The RpTOE1-RpFT Module Is Involved in Rejuvenation during Root-Based Vegetative Propagation in Robinia pseudoacacia
Abstract
1. Introduction
2. Results
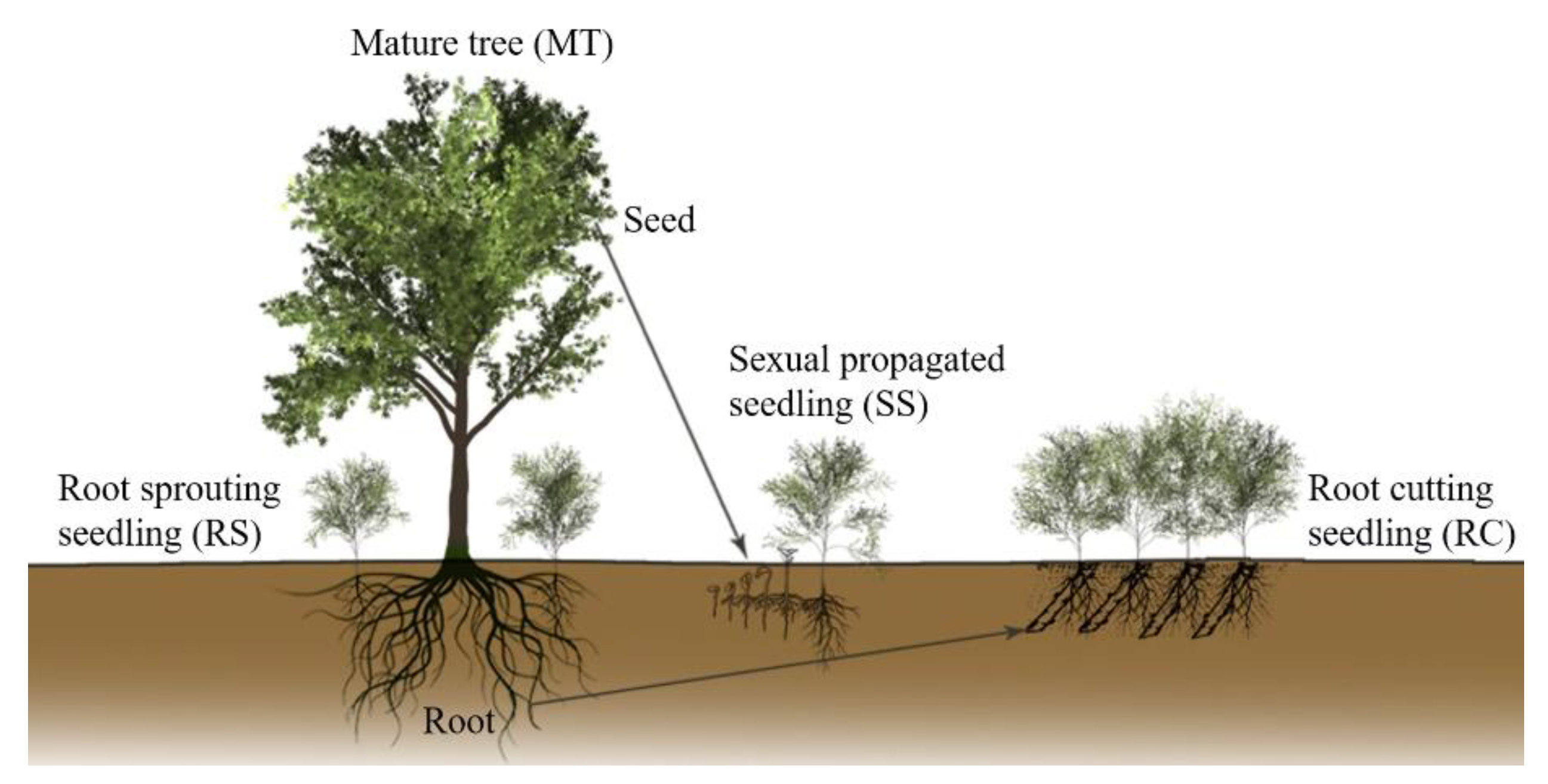
2.1. Experimental Design for Exploring the Genetic Regulation of Rejuvenation during Vegetative Propagation
2.2. Comparative Transcriptome Profiles between MTs and Seedlings over Two Years
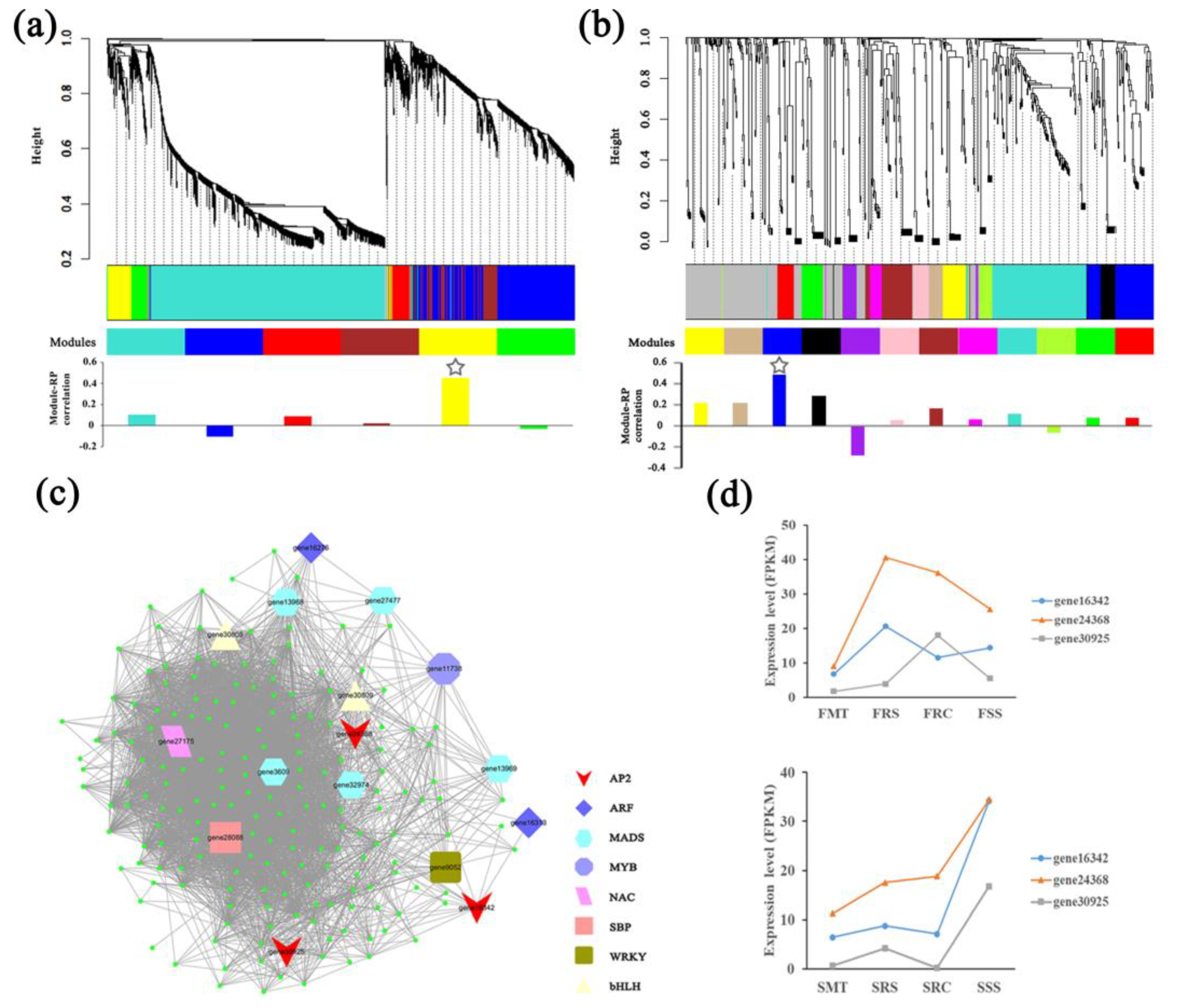
2.3. Identification of Gene and miRNA Co-Expression Modules Related to Vegetative Propagation in R. pseudoacacia
2.4. Identification and Characterization of RpTOE1
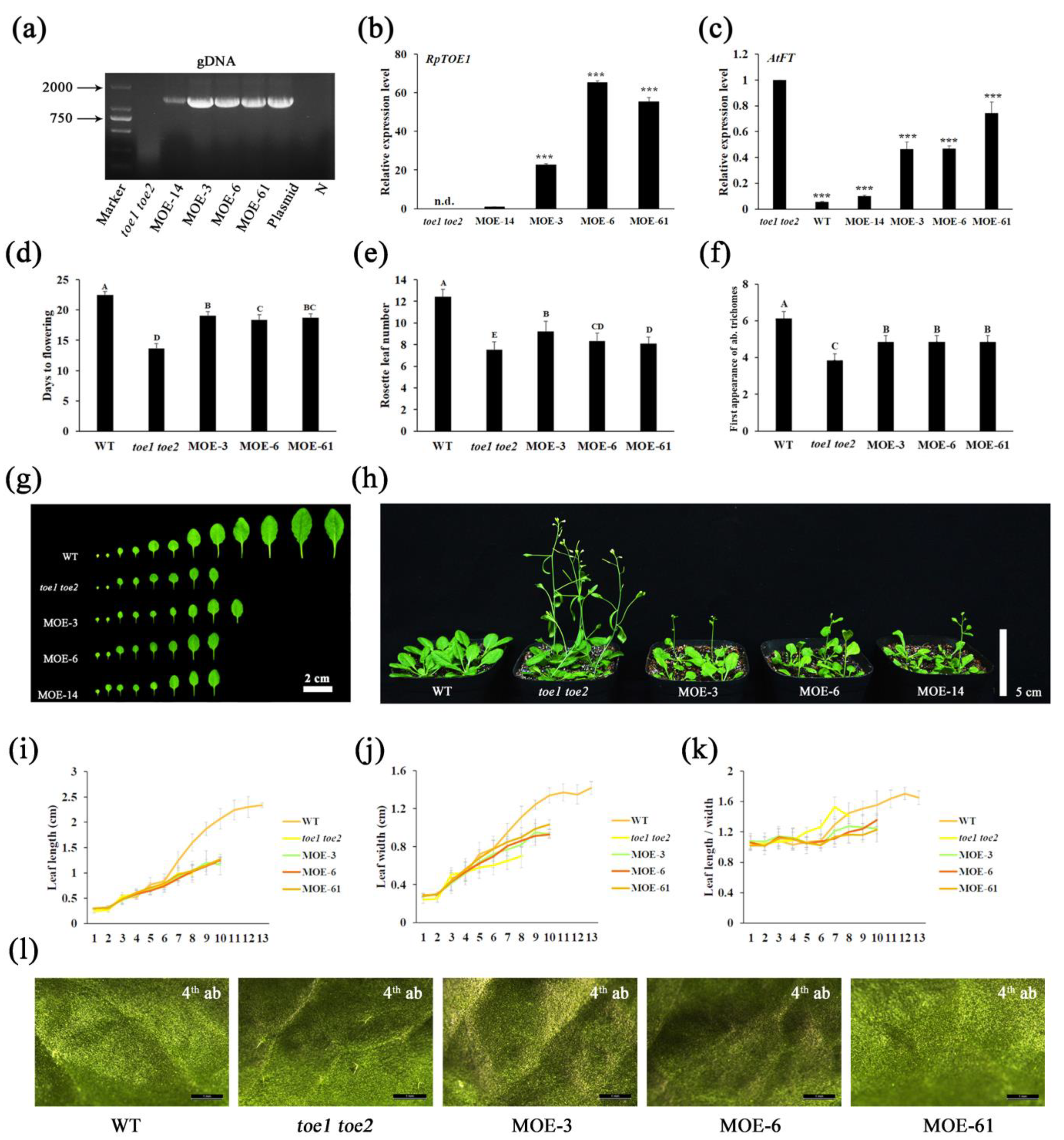
2.5. Heterologous Overexpression of RpTOE1 Prolongs the Vegetative Stage in Arabidopsis
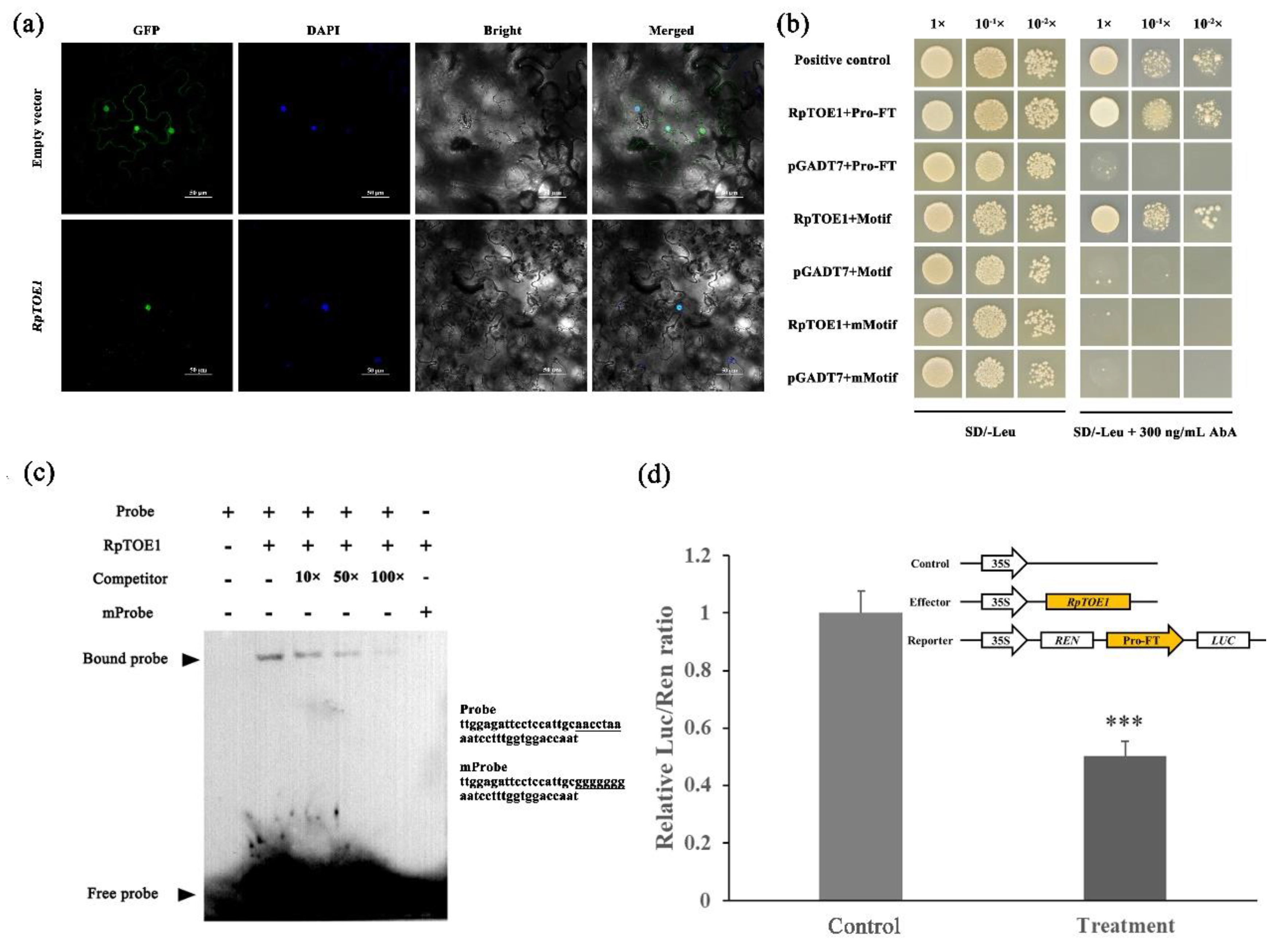
2.6. RpTOE1 Can Directly Bind to the Promoter of RpFT and Inhibit Its Expression
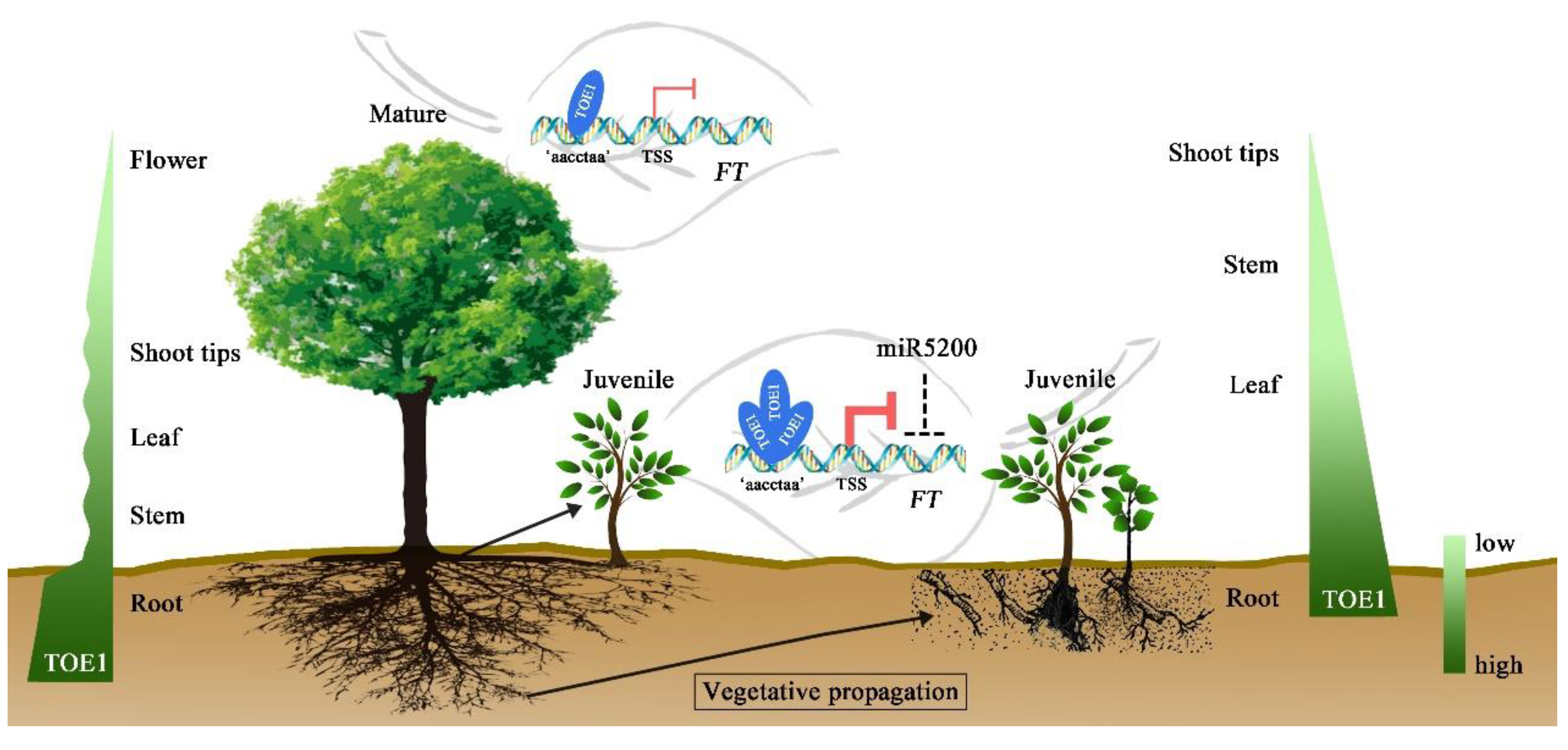
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. RNA Isolation, Sequencing, and Data Analysis
4.3. sRNA Sequencing and Bioinformatic Analysis
4.4. Weighted Gene Co-Expression Network Construction and Module Detection
4.5. Phylogenetic Analysis
4.6. Subcellular Localization Assay
4.7. Electrophoretic Mobility Shift Assay (EMSA)
4.8. Yeast One-Hybrid (Y1H) Assays
4.9. Dual-Luciferase Reporter Assays
4.10. RNA Isolation and Reverse-Transcription Quantitative PCR (qRT-PCR)
4.11. Generation of Transgenic Arabidopsis
4.12. Availability of Data and Materials
4.13. Accession Numbers
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wallace, E.R. Vegetative Propagation. Nature 1967, 215, 1424. [Google Scholar] [CrossRef][Green Version]
- Amano, R.; Nakayama, H.; Momoi, R.; Omata, E.; Gunji, S.; Takebayashi, Y.; Kojima, M.; Ikematsu, S.; Ikeuchi, M.; Iwase, A.; et al. Molecular Basis for Natural Vegetative Propagation via Regeneration in North American Lake Cress, Rorippa aquatica (Brassicaceae). Plant Cell Physiol. 2019, 61, 353–369. [Google Scholar] [CrossRef] [PubMed]
- Kharrazi, M.; Tehranifar, A.; Nemati, H.; Bagheri, A. Vegetative Propagation of Amaryllis (Hippeastrum × johnsonii ) by Different Cutting Methods. Korean J. Hortic. Sci. 2017, 35, 373–380. [Google Scholar] [CrossRef]
- Di Battista, F.; Maccario, D.; Beruto, M.; Grauso, L.; Lanzotti, V.; Curir, P.; Monroy, F. Metabolic changes associated to the unblocking of adventitious root formation in aged, rooting-recalcitrant cuttings of Eucalyptus gunnii Hook. f. (Myrtaceae). Plant Growth Regul. 2019, 89, 73–82. [Google Scholar] [CrossRef]
- Smulders, M.J.M.; de Klerk, G.J. Epigenetics in plant tissue culture. Plant Growth Regul. 2011, 63, 137–146. [Google Scholar] [CrossRef]
- Day, M.E.; Greenwood, M.S.; Diaz-Sala, C. Age- and size-related trends in woody plant shoot development: Regulatory pathways and evidence for genetic control. Tree Physiol. 2002, 22, 507–513. [Google Scholar] [CrossRef]
- Poethig, R.S. Phase Change and the Regulation of Developmental Timing in Plants. Science 2003, 301, 334–336. [Google Scholar] [CrossRef]
- Rezq, B.-S. Juvenility, maturity, and rejuvenation in woody plants. Herbron Univ. Res. J. 2007, 3, 17–43. [Google Scholar]
- Jia, X.L.; Chen, Y.K.; Xu, X.Z.; Shen, F.; Zheng, Q.B.; Du, Z.; Wang, Y.; Wu, T.; Xu, X.F.; Han, Z.H.; et al. miR156 switches on vegetative phase change under the regulation of redox signals in apple seedlings. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Huang, H.-J.; Chen, Y.; Kuo, J.-L.; Kuo, T.-T.; Tzeng, C.-C.; Huang, B.-L.; Chen, C.-M.; Huang, L.-C. Rejuvenation of Sequoia sempervirens in Vitro: Changes in Isoesterases and Isoperoxidases. Plant Cell Physiol. 1996, 37, 77–80. [Google Scholar] [CrossRef]
- Huang, L.C.; Lius, S.; Huang, B.L.; Murashige, T.; Mahdi, E.F.M.; Van Gundy, R. Rejuvenation of Sequoia sempervirens by repeated grafting of shoot tips onto juvenile rootstocks in vitro: Model for phase reversal of trees. Plant Physiol. 1992, 98, 166–173. [Google Scholar] [CrossRef]
- Zhang, Z.; Sun, Y.; Li, Y. Plant rejuvenation: From phenotypes to mechanisms. Plant Cell Rep. 2020, 39, 1249–1262. [Google Scholar] [CrossRef]
- Conner, A.J.; Searle, H.; Jacobs, J.M.E. Rejuvenation of chicory and lettuce plants following phase change in tissue culture. BMC Biotechnol. 2019, 19, 65. [Google Scholar] [CrossRef]
- Liu, H.; Gao, Y.; Song, X.; Ma, Q.; Zhang, J.; Pei, D. A novel rejuvenation approach to induce endohormones and improve rhizogenesis in mature Juglans tree. Plant Methods 2018, 14, 13. [Google Scholar] [CrossRef]
- Díaz-Sala, C. Direct reprogramming of adult somatic cells toward adventitious root formation in forest tree species: The effect of the juvenile-adult transition. Front. Plant Sci. 2014, 5, 310. [Google Scholar] [CrossRef]
- Chen, Y.T.; Shen, C.H.; Lin, W.D.; Chu, H.A.; Huang, B.L.; Kuo, C.I.; Yeh, K.W.; Huang, L.C.; Chang, I.F. Small RNAs of Sequoia sempervirens during rejuvenation and phase change. Plant Biol. 2013, 15, 27–36. [Google Scholar] [CrossRef]
- Wang, J.-W.; Czech, B.; Weigel, D. miR156-Regulated SPL Transcription Factors Define an Endogenous Flowering Pathway in Arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.-W.; Weigel, D.; Poethig, R.S. The Sequential Action of miR156 and miR172 Regulates Developmental Timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef]
- Heide, O.M. Juvenility, maturation and rejuvenation in plants: Adventitious bud formation as a novel rejuvenation process. J. Hortic. Sci. Biotechnol. 2018, 94, 2–11. [Google Scholar] [CrossRef]
- Zhang, T.-Q.; Lian, H.; Tang, H.; Dolezal, K.; Zhou, C.M.; Yu, S.; Chen, J.H.; Chen, Q.; Liu, H.; Ljung, K.; et al. An intrinsic microRNA timer regulates progressive decline in shoot regenerative capacity in plants. Plant Cell 2015, 27, 349–360. [Google Scholar] [CrossRef]
- Yu, S.; Lian, H.; Wang, J.-W. Plant developmental transitions: The role of microRNAs and sugars. Curr. Opin. Plant Biol. 2015, 27, 1–7. [Google Scholar] [CrossRef]
- Kim, J.J.; Lee, J.H.; Kim, W.; Jung, H.S.; Huijser, P.; Ahn, J.H. The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module regulates ambient temperature-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Plant Physiol. 2012, 159, 461–478. [Google Scholar] [CrossRef]
- Spanudakis, E.; Jackson, S. The role of microRNAs in the control of flowering time. J. Exp. Bot. 2014, 65, 365–380. [Google Scholar] [CrossRef]
- Teotia, S.; Tang, G. To Bloom or Not to Bloom: Role of MicroRNAs in Plant Flowering. Mol. Plant 2015, 8, 359–377. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-W. Regulation of flowering time by the miR156-mediated age pathway. J. Exp. Bot. 2014, 65, 4723–4730. [Google Scholar] [CrossRef]
- Schmid, M.; Uhlenhaut, H.; Godard, F.; Demar, M.; Bressan, R.; Weigel, D.; Lohmann, J. Dissection of floral induction pathways using global expression analysis. Development 2003, 130, 6001–6012. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Seo, Y.H.; Seo, P.J.; Reyes, J.L.; Yun, J.; Chua, N.H.; Park, C.M. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 2007, 19, 2736–2748. [Google Scholar] [CrossRef] [PubMed]
- Lian, H.; Wang, L.; Ma, N.; Zhou, C.-M.; Han, L.; Zhang, T.-Q.; Wang, J.-W. Redundant and specific roles of individual MIR172 genes in plant development. PLoS Biol. 2021, 19, e3001044. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, C.; Mai, Y.; Li, L.; Gao, J.; Shang, G.; Lian, H.; Han, L.; Zhang, T.-Q.; Tang, H.; et al. A spatiotemporally regulated transcriptional complex underlies heteroblastic development of leaf hairs in Arabidopsis thaliana. EMBO J. 2019, 38. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, L.; Zeng, L.; Zhang, C.; Ma, H. Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev. 2015, 29, 975–987. [Google Scholar] [CrossRef]
- Zhai, Q.; Zhang, X.; Wu, F.; Feng, H.; Deng, L.; Xu, L.; Zhang, M.; Wang, Q.; Li, C. Transcriptional Mechanism of Jasmonate Receptor COI1-Mediated Delay of Flowering Time in Arabidopsis. Plant Cell 2015, 27, 2814–2828. [Google Scholar] [CrossRef]
- Werner, S.; Bartrina, I.; Schmülling, T. Cytokinin regulates vegetative phase change in Arabidopsis thaliana through the miR172/TOE1-TOE2 module. Nat. Commun. 2021, 12, 5816. [Google Scholar] [CrossRef]
- Aukerman, M.J.; Sakai, H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef]
- Chen, X. A MicroRNA as a Translational Repressor of APETALA2 in Arabidopsis Flower Development. Science 2004, 303, 2022–2025. [Google Scholar] [CrossRef]
- Mathieu, J.; Yant, L.; Muerdter, F.; Küttner, F.; Schmid, M. Repression of Flowering by the miR172 Target SMZ. PLoS Biol. 2009, 7, e1000148. [Google Scholar] [CrossRef]
- Yant, L.; Mathieu, J.; Dinh, T.T.; Ott, F.; Lanz, C.; Wollmann, H.; Chen, X.; Schmid, M. Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 2010, 22, 2156–2170. [Google Scholar] [CrossRef]
- Sun, L.; Zhu, Z. The Molecular Basis of Age-Modulated Plant De Novo Root Regeneration Decline in Arabidopsis thaliana. Plant Cell Physiol. 2020, 62, 3–7. [Google Scholar] [CrossRef]
- Luo, Y.; Guo, Z.; Li, L. Evolutionary conservation of microRNA regulatory programs in plant flower development. Dev. Biol. 2013, 380, 133–144. [Google Scholar] [CrossRef]
- Wang, P.; Cheng, T.; Lu, M.; Liu, G.; Li, M.; Shi, J.; Lu, Y.; Laux, T.; Chen, J. Expansion and Functional Divergence of AP2 Group Genes in Spermatophytes Determined by Molecular Evolution and Arabidopsis Mutant Analysis. Front. Plant Sci. 2016, 7, 1383. [Google Scholar] [CrossRef]
- Wang, J.-W.; Park, M.Y.; Wang, L.-J.; Koo, Y.; Chen, X.-Y.; Weigel, D.; Poethig, R.S. MiRNA Control of Vegetative Phase Change in Trees. PLoS Genet. 2011, 7, e1002012. [Google Scholar] [CrossRef]
- Tang, M.; Bai, X.; Niu, L.-J.; Chai, X.; Chen, M.-S.; Xu, Z.-F. miR172 Regulates both Vegetative and Reproductive Development in the Perennial Woody Plant Jatropha curcas. Plant Cell Physiol. 2018, 59, 2549–2563. [Google Scholar] [CrossRef]
- Lawrence, E.H.; Leichty, A.R.; Doody, E.E.; Ma, C.; Strauss, S.H.; Poethig, R.S. Vegetative phase change in Populus tremula × alba. New Phytol. 2021, 231, 351–364. [Google Scholar] [CrossRef]
- Li, H.; Zhao, X.; Dai, H.; Wu, W.; Mao, W.; Zhang, Z. Tissue Culture Responsive MicroRNAs in Strawberry. Plant Mol. Biol. Rep. 2012, 30, 1047–1054. [Google Scholar] [CrossRef]
- Ye, B.; Zhang, K.; Wang, J. The role of miR156 in rejuvenation in Arabidopsis thaliana. J. Integr. Plant Biol. 2019, 62, 550–555. [Google Scholar] [CrossRef]
- Cheng, Y.-J.; Shang, G.-D.; Xu, Z.-G.; Yu, S.; Wu, L.-Y.; Zhai, D.; Tian, S.-L.; Gao, J.; Wang, L.; Wang, J.-W. Cell division in the shoot apical meristem is a trigger for miR156 decline and vegetative phase transition in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Vidoy-Mercado, I.; Narváez, I.; Palomo-Ríos, E.; Litz, R.; Barceló-Muñoz, A.; Pliego-Alfaro, F. Reinvigoration/Rejuvenation Induced through Micrografting of Tree Species: Signaling through Graft Union. Plants 2021, 10, 1197. [Google Scholar] [CrossRef]
- George, E.F.; Hall, M.A.; De Klerk, G.J. Stock plant physiological factors affecting growth and morphogenesis. In Plant Propagation by Tissue Culture; George, E.F., Hall, M.A., De Klerk, G., Eds.; Springer: Dordrecht, The Netherlands, 2008; pp. 403–422. [Google Scholar]
- Shanthi, K.; Bachpai, V.K.W.; Anisha, S.; Ganesan, M.; Anithaa, R.G.; Subashini, V.; Chakravarthi, M.; Sivakumar, V.; Yasodha, R. Micropropagation of Eucalyptus camaldulensis for the production of rejuvenated stock plants for microcuttings propagation and genetic fidelity assessment. New For. 2014, 46, 357–371. [Google Scholar] [CrossRef]
- Pierik, R.L.M. In Vitro Culture of Higher Plants; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1997. [Google Scholar]
- Deuber, C.G.; Farrar, J.L. Vegetative propagation of Norway spruce. J. For. 1940, 38, 578–585. [Google Scholar] [CrossRef]
- Xiao, Z.; Ji, N.; Zhang, X.; Zhang, Y.; Wang, Y.; Wu, T.; Xu, X.; Han, Z. The lose of juvenility elicits adventitious rooting recalcitrance in apple rootstocks. Plant Cell Tissue Organ Cult. 2014, 119, 51–63. [Google Scholar] [CrossRef]
- Nicolescu, V.-N.; Hernea, C.; Bakti, B.; Keserű, Z.; Antal, B.; Rédei, K. Black locust (Robinia pseudoacacia L.) as a multi-purpose tree species in Hungary and Romania: A review. J. For. Res. 2018, 29, 1449–1463. [Google Scholar] [CrossRef]
- Panchuk, I.I.; Zentgraf, U.; Volkov, R.A. Expression of the Apx gene family during leaf senescence of Arabidopsis thaliana. Planta 2005, 222, 926–932. [Google Scholar] [CrossRef]
- Selbach, M.; Schwanhäusser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008, 455, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Iwase, A.; Mitsuda, N.; Koyama, T.; Hiratsu, K.; Kojima, M.; Arai, T.; Inoue, Y.; Seki, M.; Sakakibara, H.; Sugimoto, K.; et al. The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr. Biol. 2011, 21, 508–514. [Google Scholar] [CrossRef]
- Ye, B.B.; Shang, G.D.; Pan, Y.; Xu, Z.G.; Wang, J.W. AP2/ERF transcription factors integrate age and wound signals for root regeneration. Plant Cell 2020, 32, 226–241. [Google Scholar] [CrossRef]
- Winkelmann, T.; Spethmann, W.; Seegert, A. Propagation of 285-year-old alley linden (Tilia × vulgaris) trees via long cuttings. Eur. J. Hortic. Sci. 2020, 85, 160–168. [Google Scholar] [CrossRef]
- Prewitt, S.F.; Shalit-Kaneh, A.; Maximova, S.N.; Guiltinan, M.J. Inter-species functional compatibility of the Theobroma cacao and Arabidopsis FT orthologs: 90 million years of functional conservation of meristem identity genes. BMC Plant Biol. 2021, 21, 218. [Google Scholar] [CrossRef]
- Du, S.-S.; Li, L.; Li, L.; Wei, X.; Xu, F.; Xu, P.; Wang, W.; Xu, P.; Cao, X.; Miao, L.; et al. Photoexcited Cryptochrome2 Interacts Directly with TOE1 and TOE2 in Flowering Regulation. Plant Physiol. 2020, 184, 487–505. [Google Scholar] [CrossRef]
- Wu, L.; Liu, D.; Wu, J.; Zhang, R.; Qin, Z.; Li, A.; Fu, D.; Zhai, W.; Mao, L. Regulation of FLOWERING LOCUS T by a MicroRNA in Brachypodium distachyon. Plant Cell 2013, 25, 4363–4377. [Google Scholar] [CrossRef]
- Zhu, Q.-H.; Helliwell, C.A. Regulation of flowering time and floral patterning by miR172. J. Exp. Bot. 2010, 62, 487–495. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, J.; Sun, Y.; Wang, S.; Xing, X.; Feng, X.; Pérez-Pérez, J.M.; Li, Y. Genome-wide high-resolution mapping of DNA methylation reveals epigenetic variation in the offspring of sexual and asexual propagation in Robinia pseudoacacia. Plant Cell Rep. 2021, 1–13. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Cheadle, C.; Vawter, M.P.; Freed, W.J.; Becker, K.G. Analysis of Microarray Data Using Z Score Transformation. J. Mol. Diagn. 2003, 5, 73–81. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Liu, X.; Hu, A.; Zhao, J.; Chen, F. Identification of Key Gene Modules in Human Osteosarcoma by Co-Expression Analysis Weighted Gene Co-Expression Network Analysis (WGCNA). J. Cell. Biochem. 2017, 118, 3953–3959. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2016, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Liu, J.; Cao, S.; Guo, Q.; Sun, Y.; Niu, D.; Long, C.; Fan, Y.; Li, Y. The RpTOE1-RpFT Module Is Involved in Rejuvenation during Root-Based Vegetative Propagation in Robinia pseudoacacia. Int. J. Mol. Sci. 2022, 23, 5079. https://doi.org/10.3390/ijms23095079
Zhang Z, Liu J, Cao S, Guo Q, Sun Y, Niu D, Long C, Fan Y, Li Y. The RpTOE1-RpFT Module Is Involved in Rejuvenation during Root-Based Vegetative Propagation in Robinia pseudoacacia. International Journal of Molecular Sciences. 2022; 23(9):5079. https://doi.org/10.3390/ijms23095079
Chicago/Turabian StyleZhang, Zijie, Jie Liu, Sen Cao, Qi Guo, Yuhan Sun, Dongsheng Niu, Cui Long, Yingming Fan, and Yun Li. 2022. "The RpTOE1-RpFT Module Is Involved in Rejuvenation during Root-Based Vegetative Propagation in Robinia pseudoacacia" International Journal of Molecular Sciences 23, no. 9: 5079. https://doi.org/10.3390/ijms23095079
APA StyleZhang, Z., Liu, J., Cao, S., Guo, Q., Sun, Y., Niu, D., Long, C., Fan, Y., & Li, Y. (2022). The RpTOE1-RpFT Module Is Involved in Rejuvenation during Root-Based Vegetative Propagation in Robinia pseudoacacia. International Journal of Molecular Sciences, 23(9), 5079. https://doi.org/10.3390/ijms23095079






