Metalloendopeptidase ADAM-like Decysin 1 (ADAMDEC1) in Colonic Subepithelial PDGFRα+ Cells Is a New Marker for Inflammatory Bowel Disease
Abstract
:1. Introduction
2. Results
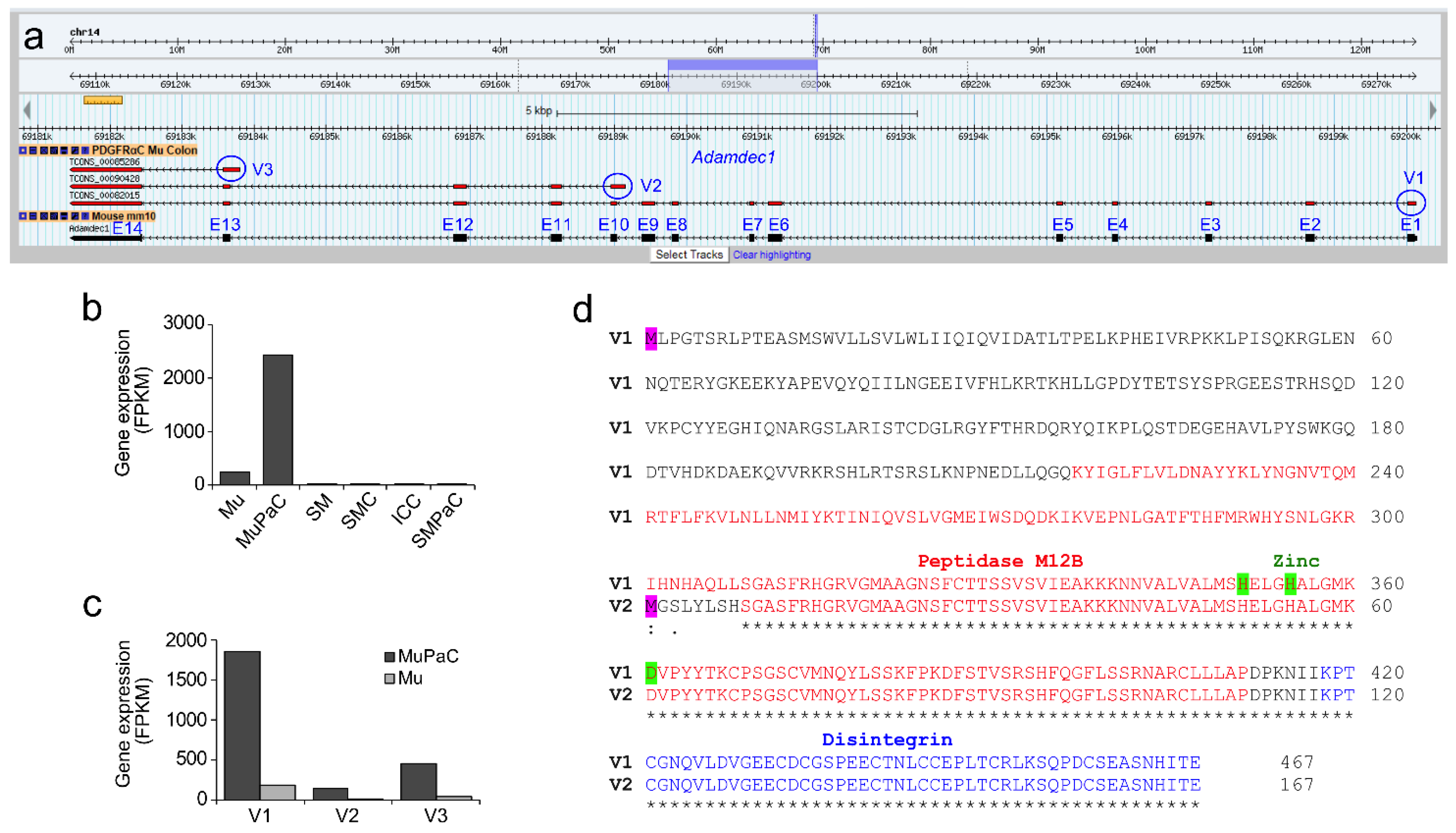
2.1. Selective Expression of Adamdec1 within Intestinal PDGFRα+ Cells and Localization of the Protein to the Mucosal Lining
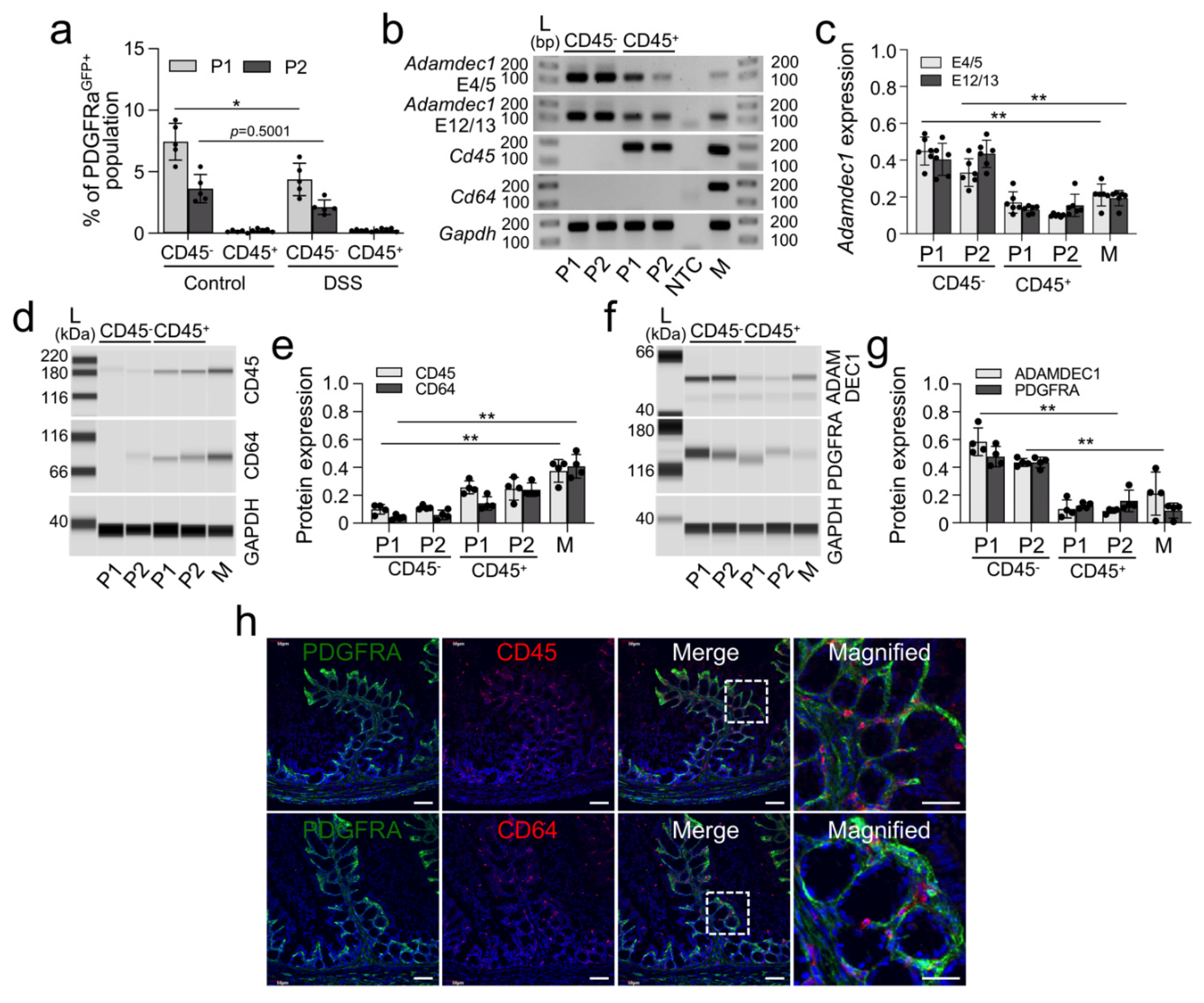
2.2. Reduction of Colonic Mucosal PDGFRα+ Cells in DSS-Induced Colitis
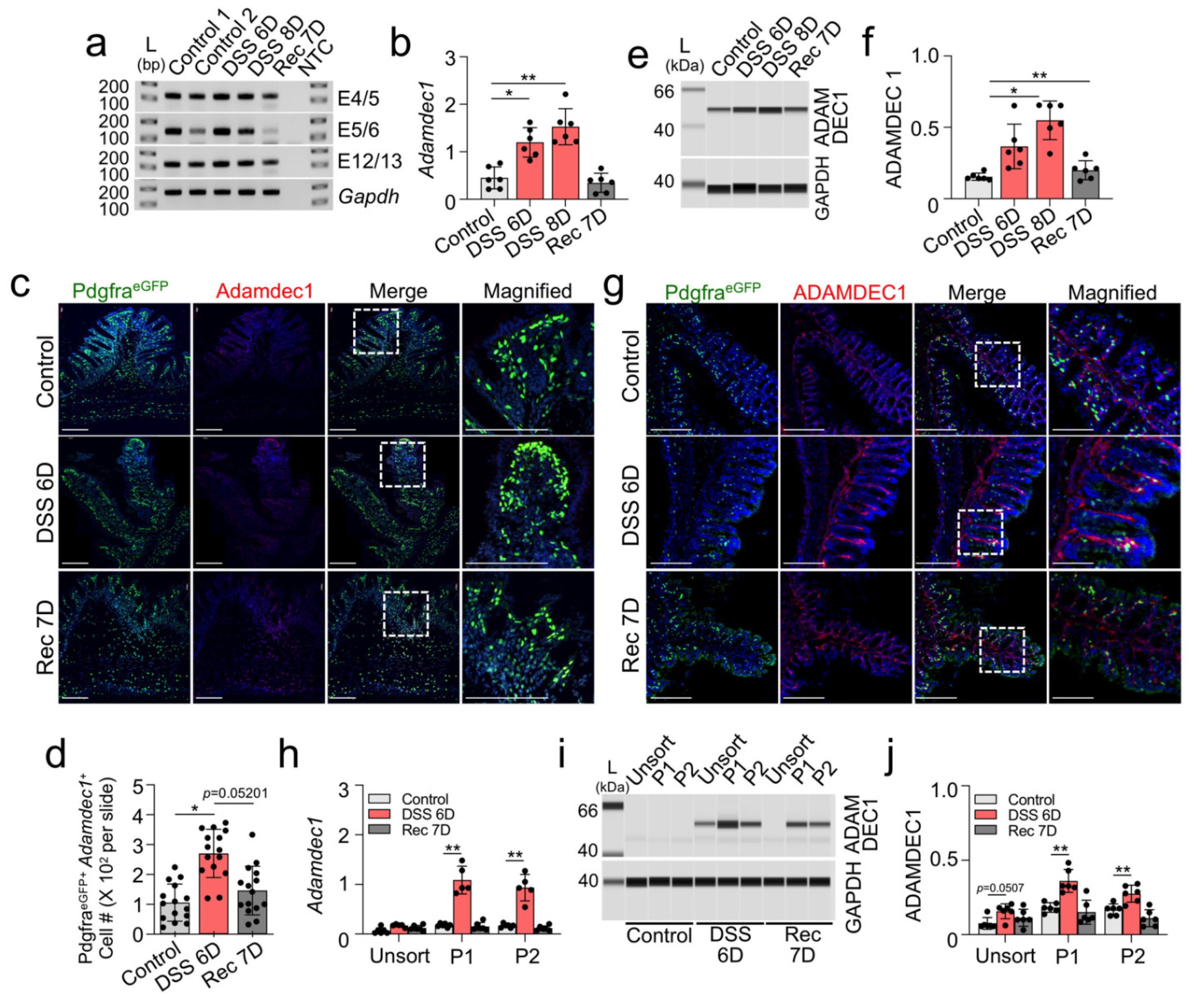
2.3. Upregulation of Adamdec1 Transcripts and Protein in Colonic Mucosal PDGFRα+ Cells in DSS-Induced Colitis
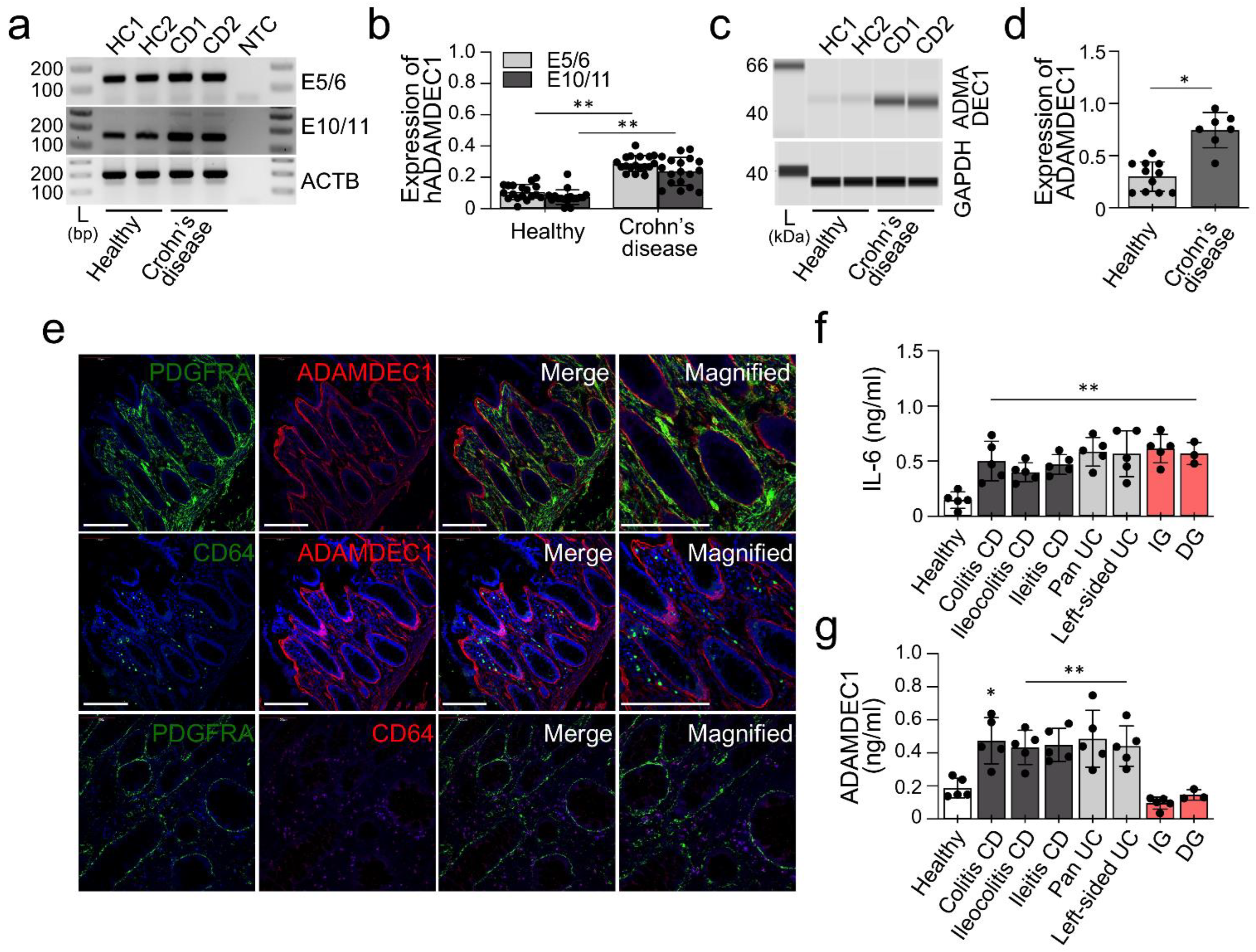
2.4. ADAMDEC1 Is Upregulated in Human Tissue Affected by IBD
3. Discussion
4. Methods and Materials
4.1. Animal and Tissue Preparation
4.2. Dextran Sulfate Sodium Treatment
4.3. Human Samples
4.4. Fluorescence-Activated Cell Sorting (FACS)
4.5. Isolation of Total RNAs
4.6. Real Time PCR
4.7. Confocal Microscopy and Immunofluorescence Analysis
4.8. Automated Immunoblot
4.9. RNA In Situ Hybridization
4.10. Enzyme-Linked Immunosorbent Assay (ELISA)
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mueller, C.G.; Rissoan, M.C.; Salinas, B.; Ait-Yahia, S.; Ravel, O.; Bridon, J.M.; Briere, F.; Lebecque, S.; Liu, Y.J. Polymerase chain reaction selects a novel disintegrin proteinase from CD40-activated germinal center dendritic cells. J. Exp. Med. 1997, 186, 655–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casewell, N.R. On the ancestral recruitment of metalloproteinases into the venom of snakes. Toxicon 2012, 60, 449–454. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, N.R.; Chew, T.S.; Dunne, J.; Marnane, R.; Nedjat-Shokouhi, B.; Smith, P.J.; Bloom, S.L.; Smith, A.M.; Segal, A.W. Critical Role of the Disintegrin Metalloprotease ADAM-like Decysin-1 [ADAMDEC1] for Intestinal Immunity and Inflammation. J. Crohn’s Colitis 2016, 10, 1417–1427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Bruyn, M.; Machiels, K.; Vandooren, J.; Lemmens, B.; Van Lommel, L.; Breynaert, C.; Van der Goten, J.; Staelens, D.; Billiet, T.; De Hertogh, G.; et al. Infliximab restores the dysfunctional matrix remodeling protein and growth factor gene expression in patients with inflammatory bowel disease. Inflamm. Bowel Dis. 2014, 20, 339–352. [Google Scholar] [CrossRef]
- Smith, A.M.; Sewell, G.W.; Levine, A.P.; Chew, T.S.; Dunne, J.; O’Shea, N.R.; Smith, P.J.; Harrison, P.J.; Macdonald, C.M.; Bloom, S.L.; et al. Disruption of macrophage pro-inflammatory cytokine release in Crohn’s disease is associated with reduced optineurin expression in a subset of patients. Immunology 2015, 144, 45–55. [Google Scholar] [CrossRef] [Green Version]
- Pasini, F.S.; Zilberstein, B.; Snitcovsky, I.; Roela, R.A.; Mangone, F.R.; Ribeiro, U., Jr.; Nonogaki, S.; Brito, G.C.; Callegari, G.D.; Cecconello, I.; et al. A gene expression profile related to immune dampening in the tumor microenvironment is associated with poor prognosis in gastric adenocarcinoma. J. Gastroenterol. 2014, 49, 1453–1466. [Google Scholar] [CrossRef] [Green Version]
- Supiot, S.; Gouraud, W.; Campion, L.; Jezequel, P.; Buecher, B.; Charrier, J.; Heymann, M.F.; Mahe, M.A.; Rio, E.; Cherel, M. Early dynamic transcriptomic changes during preoperative radiotherapy in patients with rectal cancer: A feasibility study. World J. Gastroenterol. 2013, 19, 3249–3254. [Google Scholar] [CrossRef]
- Fritsche, J.; Müller, A.; Hausmann, M.; Rogler, G.; Andreesen, R.; Kreutz, M. Inverse regulation of the ADAM-family members, decysin and MADDAM/ADAM19 during monocyte differentiation. Immunology 2003, 110, 450–457. [Google Scholar] [CrossRef]
- Lee, M.Y.; Park, C.; Berent, R.M.; Park, P.J.; Fuchs, R.; Syn, H.; Chin, A.; Townsend, J.; Benson, C.C.; Redelman, D.; et al. Smooth Muscle Cell Genome Browser: Enabling the Identification of Novel Serum Response Factor Target Genes. PLoS ONE 2015, 10, e0133751. [Google Scholar] [CrossRef] [Green Version]
- Ha, S.E.; Lee, M.Y.; Kurahashi, M.; Wei, L.; Jorgensen, B.G.; Park, C.; Park, P.J.; Redelman, D.; Sasse, K.C.; Becker, L.S.; et al. Transcriptome analysis of PDGFRalpha+ cells identifies T-type Ca2+ channel CACNA1G as a new pathological marker for PDGFRalpha+ cell hyperplasia. PLoS ONE 2017, 12, e0182265. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.Y.; Ha, S.E.; Park, C.; Park, P.J.; Fuchs, R.; Wei, L.; Jorgensen, B.G.; Redelman, D.; Ward, S.M.; Sanders, K.M.; et al. Transcriptome of interstitial cells of Cajal reveals unique and selective gene signatures. PLoS ONE 2017, 12, e0176031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breland, A.; Ha, S.E.; Jorgensen, B.G.; Jin, B.; Gardner, T.A.; Sanders, K.M.; Ro, S. Smooth Muscle Transcriptome Browser: Offering genome-wide references and expression profiles of transcripts expressed in intestinal SMC, ICC, and PDGFRalpha(+) cells. Sci. Rep. 2019, 9, 387. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.E.; Byungchang, J.; Jorgensen, B.G.; Zogg, H.; Wei, L.; Singh, R.; Park, C.; Kurahashi, M.; Kim, S.; Baek, G.; et al. Transcriptome profiling of subepithelial PDGFRα+ cells in colonic mucosa reveals several cell-selective markers. PLoS ONE, 2022; in print. [Google Scholar]
- Kurahashi, M.; Nakano, Y.; Peri, L.E.; Townsend, J.B.; Ward, S.M.; Sanders, K.M. A novel population of subepithelial platelet-derived growth factor receptor alpha-positive cells in the mouse and human colon. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 304, G823–G834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norton, S.E.; Dunn, E.T.; McCall, J.L.; Munro, F.; Kemp, R.A. Gut macrophage phenotype is dependent on the tumor microenvironment in colorectal cancer. Clin. Transl. Immunol. 2016, 5, e76. [Google Scholar] [CrossRef] [PubMed]
- Prantner, D.; Nallar, S.; Richard, K.; Spiegel, D.; Collins, K.D.; Vogel, S.N. Classically activated mouse macrophages produce methylglyoxal that induces a TLR4- and RAGE-independent proinflammatory response. J. Leukoc. Biol. 2020, 109, 605–619. [Google Scholar] [CrossRef]
- Lu, Y.C.; Yeh, W.C.; Ohashi, P.S. LPS/TLR4 signal transduction pathway. Cytokine 2008, 42, 145–151. [Google Scholar] [CrossRef]
- Takakura, N.; Yoshida, H.; Ogura, Y.; Kataoka, H.; Nishikawa, S.; Nishikawa, S.I. PDGFR alpha expression during mouse embryogenesis—Immunolocalization analyzed by whole-mount immunohistostaining using the monoclonal anti-mouse PDGFR alpha antibody APA5. J. Histochem. Cytochem. 1997, 45, 883–893. [Google Scholar] [CrossRef] [Green Version]
- Gottfried-Blackmore, A.; Namkoong, H.; Adler, E.; Martin, B.; Gubatan, J.; Fernandez-Becker, N.; Clarke, J.O.; Idoyaga, J.; Nguyen, L.; Habtezion, A. Gastric Mucosal Immune Profiling and Dysregulation in Idiopathic Gastroparesis. Clin. Transl. Gastroenterol. 2021, 12, e00349. [Google Scholar] [CrossRef]
- Yoshida, S.; Setoguchi, M.; Higuchi, Y.; Akizuki, S.; Yamamoto, S. Molecular cloning of cDNA encoding MS2 antigen, a novel cell surface antigen strongly expressed in murine monocytic lineage. Int. Immunol. 1990, 2, 585–591. [Google Scholar] [CrossRef]
- Black, R.A.; Rauch, C.T.; Kozlosky, C.J.; Peschon, J.J.; Slack, J.L.; Wolfson, M.F.; Castner, B.J.; Stocking, K.L.; Reddy, P.; Srinivasan, S.; et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature 1997, 385, 729–733. [Google Scholar] [CrossRef] [PubMed]
- Roberts, C.M.; Tani, P.H.; Bridges, L.C.; Laszik, Z.; Bowditch, R.D. MDC-L, a novel metalloprotease disintegrin cysteine-rich protein family member expressed by human lymphocytes. J. Biol. Chem. 1999, 274, 29251–29259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fritsche, J.; Moser, M.; Faust, S.; Peuker, A.; Buttner, R.; Andreesen, R.; Kreutz, M. Molecular cloning and characterization of a human metalloprotease disintegrin--a novel marker for dendritic cell differentiation. Blood 2000, 96, 732–739. [Google Scholar] [CrossRef] [PubMed]
- Mueller, C.G.; Cremer, I.; Paulet, P.E.; Niida, S.; Maeda, N.; Lebeque, S.; Fridman, W.H.; Sautes-Fridman, C. Mannose receptor ligand-positive cells express the metalloprotease decysin in the B cell follicle. J. Immunol. 2001, 167, 5052–5060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, R.; Jin, G.; McIntyre, T.M. The soluble protease ADAMDEC1 released from activated platelets hydrolyzes platelet membrane pro-epidermal growth factor (EGF) to active high-molecular-weight EGF. J. Biol. Chem. 2017, 292, 10112–10122. [Google Scholar] [CrossRef] [Green Version]
- Kurahashi, M.; Niwa, Y.; Cheng, J.; Ohsaki, Y.; Fujita, A.; Goto, H.; Fujimoto, T.; Torihashi, S. Platelet-derived growth factor signals play critical roles in differentiation of longitudinal smooth muscle cells in mouse embryonic gut. Neurogastroenterol. Motil. 2008, 20, 521–531. [Google Scholar] [CrossRef]
- Sanders, K.M.; Ordog, T.; Koh, S.D.; Torihashi, S.; Ward, S.M. Development and plasticity of interstitial cells of Cajal. Neurogastroenterol. Motil. 1999, 11, 311–338. [Google Scholar] [CrossRef]
- Park, C.; Lee, M.Y.; Park, P.J.; Ha, S.E.; Berent, R.M.; Fuchs, R.; Miano, J.M.; Becker, L.S.; Sanders, K.M.; Ro, S. Serum Response Factor Is Essential for Prenatal Gastrointestinal Smooth Muscle Development and Maintenance of Differentiated Phenotype. J. Neurogastroenterol. Motil. 2015, 21, 589–602. [Google Scholar] [CrossRef] [Green Version]
- Plaza-Diaz, J.; Robles-Sanchez, C.; Abadia-Molina, F.; Moron-Calvente, V.; Saez-Lara, M.J.; Ruiz-Bravo, A.; Jimenez-Valera, M.; Gil, A.; Gomez-Llorente, C.; Fontana, L. Adamdec1, Ednrb and Ptgs1/Cox1, inflammation genes upregulated in the intestinal mucosa of obese rats, are downregulated by three probiotic strains. Sci. Rep. 2017, 7, 1939. [Google Scholar] [CrossRef] [Green Version]
- Bates, E.E.; Fridman, W.H.; Mueller, C.G. The ADAMDEC1 (decysin) gene structure: Evolution by duplication in a metalloprotease gene cluster on chromosome 8p12. Immunogenetics 2002, 54, 96–105. [Google Scholar] [CrossRef]
- Lund, J.; Elimar Bitsch, A.M.; Gronbech Rasch, M.; Enoksson, M.; Troeberg, L.; Nagase, H.; Loftager, M.; Overgaard, M.T.; Petersen, H.H. Monoclonal antibodies targeting the disintegrin-like domain of ADAMDEC1 modulates the proteolytic activity and enables quantification of ADAMDEC1 protein in human plasma. MAbs 2018, 10, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Lund, J.; Olsen, O.H.; Sorensen, E.S.; Stennicke, H.R.; Petersen, H.H.; Overgaard, M.T. ADAMDEC1 is a metzincin metalloprotease with dampened proteolytic activity. J. Biol. Chem. 2013, 288, 21367–21375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamilton, T.G.; Klinghoffer, R.A.; Corrin, P.D.; Soriano, P. Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol. Cell Biol. 2003, 23, 4013–4025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, L.; Singh, R.; Ha, S.E.; Martin, A.M.; Jones, L.A.; Jin, B.; Jorgensen, B.G.; Zogg, H.; Chervo, T.; Gottfried-Blackmore, A.; et al. Serotonin Deficiency Is Associated with Delayed Gastric Emptying. Gastroenterology 2021, 160, 2451–2466.e19. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ha, S.E.; Jorgensen, B.G.; Wei, L.; Jin, B.; Kim, M.-S.; Poudrier, S.M.; Singh, R.; Bartlett, A.; Zogg, H.; Kim, S.; et al. Metalloendopeptidase ADAM-like Decysin 1 (ADAMDEC1) in Colonic Subepithelial PDGFRα+ Cells Is a New Marker for Inflammatory Bowel Disease. Int. J. Mol. Sci. 2022, 23, 5007. https://doi.org/10.3390/ijms23095007
Ha SE, Jorgensen BG, Wei L, Jin B, Kim M-S, Poudrier SM, Singh R, Bartlett A, Zogg H, Kim S, et al. Metalloendopeptidase ADAM-like Decysin 1 (ADAMDEC1) in Colonic Subepithelial PDGFRα+ Cells Is a New Marker for Inflammatory Bowel Disease. International Journal of Molecular Sciences. 2022; 23(9):5007. https://doi.org/10.3390/ijms23095007
Chicago/Turabian StyleHa, Se Eun, Brian G. Jorgensen, Lai Wei, Byungchang Jin, Min-Seob Kim, Sandra M. Poudrier, Rajan Singh, Allison Bartlett, Hannah Zogg, Sei Kim, and et al. 2022. "Metalloendopeptidase ADAM-like Decysin 1 (ADAMDEC1) in Colonic Subepithelial PDGFRα+ Cells Is a New Marker for Inflammatory Bowel Disease" International Journal of Molecular Sciences 23, no. 9: 5007. https://doi.org/10.3390/ijms23095007
APA StyleHa, S. E., Jorgensen, B. G., Wei, L., Jin, B., Kim, M.-S., Poudrier, S. M., Singh, R., Bartlett, A., Zogg, H., Kim, S., Baek, G., Kurahashi, M., Lee, M.-Y., Kim, Y.-S., Choi, S.-C., Sasse, K. C., Rubin, S. J. S., Gottfried-Blackmore, A., Becker, L., ... Ro, S. (2022). Metalloendopeptidase ADAM-like Decysin 1 (ADAMDEC1) in Colonic Subepithelial PDGFRα+ Cells Is a New Marker for Inflammatory Bowel Disease. International Journal of Molecular Sciences, 23(9), 5007. https://doi.org/10.3390/ijms23095007






