Effects of Drugs Formerly Proposed for COVID-19 Treatment on Connexin43 Hemichannels
Abstract
:1. Introduction
2. Results
2.1. Determination of Working Concentrations of the Drugs
2.2. Effects of the Drugs on Cx43 Hemichannel Activity
2.3. Effects of the Drugs on Cx43 mRNA and Protein Expression
3. Discussion
4. Materials and Methods
4.1. Cell Culture Set-Up and Maintenance
4.2. Cell Viability Assessment
4.3. Cx43 Hemichannel Activity Assay
4.4. Immunoblot Analysis
4.5. RT-qPCR Analysis
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cooreman, A.; Van Campenhout, R.; Ballet, S.; Annaert, P.; Van Den Bossche, B.; Colle, I.; Cogliati, B.; Vinken, M. Connexin and Pannexin (Hemi)Channels: Emerging Targets in the Treatment of Liver Disease. Hepatology 2019, 69, 1317–1323. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y. Gap Junction-Dependent and -Independent Functions of Connexin43 in Biology. Biology 2022, 11, 283. [Google Scholar] [CrossRef] [PubMed]
- Panattoni, G.; Amoriello, R.; Memo, C.; Thalhammer, A.; Ballerini, C.; Ballerini, L. Diverse Inflammatory Threats Modulate Astrocytes Ca2+ Signaling via Connexin43 Hemichannels in Organotypic Spinal Slices. Mol. Brain 2021, 14, 159. [Google Scholar] [CrossRef] [PubMed]
- Saez, J.C.; Green, C. Involvement of Connexin Hemichannels in the Inflammatory Response of Chronic Diseases. Int. J. Mol. Sci. 2018, 19, 2469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Campenhout, R.; Gomes, A.R.; De Groof, T.W.M.; Muyldermans, S.; Devoogdt, N.; Vinken, M. Mechanisms Underlying Connexin Hemichannel Activation in Disease. Int. J. Mol. Sci. 2021, 22, 3503. [Google Scholar] [CrossRef]
- Dosch, M.; Gerber, J.; Jebbawi, F.; Beldi, G. Mechanisms of ATP Release by Inflammatory Cells. Int. J. Mol. Sci. 2018, 19, 1222. [Google Scholar] [CrossRef] [Green Version]
- Kameritsch, P.; Pogoda, K. The Role of Connexin 43 and Pannexin 1 during Acute Inflammation. Front. Physiol. 2020, 11, 594097. [Google Scholar] [CrossRef]
- Wang, W.; Hu, D.; Feng, Y.; Wu, C.; Song, Y.; Liu, W.; Li, A.; Wang, Y.; Chen, K.; Tian, M.; et al. Paxillin Mediates ATP-Induced Activation of P2X7 Receptor and NLRP3 Inflammasome. BMC Biol. 2020, 18, 182. [Google Scholar] [CrossRef]
- Willebrords, J.; Yanguas, S.C.; Maes, M.; Decrock, E.; Wang, N.; Leybaert, L.; Kwak, B.R.; Green, C.R.; Cogliati, B.; Vinken, M. Connexins and Their Channels in Inflammation. Crit. Rev. Biochem. Mol. Biol. 2016, 51, 413–439. [Google Scholar] [CrossRef]
- Lai, C.-C.; Shih, T.-P.; Ko, W.-C.; Tang, H.-J.; Hsueh, P.-R. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and Coronavirus Disease-2019 (COVID-19): The Epidemic and the Challenges. Int. J. Antimicrob. Agents 2020, 55, 105924. [Google Scholar] [CrossRef]
- WHO. COVID-19 Dashboard—Global Situation. Available online: https://covid19.who.int/ (accessed on 23 March 2022).
- Lee, C.; Choi, W.J. Overview of COVID-19 Inflammatory Pathogenesis from the Therapeutic Perspective. Arch. Pharmacal Res. 2021, 44, 99–116. [Google Scholar] [CrossRef] [PubMed]
- Wiersinga, W.J.; Rhodes, A.; Cheng, A.C.; Peacock, S.J.; Prescott, H.C. Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19) A Review. JAMA-J. Am. Med. Assoc. 2020, 324, 782–793. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and Clinical Characteristics of 99 Cases of 2019 Novel Coronavirus Pneumonia in Wuhan, China: A Descriptive Study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Gujski, M.; Jankowski, M.; Rabczenko, D.; Gorynski, P.; Juszczyk, G. The Prevalence of Acute Respiratory Distress Syndrome (ARDS) and Outcomes in Hospitalized Patients with COVID-19-A Study Based on Data from the Polish National Hospital Register. Viruses 2022, 14, 76. [Google Scholar] [CrossRef]
- Hasan, S.S.; Capstick, T.; Ahmed, R.; Kow, C.S.; Mazhar, F.; Merchant, H.A.; Zaidi, S.T.R. Mortality in COVID-19 Patients with Acute Respiratory Distress Syndrome and Corticosteroids Use: A Systematic Review and Meta-Analysis. Expert Rev. Respir. Med. 2020, 14, 1149–1163. [Google Scholar] [CrossRef]
- Hendrickson, K.W.; Peltan, I.D.; Brown, S.M. The Epidemiology of Acute Respiratory Distress Syndrome before and after Coronavirus Disease 2019. Crit. Care Clin. 2021, 37, 703–716. [Google Scholar] [CrossRef]
- Welker, C.; Huang, J.; Gil, I.J.N.; Ramakrishna, H. 2021 Acute Respiratory Distress Syndrome Update, with Coronavirus Disease 2019 Focus. J. Cardiothorac. Vasc. Anesth 2022, 36, 1188–1195. [Google Scholar] [CrossRef]
- Napoli, C.; Benincasa, G.; Criscuolo, C.; Faenza, M.; Liberato, C.; Rusciano, M. Immune Reactivity during COVID-19: Implications for Treatment. Immunol. Lett. 2021, 231, 28–34. [Google Scholar] [CrossRef]
- Bennett, M.V.; Garré, J.M.; Orellana, J.A.; Bukauskas, F.F.; Nedergaard, M.; Sáez, J.C. Connexin and pannexin hemichannels in inflammatory responses of glia and neurons. Brain Res. 2012, 1487, 3–15. [Google Scholar] [CrossRef] [Green Version]
- Mugisho, O.O.; Green, C.R.; Kho, D.T.; Zhang, J.; Graham, E.S.; Acosta, M.L.; Rupenthanl, I.D. The Inflammasome Pathway Is Amplified and Perpetuated in an Autocrine Manner through Connexin43 Hemichannel Mediated ATP Release. Biochim. Biophys. Acta-Gen. Subj. 2018, 1862, 385–393. [Google Scholar] [CrossRef]
- Mugisho, O.O.; Rupenthal, I.D.; Paquet-Durand, F.; Acosta, M.L.; Green, C.R. Targeting Connexin Hemichannels to Control the Inflammasome: The Correlation between Connexin43 and NLRP3 Expression in Chronic Eye Disease. Expert Opin. Ther. Targets 2019, 23, 855–863. [Google Scholar] [CrossRef] [PubMed]
- FDA. Azithromycin. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2013/050710s039,050711s036,050784s023lbl.pdf (accessed on 28 February 2022).
- Cui, C.; Zhang, M.; Yao, X.; Tu, S.; Hou, Z.; Jie En, V.S.; Xiang, X.; Lin, J.; Cai, T.; Shen, N.; et al. Dose Selection of Chloroquine Phosphate for Treatment of COVID-19 Based on a Physiologically Based Pharmacokinetic Model. Acta Pharm. Sin. B 2020, 10, 1216–1227. [Google Scholar] [CrossRef] [PubMed]
- FDA. Clinical Pharmacology and Biopharmaceutics Review(s). Application Number: 211379 Orig1s000. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/nda/2019/211379Orig1s000ClinPharmR.pdf (accessed on 2 March 2022).
- Saglam, O.; Demiray, G.; Güney, B.; Dogan-Kurtoglu, E.; Ulusoy, M.G.; Saraner, N.; Sevici, G.; Nacak, M.; Erenmemisoglu, A.; Tüzer, V. Single Dose, Two-Way Crossover Bioequivalence Study of Favipiravir Tablet in Healthy Male Subjects. J. Pharm. Drug Dev. 2020, 2, 1–9. [Google Scholar] [CrossRef]
- Emami, J.; Kazemi, M.; Salehi, A. In Vitro and in Vivo Evaluation of Two Hydroxychloroquine Tablet Formulations: HPLC Assay Development. J. Chromatogr. Sci. 2021, 59, 71–78. [Google Scholar] [CrossRef]
- Jackson, A.; Hill, A.; Puls, R.; Else, L.; Amin, J.; Back, D.; Lin, E.; Khoo, S.; Emery, S.; Morley, R.; et al. Pharmacokinetics of Plasma Lopinavir/Ritonavir Following the Administration of 400/100 Mg, 200/150 Mg and 200/50 Mg Twice Daily in HIV-Negative Volunteers. J. Antimicrob. Chemother. 2011, 66, 635–640. [Google Scholar] [CrossRef] [Green Version]
- EMA. Summary on Compassionate Use Remdesivir Gilead. Available online: https://www.ema.europa.eu/en/documents/other/summary-compassionate-use-remdesivir-gilead_en.pdf (accessed on 2 February 2022).
- Glue, P.; Schenker, S.; Gupta, S.; Clement, R.P.; Zambas, D.; Salfi, M. The Single Dose Pharmacokinetics of Ribavirin in Subjects with Chronic Liver Disease. Br. J. Clin. Pharmacol. 2000, 49, 417–421. [Google Scholar] [CrossRef] [Green Version]
- EMA. Summary of Product Characteristics of Norvir 100 mg. Available online: https://www.ema.europa.eu/en/documents/product-information/norvir-epar-product-information_en.pdf (accessed on 4 April 2022).
- Bell, S.M.; Chang, X.; Wambaugh, J.F.; Allen, D.G.; Bartels, M.; Brouwer, K.L.R.; Casey, W.M.; Choksi, N.; Ferguson, S.S.; Fraczkiewicz, G.; et al. In Vitro to in Vivo Extrapolation for High Throughput Prioritization and Decision Making. Toxicol. Vitr. 2018, 47, 213–227. [Google Scholar] [CrossRef]
- Shebley, M.; Sandhu, P.; Riedmaier, A.E.; Jamei, M.; Narayanan, R.; Patel, A.; Peters, S.A.; Reddy, V.P.; Zheng, M.; de Zwart, L.; et al. Physiologically Based Pharmacokinetic Model Qualification and Reporting Procedures for Regulatory Submissions: A Consortium Perspective. Clin. Pharmacol. Ther. 2018, 104, 88–110. [Google Scholar] [CrossRef]
- Punt, A.; Louisse, J.; Pinckaers, N.; Fabian, E.; van Ravenzwaay, B. Predictive Performance of Next Generation Physiologically Based Kinetic (PBK) Model Predictions in Rats Based on In Vitro and In Silico Input Data. Toxicol. Sci. 2021, 186, 18–28. [Google Scholar] [CrossRef]
- Vinken, M.; Hengstler, J.G. Characterization of Hepatocyte-Based in Vitro Systems for Reliable Toxicity Testing. Arch. Toxicol. 2018, 92, 2981–2986. [Google Scholar] [CrossRef] [Green Version]
- Mehta, J.; Rolta, R.; Mehta, B.B.; Kaushik, N.; Choi, E.H.; Kaushik, N.K. Role of Dexamethasone and Methylprednisolone Corticosteroids in Coronavirus Disease 2019 Hospitalized Patients: A Review. Front. Microbiol. 2022, 13, 46. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhong, W.; Salam, A.; Tarning, J.; Zhan, Q.; Huang, J.; Weng, H.; Bai, C.; Ren, Y.; Yamada, K.; et al. Phase 2a, Open-Label, Dose-Escalating, Multi-Center Pharmacokinetic Study of Favipiravir (T-705) in Combination with Oseltamivir in Patients with Severe Influenza. eBioMedicine 2020, 62, 103125. [Google Scholar] [CrossRef] [PubMed]
- Cvetkovic, R.S.; Goa, K.L. Lopinavir/Ritonavir—A Review of Its Use in the Management of HIV Infection. Drugs 2003, 63, 769–802. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Wang, Y.; Wen, D.; Liu, W.; Wang, J.; Fan, G.; Ruan, L.; Song, B.; Cai, Y.; Wei, M.; et al. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. N. Engl. J. Med. 2020, 382, 1787–1799. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Xu, C.; Wang, S.; Zhang, Y.; Li, W. The Role of Connexin Hemichannels in Inflammatory Diseases. Biology 2022, 11, 237. [Google Scholar] [CrossRef]
- Tay, M.Z.; Poh, C.M.; Renia, L.; MacAry, P.A.; Ng, L.F.P. The Trinity of COVID-19: Immunity, Inflammation and Intervention. Nat. Rev. Immunol. 2020, 20, 363–374. [Google Scholar] [CrossRef]
- Orellana, J.A.; Carlos Saez, J.; Bennett, M.V.L.; Berman, J.W.; Morgello, S.; Eugenin, E.A. HIV Increases the Release of Dickkopf-1 Protein from Human Astrocytes by a Cx43 Hemichannel-Dependent Mechanism. J. Neurochem. 2014, 128, 752–763. [Google Scholar] [CrossRef] [Green Version]
- Hernandez-Guerra, M.; Hadjihambi, A.; Jalan, R. Gap Junctions in Liver Disease: Implications for Pathogenesis and Therapy. J. Hepatol. 2019, 70, 759–772. [Google Scholar] [CrossRef] [Green Version]
- Thimm, J.; Mechler, A.; Lin, H.; Rhee, S.; Lal, R. Calcium-Dependent Open/Closed Conformations and Interfacial Energy Maps of Reconstituted Hemichannels. J. Biol. Chem. 2005, 280, 10646–10654. [Google Scholar] [CrossRef] [Green Version]
- Willebrords, J.; Maes, M.; Yanguas, S.C.; Vinken, M. Inhibitors of Connexin and Pannexin Channels as Potential Therapeutics. Pharmacol. Ther. 2017, 180, 144–160. [Google Scholar] [CrossRef] [Green Version]
- Shen, C.; Kim, M.R.; Noh, J.M.; Kim, S.J.; Ka, S.-O.; Kim, J.H.; Park, B.-H.; Park, J.H. Glucocorticoid Suppresses Connexin 43 Expression by Inhibiting the Akt/MTOR Signaling Pathway in Osteoblasts. Calcif. Tissue Int. 2016, 99, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.-Y.; Zhang, W.-S.; Zhang, H.; Cao, Y.; Zhou, H.-Y. The Role of Connexin-43 in the Inflammatory Process: A New Potential Therapy to Influence Keratitis. J. Ophthalmol. 2019, 2019, 9312827. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Liu, B.; Zeng, L.; Yang, X.; Huang, R.; Wu, C.; Zhu, H.; Gao, Y.; Yuan, D.; Yu, J. CX43 Inhibition Attenuates Sepsis-Induced Intestinal Injury via Downregulating ROS Transfer and the Activation of the JNK1/SIRT1/FoxO3a Signaling Pathway. Mediat. Inflamm. 2019, 2019, 7854389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willebrords, J.; Maes, M.; Yanguas, S.C.; Cogliati, B.; Vinken, M. Detection of Connexins in Liver Cells Using Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis and Immunoblot Analysis. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2016; Volume 1437, pp. 37–53. [Google Scholar]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Freda, C.T.; Yin, W.; Ghebrehiwet, B.; Rubenstein, D.A. SARS-CoV-2 Structural Proteins Exposure Alter Thrombotic and Inflammatory Responses in Human Endothelial Cells. Cell. Mol. Bioeng. 2022, 15, 43–53. [Google Scholar] [CrossRef]
- Beckmann, A.; Grissmer, A.; Meier, C.; Tschernig, T. Intercellular Communication between Alveolar Epithelial Cells and Macrophages. Ann. Anat.-Anat. Anz. 2020, 227, 151417. [Google Scholar] [CrossRef]
- Isakson, B.E.; Seedorf, G.J.; Lubman, R.L.; Evans, W.H.; Boitano, S. Cell-Cell Communication in Heterocellular Cultures of Alveolar Epithelial Cells. Am. J. Respir. Cell Mol. Biol. 2003, 29, 552–561. [Google Scholar] [CrossRef]
- Eltzschig, H.K.; Eckle, T.; Mager, A.; Kueper, N.; Karcher, C.; Weissmueller, T.; Boengler, K.; Schulz, R.; Robson, S.C.; Colgan, S.P. ATP Release from Activated Neutrophils Occurs via Connexin 43 and Modulates Adenosine-Dependent Endothelial Cell Function. Circ. Res. 2006, 99, 1100–1108. [Google Scholar] [CrossRef] [Green Version]
- Swartzendruber, J.A.; Nicholson, B.J.; Murthy, A.K. The Role of Connexin 43 in Lung Disease. Life 2020, 10, 363. [Google Scholar] [CrossRef]
- Sarieddine, M.Z.R.; Scheckenbach, K.E.L.; Foglia, B.; Maass, K.; Garcia, I.; Kwak, B.R.; Chanson, M. Connexin43 Modulates Neutrophil Recruitment to the Lung. J. Cell. Mol. Med. 2009, 13, 4560–4570. [Google Scholar] [CrossRef] [Green Version]
- Kelley, N.; Jeltema, D.; Duan, Y.; He, Y. The NLRP3 Inflammasome: An Overview of Mechanisms of Activation and Regulation. Int. J. Mol. Sci. 2019, 20, 3328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Luo, X.; Poetsch, M.S.; Oertel, R.; Nichani, K.; Schneider, M.; Strano, A.; Hasse, M.; Steiner, R.-P.; Cyganek, L.; et al. Synergistic Adverse Effects of Azithromycin and Hydroxychloroquine on Human Cardiomyocytes at a Clinically Relevant Treatment Duration. Pharmaceuticals 2022, 15, 220. [Google Scholar] [CrossRef] [PubMed]
- Iyyathurai, J.; Decuypere, J.-P.; Leybaert, L.; D’hondt, C.; Bultynck, G. Connexins: Substrates and Regulators of Autophagy. BMC Cell Biol. 2016, 17, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van den Borne, B.E.; Dijkmans, B.A.C.; DeRooij, H.H.; LeCessie, S.; Verweij, C.L. Chloroquine and Hydroxychloroquine Equally Affect Tumor Necrosis Factor-Alpha, Interleukin 6, and Interferon-Gamma Production by Peripheral Blood Mononuclear Cells. J. Rheumatol. 1997, 24, 55–60. [Google Scholar]
- Berman, J.W.; Carvallo, L.; Buckner, C.M.; Luers, A.; Prevedel, L.; Bennett, M.V.; Eugenin, E.A. HIV-Tat Alters Connexin43 Expression and Trafficking in Human Astrocytes: Role in NeuroAIDS. J. Neuroinflamm. 2016, 13, 54. [Google Scholar] [CrossRef] [Green Version]
- Reyskens, K.M.S.E.; Fisher, T.-L.; Schisler, J.C.; O’Connor, W.G.; Rogers, A.B.; Willis, M.S.; Planesse, C.; Rondeau, P.; Bourdon, E.; Essop, M.F. The Maladaptive Effects of HIV Protease Inhibitors (Lopinavir/Ritonavir) on the Rat Heart. PLoS ONE 2013, 8, e73347. [Google Scholar] [CrossRef]
- Stower, H. Lopinavir-Ritonavir in Severe COVID-19. Nat. Med. 2020, 26, 465. [Google Scholar] [CrossRef] [Green Version]
- Tchesnokov, E.P.; Feng, J.Y.; Porter, D.P.; Gotte, M. Mechanism of Inhibition of Ebola Virus RNA-Dependent RNA Polymerase by Remdesivir. Viruses 2019, 11, 326. [Google Scholar] [CrossRef] [Green Version]
- Ismail, F.S.; Moinfar, Z.; Prochnow, N.; Dambach, H.; Hinkerohe, D.; Haase, C.G.; Förster, E.; Faustmann, P.M. Dexamethasone and Levetiracetam Reduce Hetero-Cellular Gap-Junctional Coupling between F98 Glioma Cells and Glial Cells in Vitro. J. Neuro-Oncol. 2017, 131, 469–476. [Google Scholar] [CrossRef] [Green Version]
- Warawdekar, U.M.; Jain, V.; Patel, H.; Nanda, A.; Kamble, V. Modifying Gap Junction Communication in Cancer Therapy. Curr. Res. Transl. Med. 2021, 69, 103268. [Google Scholar] [CrossRef]
- Martin, F.C.; Handforth, A. Carbenoxolone and Mefloquine Suppress Tremor in the Harmaline Mouse Model of Essential Tremor. Mov. Disord. 2006, 21, 1641–1649. [Google Scholar] [CrossRef] [PubMed]
- Cruikshank, S.J.; Hopperstad, M.; Younger, M.; Connors, B.W.; Spray, D.C.; Srinivas, M. Potent Block of Cx36 and Cx50 Gap Junction Channels by Mefloquine. Proc. Natl. Acad. Sci. USA 2004, 101, 12364–12369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, J.; Zhou, F.; Heybati, K.; Ali, S.; Zuo, Q.K.; Hou, W.; Dhivagaran, T.; Ramaraju, H.B.; Chang, O.; Wong, C.Y.; et al. Efficacy of Chloroquine and Hydroxychloroquine for the Treatment of Hospitalized COVID-19 Patients: A Meta-Analysis. Future Virol. 2022, 17, 95–118. [Google Scholar] [CrossRef] [PubMed]
- McChesney, E.W. Animal Toxicity and Pharmacokinetics of Hydroxychloroquine Sulfate. Am. J. Med. 1983, 75, 11–18. [Google Scholar] [CrossRef]
- Horby, P.; Lim, W.S.; Emberson, J.R.; Mafham, M.; Bell, J.L.; Linsell, L.; Staplin, N.; Brightling, C.; Ustianowski, A.; Elmahi, E.; et al. Dexamethasone in Hospitalized Patients with Covid-19. N. Engl. J. Med. 2021, 384, 693–704. [Google Scholar] [CrossRef]
- Horby, P.W.; Landray, M.J.; Mafham, M.; Bell, J.L.; Linsell, L.; Staplin, N.; Emberson, J.R.; Palfreeman, A.; Raw, J.; Elmahi, E.; et al. Lopinavir-Ritonavir in Patients Admitted to Hospital with COVID-19 (RECOVERY): A Randomised, Controlled, Open-Label, Platform Trial. Lancet 2020, 396, 1345–1352. [Google Scholar] [CrossRef]
- Horby, P.W.; Roddick, A.; Spata, E.; Staplin, N.; Emberson, J.; Pessoa-Amorim, G.; Brightling, C.; Prudon, B.; Chadwick, D.; Ustianowski, A.; et al. Azithromycin in Patients Admitted to Hospital with COVID-19 (RECOVERY): A Randomised, Controlled, Open-Label, Platform Trial. Lancet 2021, 397, 605–612. [Google Scholar] [CrossRef]
- Sinha, M.; Gupta, A.; Gupta, S.; Singh, P.; Pandit, S.; Chauhan, S.S.; Parthasarathi, R. Analogue Discovery of Safer Alternatives to HCQ and CQ Drugs for SAR-CoV-2 by Computational Design. Comput. Biol. Med. 2021, 130, 104222. [Google Scholar] [CrossRef]
- Horby, P.; Mafham, M.; Linsell, L.; Bell, J.; Staplin, N.; Emberson, J.; Wiselka, M.; Ustianowski, A.; Elmahi, E.; Prudon, B.; et al. Effect of Hydroxychloroquine in Hospitalized Patients with Covid-19. N. Engl. J. Med. 2020, 383, 2030–2040. [Google Scholar] [CrossRef]
- Corti, D.; Purcell, L.A.; Snell, G.; Veesler, D. Tackling COVID-19 with Neutralizing Monoclonal Antibodies. Cell 2021, 184, 3086–3108. [Google Scholar] [CrossRef]
- EMA. COVID-19 Treatments. Available online: https://www.ema.europa.eu/en/human-656regulatory/overview/public-health-threats/coronavirus-disease-covid-19/treatments-vaccines/covid-19-treatments (accessed on 15 March 2022).
- Simsek-Yavuz, S.; Komsuoglu Celikyurt, F.I. An Update of Anti-Viral Treatment of COVID-19. Turk. J. Med. Sci. 2021, 51, 3372–3390. [Google Scholar] [CrossRef] [PubMed]
- Lohman, A.W.; Isakson, B.E. Differentiating Connexin Hemichannels and Pannexin Channels in Cellular ATP Release. FEBS Lett. 2014, 588, 1379–1388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosa, R.B.; Dantas, W.M.; do Nascimento, J.C.F.; da Silva, M.V.; de Oliveira, R.N.; Pena, L.J. In Vitro and in Vivo Models for Studying Sars-Cov-2, the Etiological Agent Responsible for Covid-19 Pandemic. Viruses 2021, 13, 379. [Google Scholar] [CrossRef]
- Leroy, K.; Pieters, A.; Cooreman, A.; Van Campenhout, R.; Cogliati, B.; Vinken, M. Connexin-Based Channel Activity Is Not Specifically Altered by Hepatocarcinogenic Chemicals. Int. J. Mol. Sci. 2021, 22, 11724. [Google Scholar] [CrossRef] [PubMed]
- Cooreman, A.; Van Campenhout, R.; Yanguas, S.C.; Gijbels, E.; Leroy, K.; Pieters, A.; Tabernilla, A.; Van Brantegem, P.; Annaert, P.; Cogliati, B.; et al. Cholestasis Differentially Affects Liver Connexins. Int. J. Mol. Sci. 2020, 21, 6534. [Google Scholar] [CrossRef] [PubMed]
- Eaton, S.L.; Roche, S.L.; Llavero Hurtado, M.; Oldknow, K.J.; Farquharson, C.; Gillingwater, T.H.; Wishart, T.M. Total Protein Analysis as a Reliable Loading Control for Quantitative Fluorescent Western Blotting. PLoS ONE 2013, 8, e72457. [Google Scholar] [CrossRef] [PubMed]
- Maes, M.; Willebrords, J.; Crespo Yanguas, S.; Cogliati, B.; Vinken, M. Analysis of Liver Connexin Expression Using Reverse Transcription Quantitative Real-Time Polymerase Chain Reaction. Methods Mol. Biol. 2016, 1437, 1–19. [Google Scholar] [CrossRef] [Green Version]
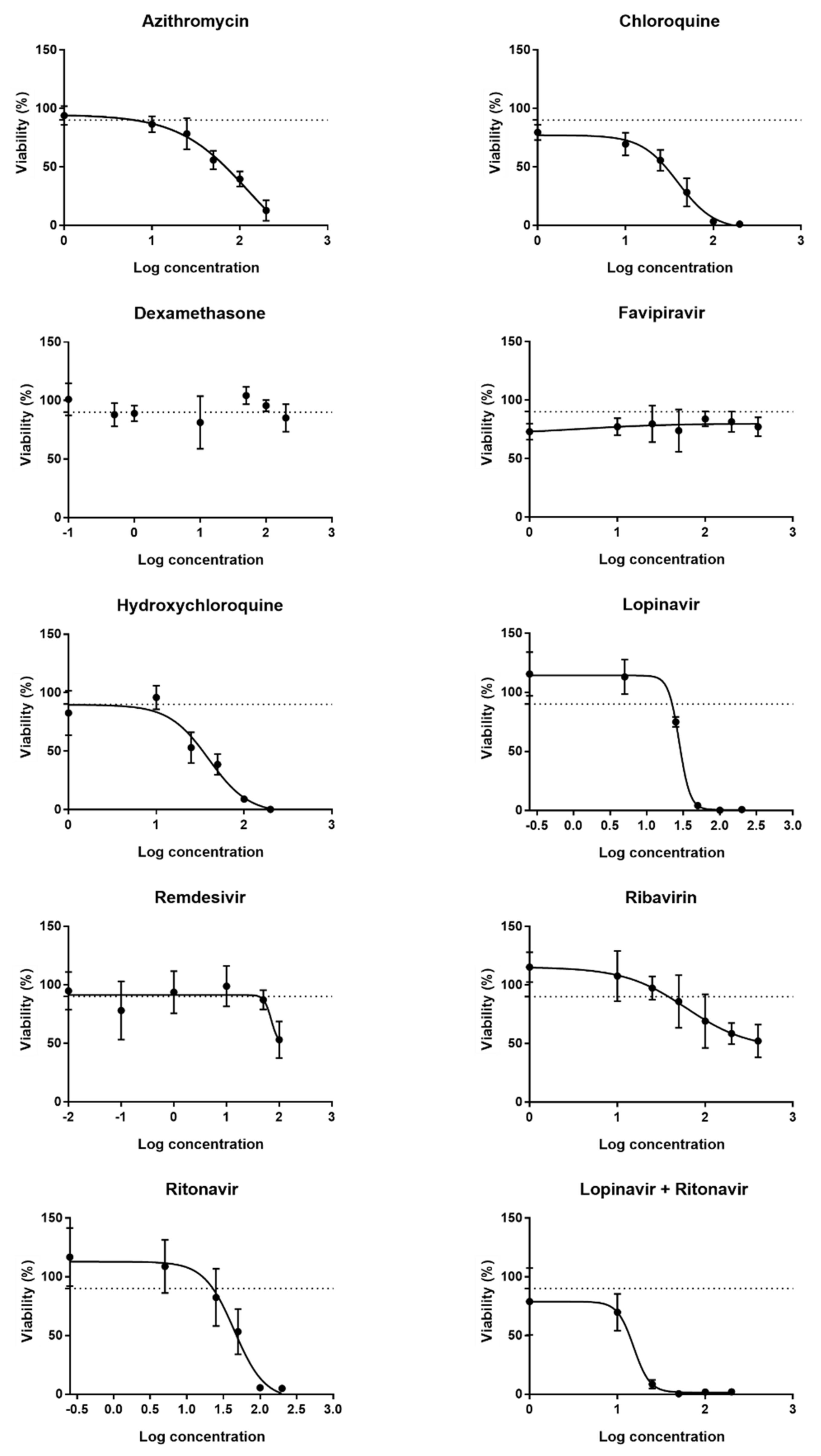
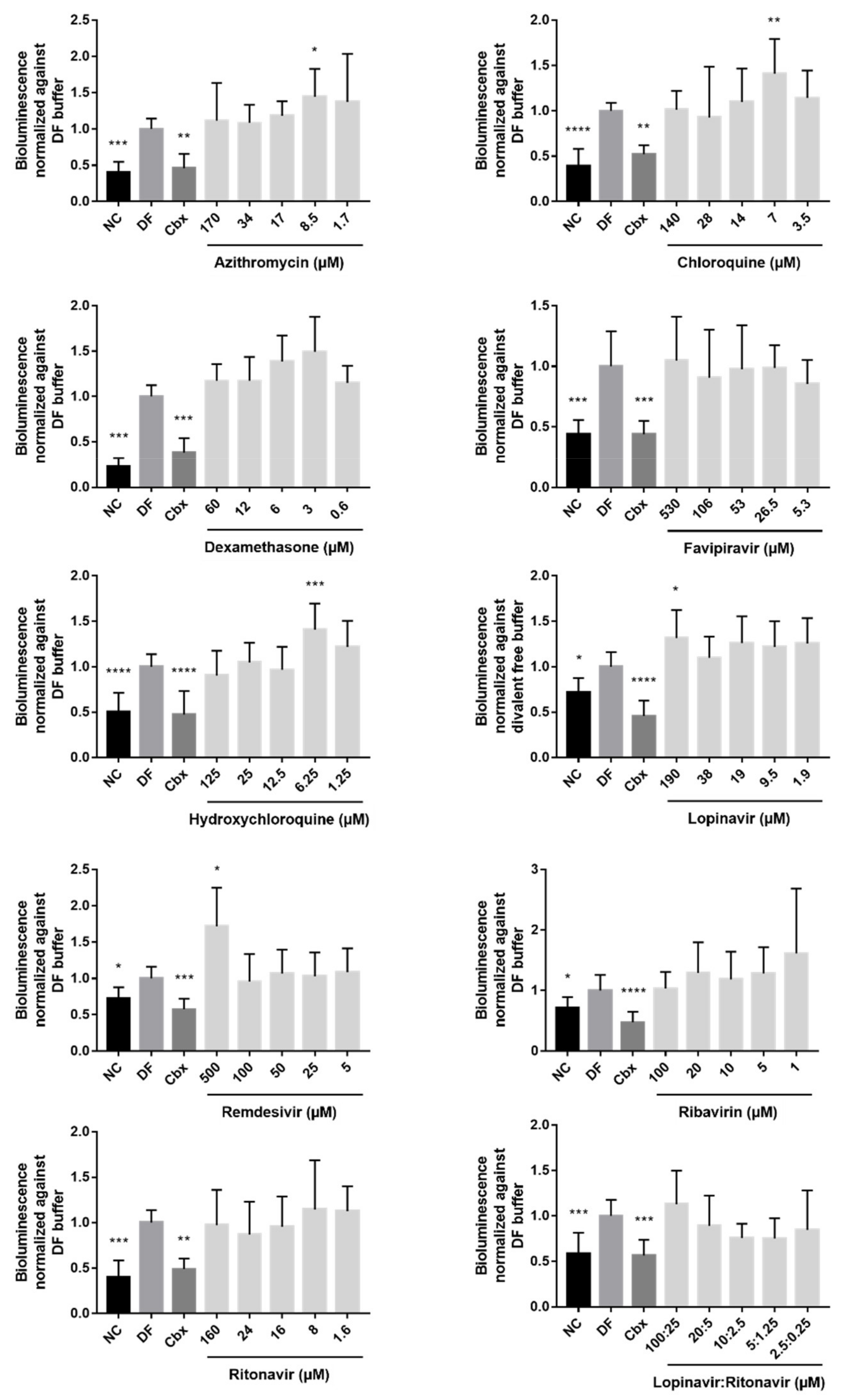
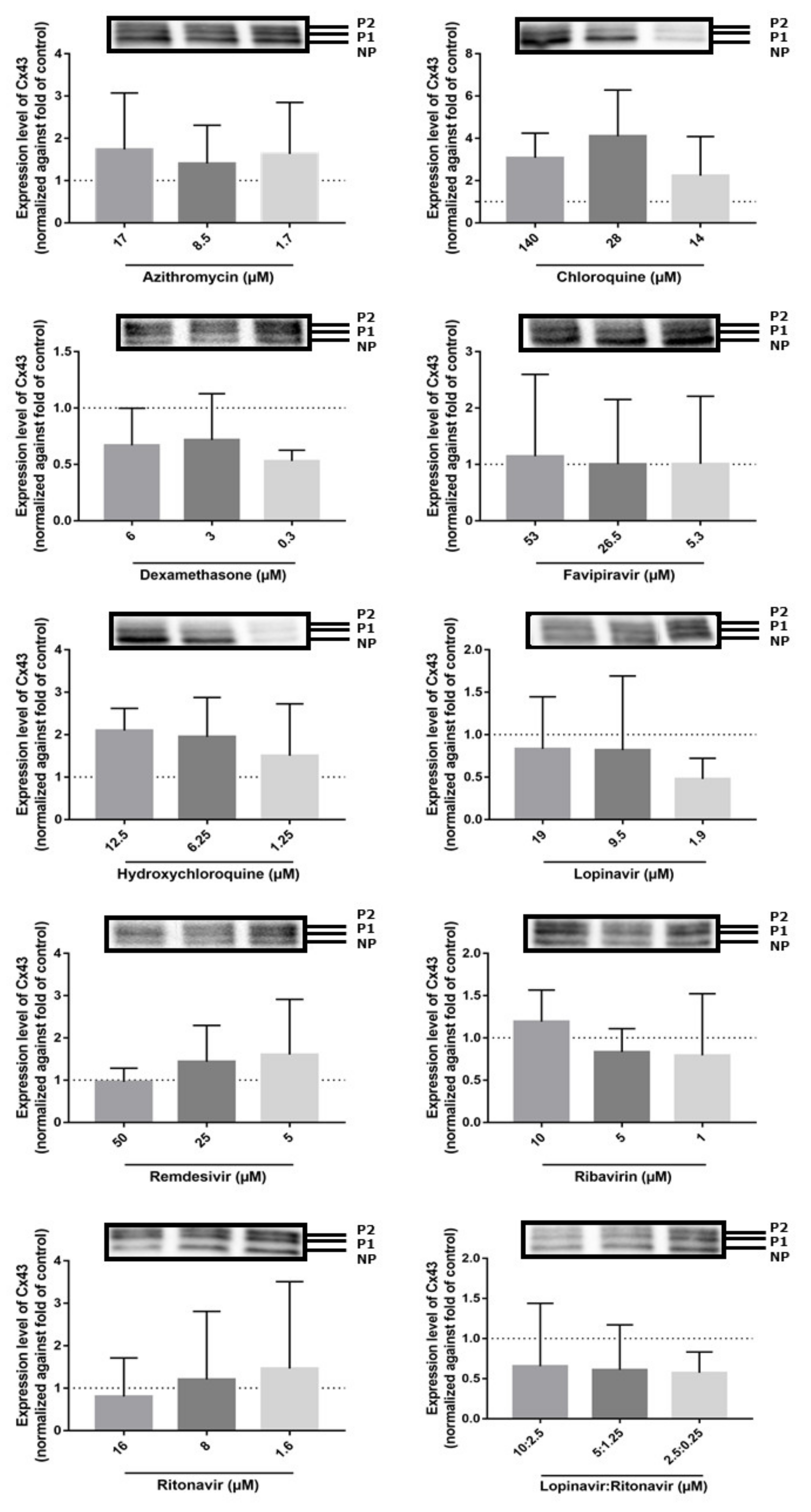
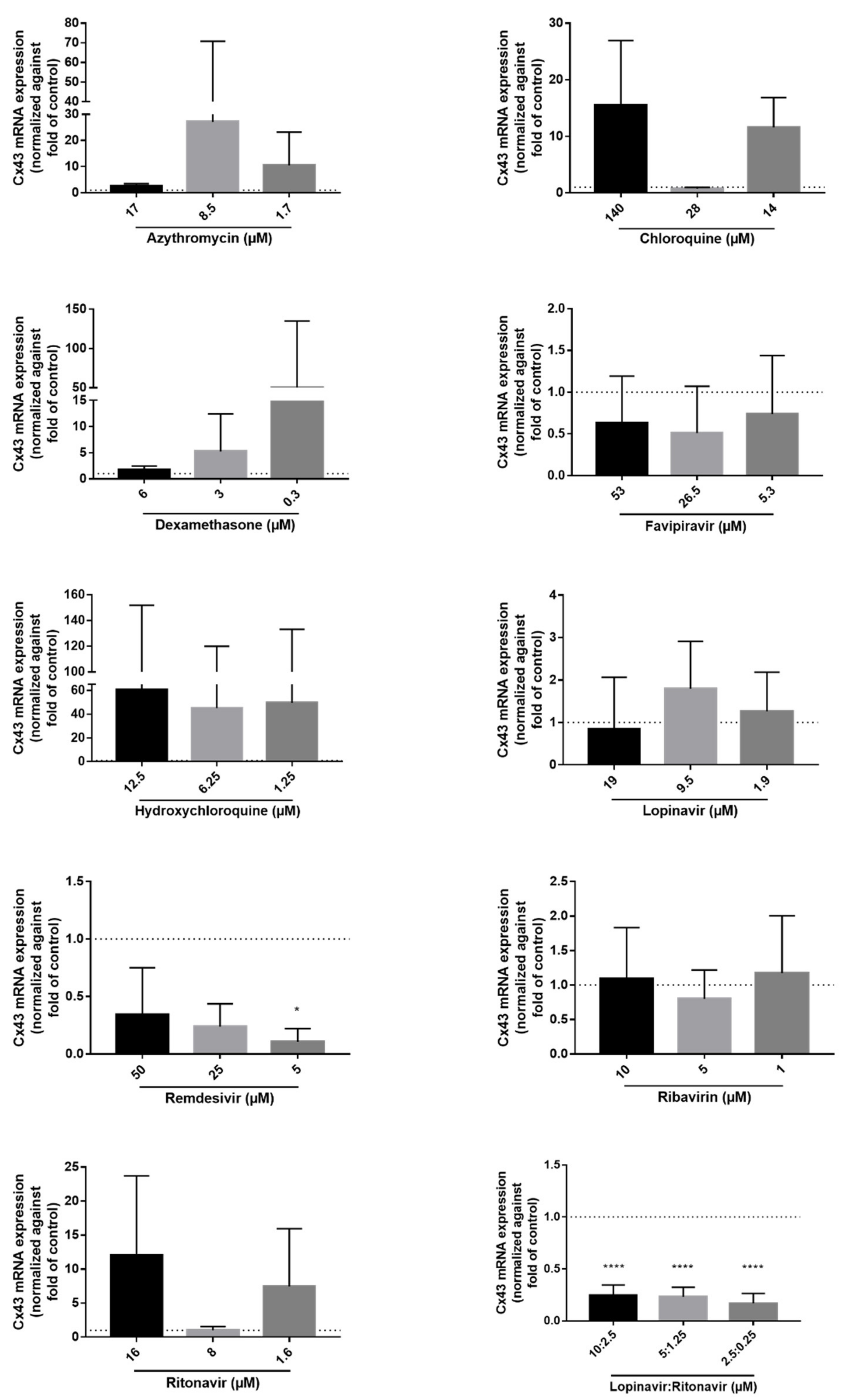
| Drug | Solvent of Stock Solutions | Cmax (µM) | CC10 (µM) | Concentration Range Tested for CC10 Determination (µM) | Concentration Range Tested for Functional Analysis (µM) | Concentration Range Tested for Expression Analysis (µM) | Reference | Supplier |
|---|---|---|---|---|---|---|---|---|
| Azithromycin (dihydrate) | DMSO | 0.52 [23] | 17 | 1–10–25–50–100–200 | 1.7–8.5–17–34–170 | 1.7–8.5–17 | A9834 | Sigma-Aldrich |
| Chloroquine (diphosphate) | Water | 0.81 [24] | 14 | 1–10–25–50–100–200 | 1.4–7–14–28–140 | 1.4–7–14 | C6628 | Sigma-Aldrich |
| Dexamethasone | DMSO | 0.63 [25] | >200 | 0.1–0.5–1–10–100–200 | 0.6–3–6–12–60 | 0.6–3–6 | D4902 | Sigma-Aldrich |
| Favipiravir | Water | 53.4 [26] | >400 | 1–10–25–50–100–200–400 | 5.3–26.5–53–106–530 | 5.3–26.5–53 | FF29069 | Bioynth Carbosynth |
| Hydroxychloroquine (sulphate) | Water | 1.36 [27] | 12.5 | 1–10–25–50–100–200 | 1.25–6.25–12.5–25–125 | 1.25–6.25–12.5 | HO915 | Sigma-Aldrich |
| Lopinavir | DMSO | 19.0 [28] | 19 | 0.25–5–25–50–100–200 | 1.9–9.5–19–38–190 | 1.9–9.5–19 | SML0491 | Sigma-Aldrich |
| Remdesivir | Water | 9.03 [29] | 50 | 0.01–0.1–1–10–50–100 | 5–25–50–100–500 | 5–25–50 | 30354–10 | Sanbio |
| Ribavirin | Water | 2.63 [30] | 10 | 1–10–25–50–100–200–400 | 1–5–10–20–100 | 1–5–10 | R9644 | Sigma-Aldrich |
| Ritonavir | DMSO | 1.17 [31] | 16 | 0.25–5–25–50–100–200 | 1.6–8–16–32–160 | 1.6–8–16 | SML1222 | Sigma-Aldrich |
| Lopinavir:Ritonavir (4:1) | DMSO | 19.0 [28] | 10:2.5 | 1–10–25–50–100–200 | 1:0.25–5:1.25–10:2.5–20:5–100:25 | 1:0.25–5:1–10:2.5 | SML1222 SML0491 | Sigma-Aldrich |
| Gene Symbol | Assay ID | Accession Number | Assay Location | Amplicon Size | Accession Number |
|---|---|---|---|---|---|
| UBC | Hs01871556-s1 | M26880.1 | 2173 | 135 | - |
| Gja1 | Hs00748445-s1 | NM_000165.5 | 1031 | 142 | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cooreman, A.; Caufriez, A.; Tabernilla, A.; Van Campenhout, R.; Leroy, K.; Kadam, P.; Sanz Serrano, J.; dos Santos Rodrigues, B.; Annaert, P.; Vinken, M. Effects of Drugs Formerly Proposed for COVID-19 Treatment on Connexin43 Hemichannels. Int. J. Mol. Sci. 2022, 23, 5018. https://doi.org/10.3390/ijms23095018
Cooreman A, Caufriez A, Tabernilla A, Van Campenhout R, Leroy K, Kadam P, Sanz Serrano J, dos Santos Rodrigues B, Annaert P, Vinken M. Effects of Drugs Formerly Proposed for COVID-19 Treatment on Connexin43 Hemichannels. International Journal of Molecular Sciences. 2022; 23(9):5018. https://doi.org/10.3390/ijms23095018
Chicago/Turabian StyleCooreman, Axelle, Anne Caufriez, Andrés Tabernilla, Raf Van Campenhout, Kaat Leroy, Prashant Kadam, Julen Sanz Serrano, Bruna dos Santos Rodrigues, Pieter Annaert, and Mathieu Vinken. 2022. "Effects of Drugs Formerly Proposed for COVID-19 Treatment on Connexin43 Hemichannels" International Journal of Molecular Sciences 23, no. 9: 5018. https://doi.org/10.3390/ijms23095018
APA StyleCooreman, A., Caufriez, A., Tabernilla, A., Van Campenhout, R., Leroy, K., Kadam, P., Sanz Serrano, J., dos Santos Rodrigues, B., Annaert, P., & Vinken, M. (2022). Effects of Drugs Formerly Proposed for COVID-19 Treatment on Connexin43 Hemichannels. International Journal of Molecular Sciences, 23(9), 5018. https://doi.org/10.3390/ijms23095018







