green Version]
- Mézeth, K.B.; Nylund, S.; Henriksen, H.; Patel, S.; Nerland, A.H.; Szilvay, A.M. RNA-dependent RNA polymerase from Atlantic halibut nodavirus contains two signals for localization to the mitochondria. Virus Res. 2007, 130, 43–52. [Google Scholar] [CrossRef]
- Souto, S.; Mérour, E.; Biacchesi, S.; Brémont, M.; Olveira, J.G.; Bandín, I. In vitro and in vivo characterization of molecular determinants of virulence in reassortant betanodavirus. J. Gen. Virol. 2015, 96, 1287–1296. [Google Scholar] [CrossRef] [Green Version]
- Souto, S.; Olveira, J.G.; Dopazo, C.P.; Bandín, I. Reassortant betanodavirus infection in turbot (Scophthalmus maximus). J. Fish Dis. 2016, 39, 1347–1356. [Google Scholar] [CrossRef] [PubMed]
- Souto, S.; Olveira, J.G.; Alonso, M.C.; Dopazo, C.P.; Bandín, I. Betanodavirus infection in bath-challenged Solea senegalensis juveniles: A comparative analysis of RGNNV, SJNNV and reassortant strains. J. Fish Dis. 2018, 41, 1571–1578. [Google Scholar] [CrossRef]
- Chaves-Pozo, E.; Valero, Y.; Lozano, M.T.; Rodríguez-Cerezo, P.; Miao, L.; Campo, V.; Esteban, M.A.; Cuesta, A. Fish granzyme A shows a greater role than granzyme B in fish innate cell-mediated cytotoxicity. Front. Immunol. 2019, 10, 2579. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Salgado, L.; Olveira, J.G.; Dopazo, C.P.; Bandín, I. Effect of rearing density on nervous necrosis virus infection in senegalese sole (Solea senegalensis). J. Fish Dis. 2021, 44, 2003–2012. [Google Scholar] [CrossRef]
- Chang, J.-S.; Chi, S.-C. GHSC70 is involved in the cellular entry of nervous necrosis virus. J. Virol. 2015, 89, 61–70. [Google Scholar] [CrossRef] [Green Version]
- Lu, M.W.; Ngou, F.H.; Chao, Y.M.; Lai, Y.S.; Chen, N.Y.; Lee, F.Y.; Chiou, P.P. Transcriptome characterization and gene expression of Epinephelus spp. in endoplasmic reticulum stress-related pathway during betanodavirus infection in vitro. BMC Genomics 2012, 13, 651. [Google Scholar] [CrossRef] [Green Version]
- De La Vega, E.; Hall, M.R.; Degnan, B.M.; Wilson, K.J. Short-term hyperthermic treatment of Penaeus monodon increases expression of heat shock protein 70 (HSP70) and reduces replication of gill associated virus (GAV). Aquaculture 2006, 253, 82–90. [Google Scholar] [CrossRef]
- Lin, Y.-R.; Hung, H.-C.; Leu, J.-H.; Wang, H.-C.; Kou, G.-H.; Lo, C.-F. The role of aldehyde dehydrogenase and Hsp70 in suppression of white spot syndrome virus replication at high temperature. J. Virol. 2011, 85, 3517–3525. [Google Scholar] [CrossRef] [Green Version]
- Pham, P.H.; Sokeechand, B.S.H.; Hamilton, M.E.; Misk, E.; Jones, G.; Lee, L.E.J.; Lumsden, J.S.; Bols, N.C. VER-155008 induced Hsp70 proteins expression in fish cell cultures while impeding replication of two RNA viruses. Antiviral Res. 2019, 162, 151–162. [Google Scholar] [CrossRef] [Green Version]
- Shan, L.P.; Chen, X.H.; Ling, F.; Zhu, B.; Wang, G.X. Targeting Heat Shock Protein 70 as an antiviral strategy against grass carp reovirus infection. Virus Res. 2018, 247, 1–9. [Google Scholar] [CrossRef]
- Poisa-Beiro, L.; Dios, S.; Ahmed, H.; Vasta, G.R.; Martínez-López, A.; Estepa, A.; Alonso-Gutiérrez, J.; Figueras, A.; Novoa, B. Nodavirus infection of sea bass (Dicentrarchus labrax) induces up-regulation of galectin-1 expression with potential anti-inflammatory activity. J. Immunol. 2009, 183, 6600–6611. [Google Scholar] [CrossRef] [Green Version]
- Poisa-Beiro, L.; Dios, S.; Montes, A.; Aranguren, R.; Figueras, A.; Novoa, B. Nodavirus increases the expression of Mx and inflammatory cytokines in fish brain. Mol. Immunol. 2008, 45, 218–225. [Google Scholar] [CrossRef] [Green Version]
- Chiang, Y.H.; Wu, Y.C.; Chi, S.C. Interleukin-1β secreted from betanodavirus-infected microglia caused the death of neurons in giant grouper brains. Dev. Comp. Immunol. 2017, 70, 19–26. [Google Scholar] [CrossRef]
- Valero, Y.; Arizcun, M.; Esteban, M.Á.; Bandín, I.; Olveira, J.G.; Patel, S.; Cuesta, A.; Chaves-Pozo, E. Nodavirus colonizes and replicates in the testis of Gilthead seabream and European sea bass modulating its immune and reproductive functions. PLoS ONE 2015, 10, e0145131. [Google Scholar] [CrossRef] [Green Version]
- Valero, Y.; Chaves-Pozo, E.; Cuesta, A. NK-Lysin is highly conserved in European sea bass and Gilthead seabream but differentially modulated during the immune response. Fish Shellfish Immunol. 2020, 99, 435–441. [Google Scholar] [CrossRef]
- Ewen, C.L.; Kane, K.P.; Bleackley, R.C. A quarter century of granzymes. Cell Death Differ. 2012, 19, 28–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voskoboinik, I.; Whisstock, J.C.; Trapani, J.A. Perforin and granzymes: Function, dysfunction and human pathology. Nat. Rev. Immunol. 2015, 15, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Praveen, K.; Leary, J.H.; Evans, D.L.; Jaso-Friedmann, L. Nonspecific cytotoxic cells of teleosts are armed with multiple granzymes and other components of the granule exocytosis pathway. Mol. Immunol. 2006, 43, 1152–1162. [Google Scholar] [CrossRef] [PubMed]
- Cordero, H.; Guzmán-Villanueva, L.T.; Chaves-Pozo, E.; Arizcun, M.; Ascencio-Valle, F.; Cuesta, A.; Esteban, M.A. Comparative ontogenetic development of two marine teleosts, Gilthead seabream and European sea bass: New insights into nutrition and immunity. Dev. Comp. Immunol. 2016, 65, 1–7. [Google Scholar] [CrossRef]
- Mulero, I.; Sepulcre, M.P.; Fuentes, I.; García-Alcázar, A.; Meseguer, J.; García-Ayala, A.; Mulero, V. Vaccination of larvae of the bony fish Gilthead seabream reveals a lack of correlation between lymphocyte development and adaptive immunocompetence. Mol. Immunol. 2008, 45, 2981–2989. [Google Scholar] [CrossRef]
- Chia, T.J.; Wu, Y.C.; Chen, J.Y.; Chi, S.C. Antimicrobial peptides (AMP) with antiviral activity against fish nodavirus. Fish Shellfish Immunol. 2010, 28, 434–439. [Google Scholar] [CrossRef]
- Iwamoto, T.; Mise, K.; Mori, K.I.; Arimoto, M.; Nakai, T.; Okuno, T. Establishment of an infectious RNA transcription system for striped jack nervous necrosis virus, the type species of the betanodaviruses. J. Gen. Virol. 2001, 82, 2653–2662. [Google Scholar] [CrossRef]
- Reed, L.J.; Müench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Pfaffl, M.W. Relative Quantification. In Real-Time PCR; Dorak, T., Ed.; Taylor & Francis Group: Abingdon, UK, 2007; Volume 63, pp. 63–82. ISBN 9780203967317. [Google Scholar]
- Cervera, L.; González-Fernández, C.; Arizcun, M.; Cuesta, A.; Chaves-Pozo, E. Severe natural outbreak of Cryptocaryon irritans in Gilthead seabream produces leukocyte mobilization and innate immunity at the gill tissue. Int. J. Mol. Sci. 2022, 23, 937. [Google Scholar] [CrossRef]
- Bradford, M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]



| Factor | dph | NNV | dpi | dph × NNV | dph × dpi | dpi × NNV | dph × dpi × NNV | |
|---|---|---|---|---|---|---|---|---|
| Gene | rdrp | 0.028 | 0.008 | 0.009 | 0.011 | 0.011 | 0.001 | 0.002 |
| cp | 0.107 | 0.003 | 0.003 | 0.140 | 0.082 | 0.000 | 0.073 | |
| mx | 0.000 | 0.036 | 0.563 | 0.036 | 0.563 | 0.246 | 0.246 | |
| mda5 | 0.000 | 0.805 | 0.278 | 0.791 | 0.297 | 0.055 | 0.047 | |
| hsp70 | 0.000 | 0.533 | 0.875 | 0.516 | 0.872 | 0.547 | 0.545 | |
| il1b | 0.000 | 0.795 | 0.000 | 0.791 | 0.000 | 0.951 | 0.950 | |
| il8 | 0.001 | 0.115 | 0.007 | 0.113 | 0.007 | 0.208 | 0.207 | |
| hamp | 0.817 | 0.122 | 0.266 | 0.607 | 0.656 | 0.441 | 0.556 | |
| nkl | 0.000 | 0.165 | 0.200 | 0.184 | 0.106 | 0.119 | 0.109 | |
| nccrp1 | 0.000 | 0.168 | 0.004 | 0.165 | 0.004 | 0.081 | 0.082 | |
| prf | 0.000 | 0.112 | 0.923 | 0.323 | 0.314 | 0.010 | 0.014 | |
| gzma | 0.000 | 0.237 | 0.968 | 0.159 | 0.717 | 0.005 | 0.012 | |
| gzmb | 0.000 | 0.093 | 0.441 | 0.090 | 0.485 | 0.044 | 0.047 | |
| Protein | Hamp | ND | 0.907 | 0.007 | ND | ND | 0.481 | ND |
| Nkl | ND | 0.004 | 0.759 | ND | ND | 0.146 | ND | |
| Viral load | ND | 0.000 | 0.000 | ND | 0.000 | 0.000 | 0.000 | |
| Survival | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.001 | 0.000 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Álvarez, M.Á.; Arizcun, M.; Chaves-Pozo, E.; Cuesta, A. Profile of Innate Immunity in Gilthead Seabream Larvae Reflects Mortality upon Betanodavirus Reassortant Infection and Replication. Int. J. Mol. Sci. 2022, 23, 5092. https://doi.org/10.3390/ijms23095092
García-Álvarez MÁ, Arizcun M, Chaves-Pozo E, Cuesta A. Profile of Innate Immunity in Gilthead Seabream Larvae Reflects Mortality upon Betanodavirus Reassortant Infection and Replication. International Journal of Molecular Sciences. 2022; 23(9):5092. https://doi.org/10.3390/ijms23095092
Chicago/Turabian StyleGarcía-Álvarez, Miguel Ángel, Marta Arizcun, Elena Chaves-Pozo, and Alberto Cuesta. 2022. "Profile of Innate Immunity in Gilthead Seabream Larvae Reflects Mortality upon Betanodavirus Reassortant Infection and Replication" International Journal of Molecular Sciences 23, no. 9: 5092. https://doi.org/10.3390/ijms23095092
APA StyleGarcía-Álvarez, M. Á., Arizcun, M., Chaves-Pozo, E., & Cuesta, A. (2022). Profile of Innate Immunity in Gilthead Seabream Larvae Reflects Mortality upon Betanodavirus Reassortant Infection and Replication. International Journal of Molecular Sciences, 23(9), 5092. https://doi.org/10.3390/ijms23095092







