Machine Learning and Pathway Analysis-Based Discovery of Metabolomic Markers Relating to Chronic Pain Phenotypes
Abstract
:1. Introduction
2. Results
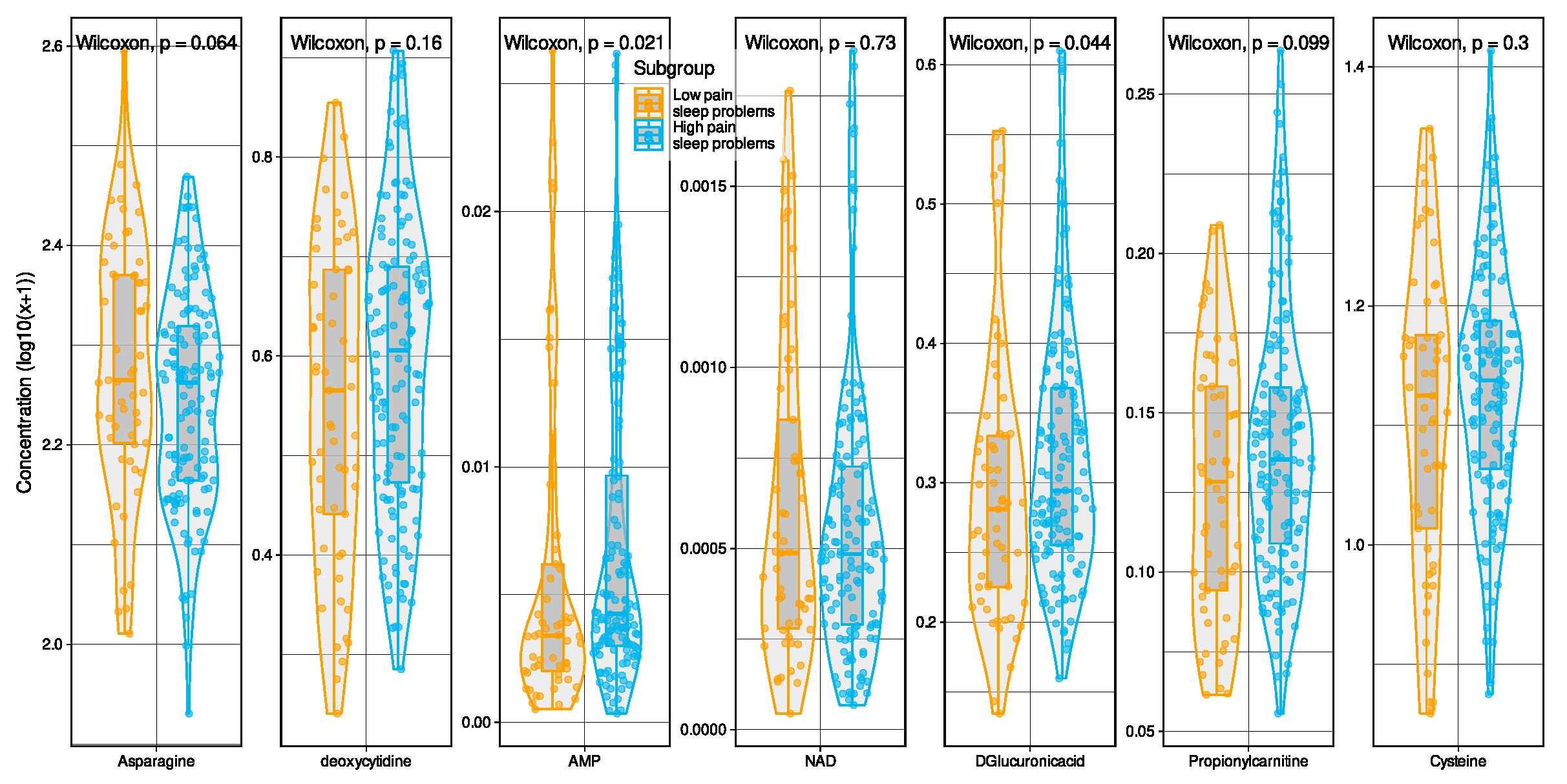
2.1. Metabolomic Markers Informative for Pain Phenotype Assignment
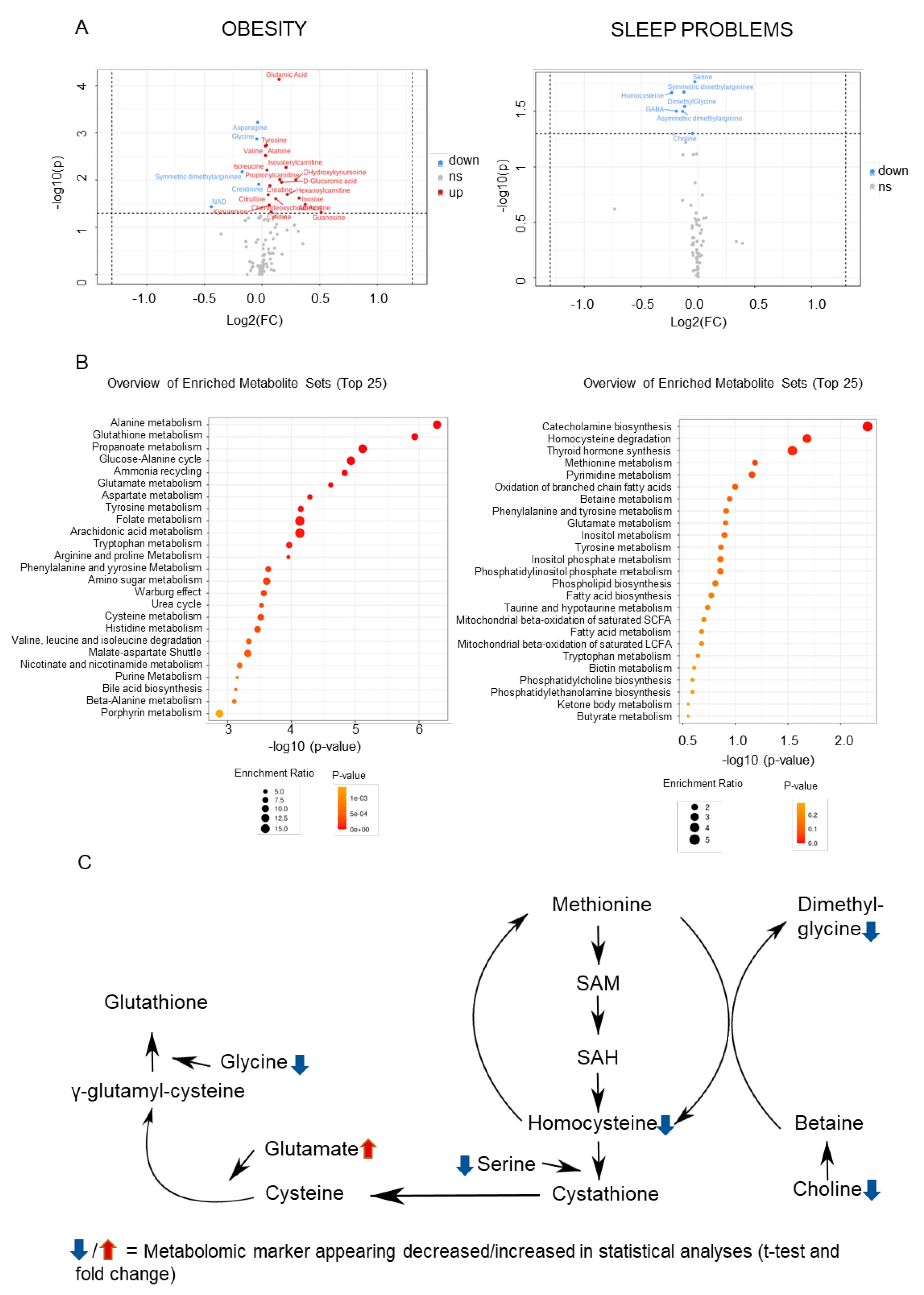
2.2. Metabolomic Markers Relevant to Obesity and Sleep
2.3. Convergences in the Findings between the Machine-Learning Approach and Pathway Analyses
3. Discussion
Strengths and Limitations
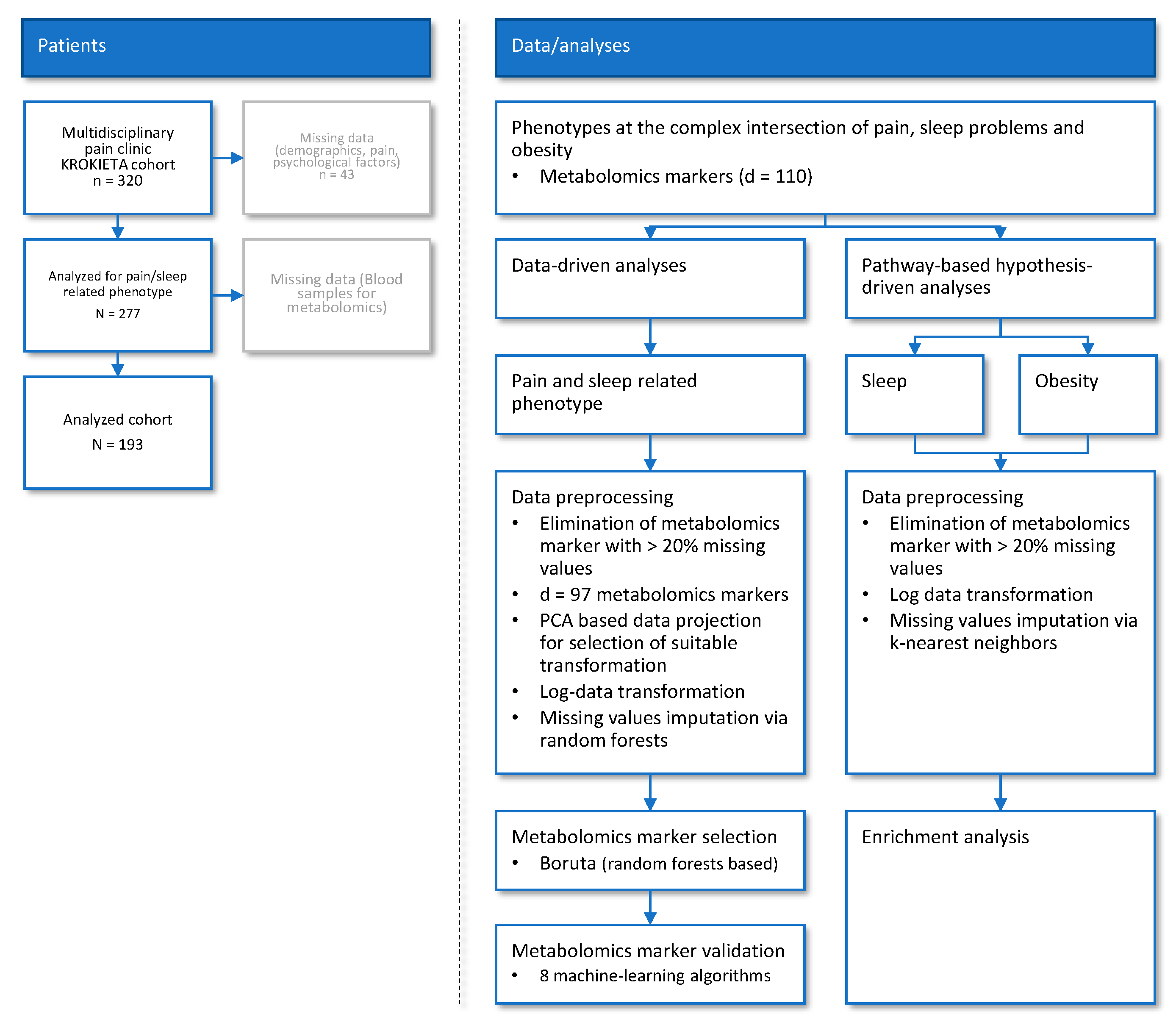
4. Material and Methods
4.1. Subjects and Study Design
4.2. Pain-Related Phenotypes
4.3. Sleep and Obesity Parameters
4.4. Serum Metabolomic Markers
4.5. Data Analysis
4.5.1. Data and Analysis Strategy
4.5.2. Data-Driven Association of Metabolomic Markers and Pain Phenotypes
Data Preprocessing and Transformation
Selection of Metabolomic Markers Informative for Pain−Phenotype Assignment
Validation of Metabolomic Markers Informative for Pain Phenotype Assignment
4.5.3. Pathway-Based Assessment of Metabolomic Markers Relevant to Sleep and Obesity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wishart, D.S. Metabolomics for Investigating Physiological and Pathophysiological Processes. Physiol. Rev. 2019, 99, 1819–1875. [Google Scholar] [CrossRef]
- Adams, S.B., Jr.; Setton, L.A.; Kensicki, E.; Bolognesi, M.P.; Toth, A.P.; Nettles, D.L. Global metabolic profiling of human osteoarthritic synovium. Osteoarthr. Cartil. 2012, 20, 64–67. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Chen, J.; Hu, C.; Xie, Z.; Li, H.; Wei, S.; Wang, D.; Wen, C.; Xu, G. Exploration of the serum metabolite signature in patients with rheumatoid arthritis using gas chromatography-mass spectrometry. J. Pharm. Biomed. Anal. 2016, 127, 60–67. [Google Scholar] [CrossRef]
- Wen, H.; Lee, T.; You, S.; Park, S.H.; Song, H.; Eilber, K.S.; Anger, J.T.; Freeman, M.R.; Park, S.; Kim, J. Urinary metabolite profiling combined with computational analysis predicts interstitial cystitis-associated candidate biomarkers. J. Proteome Res. 2015, 14, 541–548. [Google Scholar] [CrossRef] [Green Version]
- Clos-Garcia, M.; Andres-Marin, N.; Fernandez-Eulate, G.; Abecia, L.; Lavin, J.L.; van Liempd, S.; Cabrera, D.; Royo, F.; Valero, A.; Errazquin, N.; et al. Gut microbiome and serum metabolome analyses identify molecular biomarkers and altered glutamate metabolism in fibromyalgia. EBioMedicine 2019, 46, 499–511. [Google Scholar] [CrossRef] [Green Version]
- Malatji, B.G.; Meyer, H.; Mason, S.; Engelke, U.F.H.; Wevers, R.A.; van Reenen, M.; Reinecke, C.J. A diagnostic biomarker profile for fibromyalgia syndrome based on an NMR metabolomics study of selected patients and controls. BMC Neurol. 2017, 17, 88. [Google Scholar] [CrossRef]
- Menzies, V.; Starkweather, A.; Yao, Y.; Thacker, L.R., II; Garrett, T.J.; Swift-Scanlan, T.; Kelly, D.L.; Patel, P.; Lyon, D.E. Metabolomic Differentials in Women With and Without Fibromyalgia. Clin. Transl. Sci. 2020, 13, 67–77. [Google Scholar] [CrossRef] [Green Version]
- Finco, G.; Locci, E.; Mura, P.; Massa, R.; Noto, A.; Musu, M.; Landoni, G.; d’Aloja, E.; De-Giorgio, F.; Scano, P.; et al. Can Urine Metabolomics Be Helpful in Differentiating Neuropathic and Nociceptive Pain? A Proof-of-Concept Study. PLoS ONE 2016, 11, e0150476. [Google Scholar] [CrossRef] [Green Version]
- Mantyselka, P.; Ali-Sisto, T.; Kautiainen, H.; Niskanen, L.; Viinamaki, H.; Velagapudi, V.; Lehto, S.M. The Association between Musculoskeletal Pain and Circulating Ornithine: A Population-Based Study. Pain Med. 2017, 18, 1145–1151. [Google Scholar] [CrossRef] [Green Version]
- Hadrevi, J.; Bjorklund, M.; Kosek, E.; Hallgren, S.; Antti, H.; Fahlstrom, M.; Hellstrom, F. Systemic differences in serum metabolome: A cross sectional comparison of women with localised and widespread pain and controls. Sci. Rep. 2015, 5, 15925. [Google Scholar] [CrossRef] [Green Version]
- Livshits, G.; Macgregor, A.J.; Gieger, C.; Malkin, I.; Moayyeri, A.; Grallert, H.; Emeny, R.T.; Spector, T.; Kastenmuller, G.; Williams, F.M.K. An omics investigation into chronic widespread musculoskeletal pain reveals epiandrosterone sulfate as a potential biomarker. Pain 2015, 156, 1845–1851. [Google Scholar] [CrossRef] [Green Version]
- Teckchandani, S.; Nagana Gowda, G.A.; Raftery, D.; Curatolo, M. Metabolomics in chronic pain research. Eur. J. Pain 2021, 25, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, T.; Mäntyselkä, P.; Hagelberg, N.; Mustola, S.; Kalso, E.; Lötsch, J. Machine learning suggests sleep as a core factor in chronic pain. Pain 2021, 162, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Sivertsen, B.; Krokstad, S.; Overland, S.; Mykletun, A. The epidemiology of insomnia: Associations with physical and mental health: The HUNT-2 study. J. Psychosom. Res. 2009, 67, 109–116. [Google Scholar] [CrossRef]
- Finan, P.H.; Goodin, B.R.; Smith, M.T. The association of sleep and pain: An update and a path forward. J. Pain Off. J. Am. Pain Soc. 2013, 14, 1539–1552. [Google Scholar] [CrossRef] [Green Version]
- Husak, A.J.; Bair, M.J. Chronic Pain and Sleep Disturbances: A Pragmatic Review of Their Relationships, Comorbidities, and Treatments. Pain Med. 2020, 21, 1142–1152. [Google Scholar] [CrossRef]
- Humer, E.; Pieh, C.; Brandmayr, G. Metabolomics in Sleep, Insomnia and Sleep Apnea. Int. J. Mol. Sci. 2020, 21, 7244. [Google Scholar] [CrossRef]
- Miettinen, T.; Kautiainen, H.; Mäntyselkä, P.; Linton, S.J.; Kalso, E. Pain interference type and level guide the assessment process in chronic pain: Categorizing pain patients entering tertiary pain treatment with the Brief Pain Inventory. PLoS ONE 2019, 14, e0221437. [Google Scholar] [CrossRef]
- Rangel-Huerta, O.D.; Pastor-Villaescusa, B.; Gil, A. Are we close to defining a metabolomic signature of human obesity? A systematic review of metabolomics studies. Metabolomics 2019, 15, 93. [Google Scholar] [CrossRef] [Green Version]
- Nandania, J.; Peddinti, G.; Pessia, A.; Kokkonen, M.; Velagapudi, V. Validation and Automation of a High-Throughput Multitargeted Method for Semiquantification of Endogenous Metabolites from Different Biological Matrices Using Tandem Mass Spectrometry. Metabolites 2018, 8, 44. [Google Scholar] [CrossRef] [Green Version]
- Murphy, K.P. Machine Learning: A Probabilistic Perspective; The MIT Press: Cambridge, MA, USA, 2012; p. 1096. [Google Scholar]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. 2008. Available online: https://CRAN.R-project.org/ (accessed on 1 March 2022).
- Kursa, M.B.; Rudnicki, W.R. Feature Selection with the Boruta Package. J. Stat. Softw. 2010, 36, 13. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Kuhn, M. Caret: Classification and Regression Training. 2018. Available online: https://cran.r-project.org/package=caret (accessed on 1 March 2022).
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Okifuji, A.; Hare, B.D. The association between chronic pain and obesity. J. Pain Res. 2015, 8, 399–408. [Google Scholar] [CrossRef] [Green Version]
- Vanderwall, A.G.; Milligan, E.D. Cytokines in Pain: Harnessing Endogenous Anti-Inflammatory Signaling for Improved Pain Management. Front. Immunol. 2019, 10, 3009. [Google Scholar] [CrossRef] [Green Version]
- Newgard, C.B.; An, J.; Bain, J.R.; Muehlbauer, M.J.; Stevens, R.D.; Lien, L.F.; Haqq, A.M.; Shah, S.H.; Arlotto, M.; Slentz, C.A.; et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009, 9, 311–326. [Google Scholar] [CrossRef] [Green Version]
- Rockel, J.S.; Kapoor, M. The Metabolome and Osteoarthritis: Possible Contributions to Symptoms and Pathology. Metabolites 2018, 8, 92. [Google Scholar] [CrossRef] [Green Version]
- Zhenyukh, O.; Civantos, E.; Ruiz-Ortega, M.; Sanchez, M.S.; Vazquez, C.; Peiro, C.; Egido, J.; Mas, S. High concentration of branched-chain amino acids promotes oxidative stress, inflammation and migration of human peripheral blood mononuclear cells via mTORC1 activation. Free Radic. Biol. Med. 2017, 104, 165–177. [Google Scholar] [CrossRef]
- Lewis, S.S.; Hutchinson, M.R.; Zhang, Y.; Hund, D.K.; Maier, S.F.; Rice, K.C.; Watkins, L.R. Glucuronic acid and the ethanol metabolite ethyl-glucuronide cause toll-like receptor 4 activation and enhanced pain. Brain Behav. Immun. 2013, 30, 24–32. [Google Scholar] [CrossRef] [Green Version]
- Andersen, M.L.; Martins, P.J.; D’Almeida, V.; Santos, R.F.; Bignotto, M.; Tufik, S. Effects of paradoxical sleep deprivation on blood parameters associated with cardiovascular risk in aged rats. Exp. Gerontol. 2004, 39, 817–824. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, M.S.; Holger, D.; Bui, A.T.; Craddock, T.J.A.; Tartar, J.L. Short-term sleep deprivation leads to decreased systemic redox metabolites and altered epigenetic status. PLoS ONE 2017, 12, e0181978. [Google Scholar] [CrossRef] [PubMed]
- Kervezee, L.; Cermakian, N.; Boivin, D.B. Individual metabolomic signatures of circadian misalignment during simulated night shifts in humans. PLoS Biol. 2019, 17, e3000303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seshadri, S.; Beiser, A.; Selhub, J.; Jacques, P.F.; Rosenberg, I.H.; D’Agostino, R.B.; Wilson, P.W.; Wolf, P.A. Plasma homocysteine as a risk factor for dementia and Alzheimer’s disease. N. Engl. J. Med. 2002, 346, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Lebkuchen, A.; Freitas, L.S.; Cardozo, K.H.M.; Drager, L.F. Advances and challenges in pursuing biomarkers for obstructive sleep apnea: Implications for the cardiovascular risk. Trends Cardiovasc. Med. 2021, 31, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.Y.; Winkelman, J.W.; Mao, W.C.; Yeh, C.B.; Huang, S.Y.; Kao, T.W.; Yang, C.C.H.; Kuo, T.B.J.; Chen, W.L. Short Sleep Duration Is Associated with Increased Serum Homocysteine: Insights from a National Survey. J. Clin. Sleep Med. 2019, 15, 139–148. [Google Scholar] [CrossRef]
- Suzuki, K.; Miyamoto, M.; Miyamoto, T.; Numao, A.; Suzuki, S.; Sakuta, H.; Iwasaki, A.; Watanabe, Y.; Fujita, H.; Hirata, K. Sleep apnoea headache in obstructive sleep apnoea syndrome patients presenting with morning headache: Comparison of the ICHD-2 and ICHD-3 beta criteria. J. Headache Pain 2015, 16, 56. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.M.; Wang, S.M.; Xu, H.J.; Yi, H.L.; Guan, J.; Yin, S.K. Metabolomics and microbiome profiling as biomarkers in obstructive sleep apnoea: A comprehensive review. Eur. Respir. Rev. 2021, 30, 200220. [Google Scholar] [CrossRef]
- Ligthart-Melis, G.C.; Engelen, M.; Simbo, S.Y.; Ten Have, G.A.M.; Thaden, J.J.; Cynober, L.; Deutz, N.E.P. Metabolic Consequences of Supplemented Methionine in a Clinical Context. J. Nutr. 2020, 150, 2538S–2547S. [Google Scholar] [CrossRef]
- Reed, M.C.; Thomas, R.L.; Pavisic, J.; James, S.J.; Ulrich, C.M.; Nijhout, H.F. A mathematical model of glutathione metabolism. Biol. Med. Model. 2008, 5, 8. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Fang, Y.Z.; Yang, S.; Lupton, J.R.; Turner, N.D. Glutathione metabolism and its implications for health. J. Nutr. 2004, 134, 489–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okabe, K.; Yaku, K.; Tobe, K.; Nakagawa, T. Implications of altered NAD metabolism in metabolic disorders. J. Biomed. Sci. 2019, 26, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, N.; Zhang, L.; Gao, W.; Huang, C.; Huber, P.E.; Zhou, X.; Li, C.; Shen, G.; Zou, B. NAD(+) metabolism: Pathophysiologic mechanisms and therapeutic potential. Signal Transduct. Target. Ther. 2020, 5, 227. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Sadanala, K.C.; Kim, E.K. A Metabolomic Approach to Understanding the Metabolic Link between Obesity and Diabetes. Mol. Cells 2015, 38, 587–596. [Google Scholar] [CrossRef] [Green Version]
- Zylka, M.J. Pain-relieving prospects for adenosine receptors and ectonucleotidases. Trends Mol. Med. 2011, 17, 188–196. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Adebiyi, M.G.; Luo, J.; Sun, K.; Le, T.T.; Zhang, Y.; Wu, H.; Zhao, S.; Karmouty-Quintana, H.; Liu, H.; et al. Sustained Elevated Adenosine via ADORA2B Promotes Chronic Pain through Neuro-immune Interaction. Cell Rep. 2016, 16, 106–119. [Google Scholar] [CrossRef] [Green Version]
- Lo, A.; Chernoff, H.; Zheng, T.; Lo, S.-H. Why significant variables aren’t automatically good predictors. Proc. Natl. Acad. Sci. USA 2015, 112, 13892–13897. [Google Scholar] [CrossRef] [Green Version]
- Lötsch, J.; Ultsch, A. Random Forests Followed by Computed ABC Analysis as a Feature Selection Method for Machine Learning in Biomedical Data. In Advanced Studies in Classification and Data Science; Imaizumi, T., Okada, A., Miyamoto, S., Sakaori, F., Yamamoto, Y., Vichi, M., Eds.; Springer: Singapore, 2020. [Google Scholar]
- Fonti, V.; Belitser, E. Feature selection using lasso. VU Amst. Res. Pap. Bus. Anal. 2017, 30, 1–25. [Google Scholar]
- Cleeland, C.S.; Ryan, K.M. Pain assessment: Global use of the Brief Pain Inventory. Ann. Acad. Med. Singap. 1994, 23, 129–138. [Google Scholar]
- Lötsch, J.; Malkusch, S. Interpretation of cluster structures in pain-related phenotype data using explainable artificial intelligence (XAI). Eur. J. Pain 2021, 25, 442–465. [Google Scholar] [CrossRef]
- Miettinen, T.; Sverloff, J.; Lappalainen, O.P.; Linton, S.J.; Sipila, K.; Kalso, E. Sleep problems in pain patients entering tertiary pain care: The role of pain-related anxiety, medication use, self-reported diseases, and sleep disorders. Pain 2021, 00, 1–9. [Google Scholar] [CrossRef]
- Sintonen, H. The 15D instrument of health-related quality of life: Properties and applications. Ann. Med. 2001, 33, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Partinen, M.; Gislason, T. Basic Nordic Sleep Questionnaire (BNSQ): A quantitated measure of subjective sleep complaints. J. Sleep Res. 1995, 4, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Lötsch, J.; Schiffmann, S.; Schmitz, K.; Brunkhorst, R.; Lerch, F.; Ferreiros, N.; Wicker, S.; Tegeder, I.; Geisslinger, G.; Ultsch, A. Machine-learning based lipid mediator serum concentration patterns allow identification of multiple sclerosis patients with high accuracy. Sci. Rep. 2018, 8, 14884. [Google Scholar] [CrossRef] [PubMed]
- Wilcoxon, F. Individual comparisons by ranking methods. Biometrics 1945, 1, 80–83. [Google Scholar] [CrossRef]
- Mann, H.B.; Whitney, D.R. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar] [CrossRef]
- Pearson, K. On the criterion that a given system of deviations from the probable in the case of a correlated system of variables is such that it can be reasonably supposed to have arisen from random sampling. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1900, 50, 157–175. [Google Scholar] [CrossRef] [Green Version]
- Bonferroni, C.E. Teoria statistica delle classi e calcolo delle probabilita. Pubbl. Del R Ist. Super. Di Sci. Econ. E Commer. Di Firenze 1936, 8, 3–62. [Google Scholar]
- Ihaka, R.; Gentleman, R. R: A Language for Data Analysis and Graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar] [CrossRef]
- Ciucci, S.; Ge, Y.; Duran, C.; Palladini, A.; Jimenez-Jimenez, V.; Martinez-Sanchez, L.M.; Wang, Y.; Sales, S.; Shevchenko, A.; Poser, S.W.; et al. Enlightening discriminative network functional modules behind Principal Component Analysis separation in differential-omic science studies. Sci. Rep. 2017, 7, 43946. [Google Scholar] [CrossRef] [Green Version]
- Hotelling, H. Analysis of a complex of statistical variables into principal components. J. Educ. Psychol. 1933, 24, 498–520. [Google Scholar] [CrossRef]
- Pearson, K. LIII. On lines and planes of closest fit to systems of points in space. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1901, 2, 559–572. [Google Scholar] [CrossRef] [Green Version]
- Ho, T.K. Random decision forests. In Proceedings of the Third International Conference on Document Analysis and Recognition, Montreal, QC, Canada, 14–16 August 1995; Volume 1, p. 278. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Stekhoven, D.J.; Bühlmann, P. MissForest--non-parametric missing value imputation for mixed-type data. Bioinformatics 2012, 28, 112–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Good, P.I. Resampling Methods: A Practical Guide to Data Analysis; Birkhäuser: Boston, MA, USA, 2006. [Google Scholar]
- Tillé, Y.; Matei, A. Sampling: Survey Sampling. 2016. Available online: https://cran.r−project.org/package=sampling (accessed on 1 March 2022).
- Brodersen, K.H.; Ong, C.S.; Stephan, K.E.; Buhmann, J.M. The Balanced Accuracy and Its Posterior Distribution. In Proceedings of the 20th International Conference on Pattern Recognition (ICPR), Istanbul, Turkey, 23–26 August 2010; pp. 3121–3124. [Google Scholar]
- Peterson, W.; Birdsall, T.; Fox, W. The theory of signal detectability. Trans. IRE Prof. Group Inf. Theory 1954, 4, 171–212. [Google Scholar] [CrossRef]
- Altman, D.G.; Bland, J.M. Diagnostic tests 2: Predictive values. BMJ 1994, 309, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altman, D.G.; Bland, J.M. Diagnostic tests. 1: Sensitivity and specificity. BMJ 1994, 308, 1552. [Google Scholar] [CrossRef] [Green Version]
- Sørensen, T.J. A Method of Establishing Groups of Equal Amplitude in Plant Sociology Based on Similarity of Species Content and Its Application to Analyses of the Vegetation on Danish Commons. Kongelige Danske Videnskabernes Selskab, Biologiske Skrifter 1948, 5, 1–34. [Google Scholar]
- Jardine, N.; van Rijsbergen, C.J. The use of hierarchic clustering in information retrieval. Inf. Storage Retr. 1971, 7, 217–240. [Google Scholar] [CrossRef]
- Lotsch, J.; Ultsch, A. Machine learning in pain research. Pain 2017, 159, 623–630. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Schapire, R.E.; Freund, Y. A short introduction to boosting. J. Jpn. Soc. Artif. Intell. 1999, 14, 771–780. [Google Scholar]
- Cover, T.; Hart, P. Nearest neighbor pattern classification. IEEE Trans. Inf. Theor. 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Hothorn, T.; Hornik, K.; Zeileis, A. Unbiased Recursive Partitioning: A Conditional Inference Framework. J. Comput. Graph. Stat. 2006, 15, 651–674. [Google Scholar] [CrossRef] [Green Version]
- Breimann, L.; Friedman, J.H.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees; Chapman and Hall: Boca Raton, FL, USA, 1993. [Google Scholar]
- Quinlan, J.R. Induction of Decision Trees. Mach. Learn. 1986, 1, 81–106. [Google Scholar] [CrossRef] [Green Version]
- Frank, E.; Witten, I.H. Generating Accurate Rule Sets Without Global Optimization. In Proceedings of the ICML, Madison, WI, USA, 24–27 July 1998. [Google Scholar]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Karatzoglou, A.; Smola, A.; Hornik, K.; Zeileis, A. kernlab—An S4 Package for Kernel Methods in R. J. Stat. Softw. 2004, 11, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; He, T.; Benesty, M.; Khotilovich, V.; Tang, Y.; Cho, H.; Chen, K.; Mitchell, R.; Cano, I.; Zhou, T.; et al. xgboost: Extreme Gradient Boosting. 2020. Available online: https://cran.r−project.org/package=xgboost (accessed on 1 March 2022).
- Therneau, T.; Atkinson, B. rpart: Recursive Partitioning and Regression Trees. 2019. Available online: https://cran.r−project.org/package=rpart (accessed on 1 March 2022).
- Kuhn, M.; Quinlan, R. C50: C5.0 Decision Trees and Rule-Based Models. 2018. Available online: https://CRAN.R−project.org/package=C50 (accessed on 1 March 2022).
- Hornik, K.; Buchta, C.; Zeileis, A. Open-source machine learning: R meets Weka. Comput. Stat. 2009, 24, 225–232. [Google Scholar] [CrossRef] [Green Version]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef] [Green Version]
- Stacklies, W.; Redestig, H.; Scholz, M.; Walther, D.; Selbig, J. pcaMethods—A bioconductor package providing PCA methods for incomplete data. Bioinformatics 2007, 23, 1164–1167. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. MSEA: A web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010, 38, W71–W77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Category | Variable | n | Median | Interquartile Range | Categories and n Per Category |
|---|---|---|---|---|---|
| Demographics | Age | 193 | 48 | 38–56 | - |
| Sex | 193 | - | - | Men = 71 Women = 122 | |
| Living situation | No. of children | 193 | 2 | 0–2 | - |
| Civil status | 192 | - | - | Married = 75 Registered relationship = 0 Cohabiting = 38 Unmarried = 49 Separated = 25 Widow = 5 | |
| Education in years | 188 | 13 | 11–15.13 | - | |
| Type of work | 193 | - | - | Agriculture = 2 Manual work = 15 Office work = 94 Studying or at school = 9 Housewife = 2 Pensioner = 40 Unemployed = 31 | |
| Household income | 184 | 4 | 3–6 | - | |
| Missed workdays within previous 12 mo | 173 | 39 | 2–180 | - | |
| Pain related | No. of pain areas | 193 | 3 | 2–5 | - |
| Duration of pain | 193 | - | - | <1 mo = 0 1–3 mo = 2 3–6 mo = 5 6–12 mo = 23 1–2 y = 30 >2 y = 123 | |
| Pain intensity | 193 | 6 | 5–6.75 | - | |
| Affective pain interference | 193 | 7 | 4.75–8.25 | - | |
| Activity pain interference | 193 | 6.67 | 5.67–8 | - | |
| Any neuropathic pain | 188 | - | - | No = 117, yes = 71 | |
| Low back pain | 188 | - | - | No = 132, yes = 56 | |
| Musculoskeletal pain other than back pain | 188 | - | - | No = 145, yes = 43 | |
| Facial pain | 188 | - | - | No = 178, yes = 10 | |
| Abdominal pain | 188 | - | - | No = 181, yes = 7 | |
| Complex regional pain syndrome | 188 | - | - | No = 177, yes = 11 | |
| Headache | 188 | - | - | No = 184, 1 = 4 | |
| Phantom pain | 188 | - | - | No = 188 | |
| Fibromyalgia | 188 | - | - | No = 170, yes = 18 | |
| Chronic pain syndrome | 188 | - | - | No = 184, yes = 4 | |
| Other pain diagnosis | 188 | - | - | No = 168, yes = 20 | |
| Previous treatments | Negative treatment experiences | 193 | 3 | 1–4 | - |
| Positive treatment experiences | 193 | 4 | 2–6 | - | |
| Physician visits within previous 12 mo | 181 | 10 | 5–14 | - | |
| Comorbidities | Hypertension | 192 | - | - | No = 135, Yes = 57 |
| Heart failure | 192 | - | - | No = 187, Yes = 5 | |
| Angina pectoris | 192 | - | - | No = 180, Yes = 12 | |
| Diabetes | 191 | - | - | No = 175, Yes = 16 | |
| Asthma | 192 | - | - | No = 160, Yes = 32 | |
| Chronic obstructive pulmonary disease | 192 | - | - | No = 186, Yes = 6 | |
| Rheumatoid arthritis | 192 | - | - | No = 190, Yes = 2 | |
| Joint disease other than rheumatoid arthritis | 192 | - | - | No = 141, Yes = 51 | |
| Low back pain | 192 | - | - | No = 91, Yes = 101 | |
| Depression | 190 | - | - | No = 135, Yes = 55 | |
| Psychiatric disorder other than depression | 192 | - | - | No = 181, Yes = 11 | |
| Hypercholesterolemia ever in life | 166 | - | - | No = 94, Yes = 72 | |
| Using cholesterol medication | 168 | - | - | No = 143, Yes = 25 | |
| High blood pressure ever in life | 190 | - | - | No = 107, Yes = 83 | |
| Blood pressure medication use ever in life | 85 | - | - | No = 28, Yes = 57 | |
| Diabetes type | 159 | - | - | No = 130 No, but elevated blood sugar = 7 Yes, type 1 diabetes = 4 Yes, type 2 diabetes = 14 Yes, but don’t know type = 1 Yes, diabetes during pregnancy = 3 | |
| Lifestyle | Smoking currently | 193 | - | - | No = 118, yes = 75 |
| Exercise periods of >20 min per week | 190 | 2 | 0–3 | - | |
| Hours spent sitting per day | 185 | 6 | 3.5–9.5 | - | |
| Sleep problems index | 190 | 17 | 14–20 | - | |
| Nutritional index | 135 | 1 | 1–2 | - | |
| Drug abuse | 135 | 0 | 0–0 | No = 124 Has used = 10 Dependent = 1 | |
| Alcohol consumption frequency | 126 | - | - | Never = 19 Once a month or less = 43 2–4 times a month = 40 2–3 times a week = 20 4 times a week or more = 4 | |
| Body mass index | 192 | 27.82 | 24.23–32.71 | - | |
| Systolic blood pressure, mm Hg | 193 | 135 | 124–150 | - | |
| Diastolic blood pressure, mm Hg | 193 | 86 | 80–94 | - | |
| Waist circumference | 192 | 95.25 | 84.5–106.25 | - |
| Parameter | Full Feature Set | Reduced Feature Set | ||
|---|---|---|---|---|
| Feature set | Original | Permuted | Original | Permuted |
| Sensitivity, recall | 0 (0–0) | 0 (0–0) | 31.6 (26.3–36.8) | 10.5 (5.3–15.8) |
| Specificity | 100 (97.8–100) | 100 (100–100) | 88.9 (84.4–91.1) | 91.1 (86.7–93.3) |
| Positive predictive value, precision | 0 (0–50) | 50 (0–100) | 53.6 (45.5–60) | 33.3 (22.2–45.5) |
| Negative predictive value | 70.3 (70.3–70.3) | 70.3 (70.3–70.3) | 75 (73.7–76.9) | 70.5 (69.4–71.9) |
| F1 | 10 (9.5–10) | 10 (10–10) | 38.8 (33–45.2) | 16.7 (14.3–25) |
| Balanced Accuracy | 50 (49.9–50) | 50 (50–50) | 59.1 (57.1–62.9) | 50.4 (47.8–53.5) |
| ROC-AUC | 50.7 (46.5–56.1) | 51.3 (46.7–55.1) | 70 (66.3–75.2) | 56.1 (49.3–61.6) |
| Metabolomic Marker | FC | log2(FC) | Raw.Pval | −log10(p) |
|---|---|---|---|---|
| Obesity | ||||
| Glutamate | 1.1076 | 0.14741 | 7.385 × 10−5 | 4.1317 |
| Asparagine | 0.97389 | −0.038168 | 0.00060007 | 3.2218 |
| Glycine | 0.96871 | −0.045858 | 0.0013494 | 2.8698 |
| Tyrosine | 1.0282 | 0.040139 | 0.0018034 | 2.7439 |
| Valine | 1.0209 | 0.029846 | 0.0019009 | 2.721 |
| Alanine | 1.0211 | 0.030172 | 0.0030191 | 2.5201 |
| Isovalerylcarnitine | 1.155 | 0.2079 | 0.0053701 | 2.27 |
| Isoleucine | 1.0301 | 0.042839 | 0.0061138 | 2.2137 |
| Symmetric dimethylargininee | 0.88753 | −0.17213 | 0.0066633 | 2.1763 |
| Propionylcarnitine | 1.1127 | 0.15403 | 0.0097422 | 2.0113 |
| Hydroxykynurenine | 1.2256 | 0.29344 | 0.009839 | 2.007 |
| Glucuronic acid | 1.1245 | 0.16928 | 0.011138 | 1.9532 |
| Creatinine | 0.98053 | −0.028359 | 0.012257 | 1.9116 |
| Creatine | 1.0483 | 0.068066 | 0.013068 | 1.8838 |
| Hexanoylcarnitine | 1.1638 | 0.21882 | 0.020064 | 1.6976 |
| Citrulline | 1.0376 | 0.053191 | 0.02039 | 1.6906 |
| Inosine | 1.2492 | 0.32101 | 0.02406 | 1.6187 |
| Chenodeoxycholic Acid | 1.0856 | 0.11852 | 0.024663 | 1.6079 |
| Adenosine | 1.2961 | 0.37413 | 0.032691 | 1.4856 |
| Kynurenine | 1.0443 | 0.062527 | 0.034 | 1.4685 |
| NAD | 0.73752 | −0.43924 | 0.036641 | 1.436 |
| Cytidine | 1.0567 | 0.079523 | 0.047004 | 1.3279 |
| Guanosine | 1.4269 | 0.51284 | 0.047952 | 1.3192 |
| Sleep problems | ||||
| Serine | 0.98126 | −0.027298 | 0.017081 | 1.7675 |
| Symmetric dimethylarginine | 0.91811 | −0.12326 | 0.021126 | 1.6752 |
| Homocysteine | 0.85203 | −0.23103 | 0.021403 | 1.6695 |
| Dimethylglycine | 0.9218 | −0.11747 | 0.028466 | 1.5457 |
| GABA | 0.87712 | −0.18915 | 0.03143 | 1.5027 |
| Asymmetric dimethylarginine | 0.91048 | −0.1353 | 0.031587 | 1.5005 |
| Choline | 0.96778 | −0.047256 | 0.049881 | 1.3021 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miettinen, T.; Nieminen, A.I.; Mäntyselkä, P.; Kalso, E.; Lötsch, J. Machine Learning and Pathway Analysis-Based Discovery of Metabolomic Markers Relating to Chronic Pain Phenotypes. Int. J. Mol. Sci. 2022, 23, 5085. https://doi.org/10.3390/ijms23095085
Miettinen T, Nieminen AI, Mäntyselkä P, Kalso E, Lötsch J. Machine Learning and Pathway Analysis-Based Discovery of Metabolomic Markers Relating to Chronic Pain Phenotypes. International Journal of Molecular Sciences. 2022; 23(9):5085. https://doi.org/10.3390/ijms23095085
Chicago/Turabian StyleMiettinen, Teemu, Anni I. Nieminen, Pekka Mäntyselkä, Eija Kalso, and Jörn Lötsch. 2022. "Machine Learning and Pathway Analysis-Based Discovery of Metabolomic Markers Relating to Chronic Pain Phenotypes" International Journal of Molecular Sciences 23, no. 9: 5085. https://doi.org/10.3390/ijms23095085
APA StyleMiettinen, T., Nieminen, A. I., Mäntyselkä, P., Kalso, E., & Lötsch, J. (2022). Machine Learning and Pathway Analysis-Based Discovery of Metabolomic Markers Relating to Chronic Pain Phenotypes. International Journal of Molecular Sciences, 23(9), 5085. https://doi.org/10.3390/ijms23095085







