Abstract
The ETS-related gene (ERG) is proto-oncogene that is classified as a member of the ETS transcription factor family, which has been found to be consistently overexpressed in about half of the patients with clinically significant prostate cancer (PCa). The overexpression of ERG can mostly be attributed to the fusion of the ERG and transmembrane serine protease 2 (TMPRSS2) genes, and this fusion is estimated to represent about 85% of all gene fusions observed in prostate cancer. Clinically, individuals with ERG gene fusion are mostly documented to have advanced tumor stages, increased mortality, and higher rates of metastasis in non-surgical cohorts. In the current review, we elucidate ERG’s molecular interaction with downstream genes and the pathways associated with PCa. Studies have documented that ERG plays a central role in PCa progression due to its ability to enhance tumor growth by promoting inflammatory and angiogenic responses. ERG has also been implicated in the epithelial–mesenchymal transition (EMT) in PCa cells, which increases the ability of cancer cells to metastasize. In vivo, research has demonstrated that higher levels of ERG expression are involved with nuclear pleomorphism that prompts hyperplasia and the loss of cell polarity.
1. Background
In the past four years, the annual diagnostic rate of PCa cases have risen from 161,360 to 248,530 patients in the US alone, which is indicative of a 54% increase. Consequently, prostate cancer (PCa) has become increasingly prevalent within men, especially in western populations [1]. Currently, PCa is now the most common type of cancer and is the second leading cause of death among male cancer patients [2,3].
The ETS-related gene (ERG) is a transcription factor that is encoded by ERG and has been identified to be consistently overexpressed in the malignant epithelial cells found in PCa, demonstrating its potential as a biomarker or therapeutic target [4,5].
ERG was first discovered in 1987 when a cDNA clone was isolated from a library of colon cancer cells and was labeled as a new member of the E-26 transformation-specific (ETS) oncogene family [5]. ETS and ETS-related genes have been demonstrated to behave as major regulators of transcriptional activity and are directly involved in proliferation, differentiation, vasculogenesis, and angiogenesis [6,7]. Bioinformatic analysis has uncovered various fusions in the 5’ untranslated region of TMPRSS2 (21q22) when it is fused to ERG (21q22), ETV1 (7p21), ETV4 (17q21), or ETV5 (3q27), resulting in the overexpression of ETS and ETS-related genes in prostate cancer [8,9,10,11]. TMPRSS2-ERG fusion appears in approximately 50% of PCa peripheral zone tumors, while this fusion is only apparent in 12% of PCa transition zone tumors [12,13]. Additionally, ERG rearrangements seem to occur at variable frequencies depending on the studied population, with Caucasian Americans expressing ERG at a frequency of 50–55%, and African Americans showing an ERG frequency of 28% [14]. It was also observed that populations of Asian descent had varying ERG frequencies, ranging from 23% in Chinese PCa patients to 49% in Indian PCa patients [15]. We illustrate the fusion mechanism and its prevalence among different ethnicities in Figure 1.
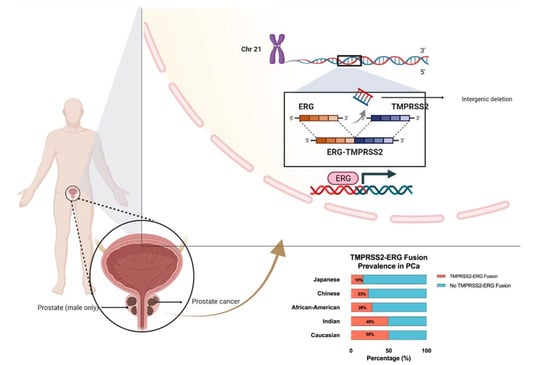
Figure 1.
Schematic diagram of TMPRSS2-ERG gene fusion on chromosome 21 and the ethnic prevalence of TMPRSS2-ERG gene fusion in prostate cancer patients.
Under normal physiological conditions, ERG has many developmental functions that vary depending on the cell type or the organism’s developmental stage. During embryogenesis, ERG has been shown to be highly expressed in the mesoderm and endothelium, where it plays an important role in vasculogenisis and in the development of bone [16]. Other endothelial genes that are positively regulated by ERG include vascular endothelial growth factor (VEGF), the von Willebrand factor, and Endoglin, which are all involved in endothelial cell differentiation and angiogenesis [17,18].
It appears that TMPRSS2-ERG gene fusion plays a role in inhibiting androgen receptor-driven differentiation, therefore producing de-differentiated cells with stem-like properties that are correlated with the EMT pathway [19,20]. The overexpression of ERG has also been observed in other cancer subtypes, such as leukemia and Ewing’s sarcoma, which indicate its potent potential as an oncogene [21]. In vivo, it has been demonstrated that the overexpression of ERG induces neoplastic changes, and its overexpression has also been correlated with epithelial nuclear pleomorphism and the loss of cell polarity [21,22]. Clinically, individuals with higher ERG expression have been found to have more advanced tumor stages, elevated Gleason scores, increased mortality, and metastasis. The overexpression of ERG has consistently been identified in the malignant epithelial cells found in PCa, demonstrating its potential as a possible biomarker and therapeutic target [23]. In this study, we reviewed and elucidated the oncogenic role of TMPRSS2-ERG fusion in PCa and the mechanism underlining its association with PCa progression and metastasis.
2. Structural Characteristics and Allosteric Autoinhibition of ERG
The ERG transcription factor has a primary structure that consists of 486 amino acids and a corresponding molecular weight of 54kDa [21]. A distinguishing characteristic of ETS family proteins is the presence of a DNA-binding domain called the ETS DNA binding domain (EBD). This EBD domain is made of 85 amino acids and contains 3 alpha-helices that are further supported by a 4-stranded anti-parallel beta sheet [24]. The EBD domain plays a critical role in DNA recognition as well as in AP-1 and co-activator recruitment [25]. Within the EBD, there are three highly conserved tryptophan residues that serve as a hydrophobic core to facilitate the helix–turn–helix binding domain in proteins [26]. ERG analysis by means peptide sequencing has also predicted the presence of phosphorylation sites for protein kinase C and a pointed (PNT) domain in the N-terminus [21]. The PNT domains are a part of a larger sterile alpha motif (SAM) family that is involved in many diverse protein–protein interactions that can allow for self-association [27]. This PNT domain has also been shown to facilitate the heterodimerization of ERG with other proteins, including other members of the ETS family, DNA-dependent kinases, AP-1 complex, and the androgen receptor (AR) [28].
3. The Function of ETS-Related Gene (ERG) in Normal Cell Types
ERG has a multitude of physiological functions that differ based on the type of cells or the organism’s developmental stage. During embryogenesis, ERG has been observed to be highly expressed in the mesoderm and endothelium, where it plays a crucial role in vasculogenesis and in the development of bone [29]. In adults, it regulates vascular homeostasis and angiogenesis by activating the transcription of endothelial specific genes, such as vascular endothelial (VE)-cadherin, an adhesion molecule that promotes vascular stability by maintaining and controlling endothelial cell contact [30]. In addition to this, VE-cadherin also plays a central role in cell proliferation and apoptosis and modulates endothelial growth factor receptor functions [31]. Other endothelial genes that are positively regulated by ERG include the vascular endothelial growth factor (VEGF), von Willebrand factor, and endoglin, which are all involved in endothelial cell differentiation and angiogenesis [26,32].
4. Prominent TMPRSS2-ERG Gene Fusion Found in PCa
In prostate cancer cells, a surprisingly common occurrence involves the fusion of ERG to TMPRSS2, which forms the fusion product of TMPRSS2-ERG. The most common mechanism by which these two genes fuse involves the deletion of intronic sequences on the long arm of chromosome 21 via an intron deletion between TMPRSS2 and ERG on chromosome 21q22.2-3 (Figure 1). This fusion mechanism has been identified as being prevalent in approximately 50% of prostate cancer patients [33]. The frequent occurrence of this fusion protein can be attributed to the presence of a homogenous deletion site that is present between ERG and TMPRSS2 [34]. Moreover, this deletion site is separated into two different classifications according to various start sites. In both of the deletion products, the 5′ end of the TMPRSS2 gene has been ligated to the 3′ end of ERG. TMPRSS2-ERG fusion results in ERG overexpression due to the androgen responsive promoter of the TMPSS2 gene allowing for the constitutive transcription of ERG, which has been shown to be correlated with increased cell proliferation, cell invasion, angiogenesis, and invasiveness in PCa cells [35,36]. In addition, this TMPRSS2-ERG fusion enhances the transcription and activates downstream oncogenes [37].
5. Functional ERG Overexpression in Prostate Cancer Cells
In prostate cancer cells, ERG overexpression increases the rate of epithelial to mesenchymal transitions via the EMT pathway, enhancing the ability of PCa cells to invade and metastasize. ERG achieves this by upregulating matrix metalloproteinases (MMPs), CXCR4, and Osteopontin (OPN), which have been correlated with higher rates of cell invasion and metastasis among patients [38]. Additionally, the TMPRSS2-ERG pathway reveals the epithelial to mesenchymal transition via the ZEB1/ZEB2 axis in PCa [39]. The constitutive expression of ERG also hyperactivates the inflammatory pathway in PCa cells by binding to Toll-like receptor 4; this activates the NF-kb pathway, increasing the transcription of target genes such as TNFA, IL6, BCLXL, BCL2, BCLXS, XIAP, and VEGF [40,41]. These proteins trigger tumor growth and progression by enhancing cell proliferation, survival, and angiogenesis. Tumor growth is further accentuated by the activation of the EZH2 promoter by ERG. This relieves the epigenetic inhibition of tumor suppressor genes such as NKX3.1, resulting in the constitutive expression of the TMPRSS2-ERG fusion gene. In addition to its role in regulating tumor cell invasion and proliferation, ERG also plays an important role in negatively regulating tumor cell differentiation by inhibiting the transcription of genes such as KLK3/PSA and SLC45A3/Prostein [42]. Altogether, the overexpression of ERG in prostate cancer is a key modulator of tumor progression and aggressiveness, as it can regulate the transcription of the proteins that mediate inflammation, cell invasion, differentiation, and oncogenesis [43].
6. ERG Ameliorates Cell Cycle Driving Genes
In ERG knockout mice, there was significant decrease in the number of cells arrested at G0 and an increase in cells at G1. This demonstrates ERG’s role in maintaining the homeostasis of the hematopoietic stem cell (HSC) population. This suggests that ERG is a major cell cycle regulator in HSC niches and that it works to maintain a constant balance between cell differentiation and renewal. It is revelated that in the absence of Phosphate and Tensin Homolog (PTEN)/TP53, ERG binds directly to the chromatin loci of various cell cycle-driving genes and reduces their expression while activating the RB gene [44,45]. This leads to E2F1 inhibition and, interestingly, the stability of luminal epithelial cell identity, antiandrogen sensitivity, and CDK4/6 inhibitor resistance.
7. ERG and the Androgen Receptor (AR)
The androgen receptor (AR) is a transcription factor for a nuclear hormone receptor that regulates growth and development in normal prostate cells [46]. In normal cells, there is a certain combination of transcription factors and histone modifications that are specific to AR known as the AR cistrome. When this AR cistrome remains unchanged, it facilitates the normal development of prostate tissue. However, modifications to this cistrome have been implicated in enhancing tumor growth and progression in PCa [47]. Two factors that have been associated with reprogramming the AR cistrome include FOXA1 and HOXB13 [48]. ERG overexpression and PTEN loss have been associated with inducing the expression of these factors [49]. Furthermore, ERG can form a complex with AR through an AR-interacting motif (AIM), and a cysteine residue that is utilized to form cross-linkers between AR and ERG exists in this domain [46]. This allows it to act as a cofactor for AR to enhance DNA binding in both high- and low- affinity conditions [50]. Quantitatively, data have shown that the formation of this complex results in a three-fold increase in AR’s DNA-binding ability [46]. With this enhanced binding ability, AR sites have been observed to be concentrated at FOXA1 and HOXB13, along with AP1 and androgen response elements (ARE) [48]. Therefore, ERG overexpression results in AR site enrichment at the FOXA1 and HOXB13 motifs, resulting in increased expression. The increase in FOXA1 and HOXB13 is then found to modify the AR cistrome, further worsening tumorigenesis. PTEN loss also contributes to this, as PI3K and AR inhibition will be lost, resulting in constitutive AR and PI3K signalling [51]. ERG overexpression has been correlated to the relative resistance of AR-targeted therapeutics [52]. Furthermore, the overexpression of ERG in MSKPCa2 cells demonstrating high AR expression demonstrated increased cell growth in the presence of enzalutamide, which is a PCa treatment that works as an AR inhibitor [50]. Therefore, ERG and AR overexpression result in prostate cancer cells having increased tolerance to androgen receptor antagonists.
8. ERG PTEN and TP53 Crosstalk
It is important to note that the overexpression of ERG via TMPRSS2-ERG fusion is not a definitive indicator of biochemical recurrence or survival and requires information regarding the PTEN and TP53 status [44,53]. PTEN is a tumor suppressor, and its inactivation is one of the most significant prognostic biomarkers in prostate cancer [54]. Moreover, the PI3K pathway is directly controlled by PTEN, and PTEN loss has been correlated with a hyperactive PI3K signalling pathway, which regulates cell survival and proliferation, in various cancers [53]. Furthermore, the combination of ERG activation and PTEN or TP53 loss can induce cell migration and transform prostatic intraepithelial neoplasia into invasive carcinoma [55].
9. ERG Stimulates Endothelial Gene Expression Utilizing the Canonical Wnt Signalling Pathway
ERG stimulates endothelial gene expression by utilizing the canonical Wnt signalling pathway. When Wnt ligands bind to frizzled receptors, it triggers a signalling cascade that prevents phosphorylation and therefore acts as a β-catenin stabilizer. The stability β-catenin allows it to translocate into the nucleus and promote the transcription of various other genes. Patients who have been identified as having TMPRSS2-ERG gene fusion have been shown to exhibit increased ERG activity, which has been directly correlated to mRNA levels with various Wnt ligands, including WNT2, WNT3A, and WNT11 [56]. Frizzled receptors such as FZD4, which has been found to mediate EMT in prostate cancer cells [57,58], have been found to also be upregulated by ERG and to contribute to increased Wnt signalling activity (Figure 2).
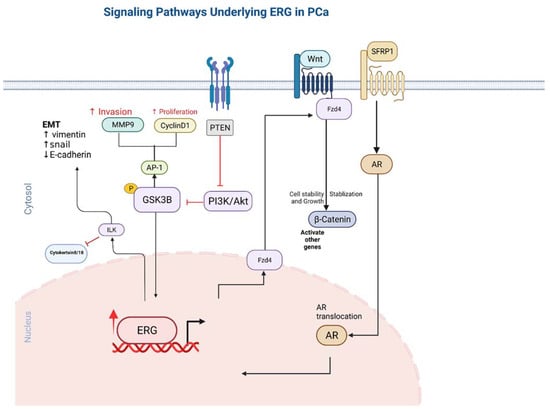
Figure 2.
The pathways involved in ERG overexpression and related outcomes in tumor cells. Directional arrows indicate activation.
FZD4 is known as an Wnt receptor in the Wnt signalling pathway and is directly related to the B-catenin signalling pathway [58]. FZD4 is required for the oncogenic effects of ERG fusion. Interestingly, FZD4 knockdown induces phenotype results in active B-integrin and E-cadherin expression that are similar to those of ERG knockdown. On the other hand, FZD4 overexpression clearly reverses the impact of ERG knockdown in PCa cell lines, showing the direct correlation and co-regulation of these two genes.
In the Wnt signaling pathway, the binding of Wnts to various receptors results in the accumulation of ß-catenin, which, when recruited by the LEF1 transcription factor, targets multiple oncogenes such as c-MYC and MMPs [59]. Studies have shown that the LEF1 promoter is the most important TMPRSS2-ERG target of the Wnt signaling pathway, as LEF1 is knockdown results in the complete inhibition of the ERG-induced Wnt pathway [59].
Another study also showed that Wnt signalling is involved in the self-renewal and differentiation of cancer stem cells. Moreover, in LNCap and C4-2B PCa cells, WNT3A levels have been positively correlated with prostasphere size and self-renewal, with increased expression resulting in a 1.5× increase in sphere formations [60]. Additionally, WNT3A has been shown to support the progression of PIN lesions into more advanced adenocarcinomas and to increase cell resistance to androgen deprivation [17]. Previous studies have shown that the increased expression of WNT1 and LEF1, especially through the Wnt signaling pathway, is correlated with lethality, hormone resistance PCa, and the EMT pathway [61].
SFRP proteins are a group composed of five proteins that are structurally and functionally similar to Wnt ligands and that act as Wnt pathway antagonists by binding to Wnt proteins or Frizzled receptors [62]. There have been previous studies that present the low expression of SFRP1 proteins in PCa; however, the role of SFRPs in PCa remain controversial [62,63,64]. In VCap cells, SFRP1 appears to promote the transcriptional activity of AR and to cause an increase in the expression of TMPRSS2-ERG. This co-expression results in the increased migration and invasion of tumor xerographs [62].
10. Integrin Linked Kinase Pathway
Previous studies have demonstrated that ERG expression results in the upregulation of the Integrin-linked kinase (ILK), which further activates Snail via poly (ADP ribose) polymerase-1 (PARP-1), resulting results in the downregulation of E-cadherin and the activation of the EMT pathway [65,66]. Furthermore, ILK activation due to ERG activation results in the suppression of cytokeratin 8/18 and increased expression of N-cadherin and vimentin, which further induces EMT pathway activity [65]. ILK has been shown to be involved in the regulation of the EMT and proliferation pathways (Figure 2), and it has been shown to have increased expression in various cancers [65,66].
11. ERG Overexpression Upregulates PI3KB-PKB/Akt Pathway
ERG overexpression is often accompanied by the loss of PTEN in PCa, which further upregulates the Akt pathway due to a lack of inhibition [67]. The upregulation of the AKT signalling pathways has been observed to work in tandem with downstream ERG targets to make cancer progression more aggressive [68]. PTEN is a known inhibitor of AKT and acts by converting PIP3 to PIP2, directly opposing AKT signalling [69]. The AKT pathway is a key regulatory step in controlling the transcription of many genes that induce cell proliferation, metabolism, anti-apoptosis, genomic instability, and differentiation. In hindsight, the binding of a substrate leads to the activation of PI3K, resulting in the catalytic conversion of PIP2 to PIP3 [70]. Akt then interacts with PIP3 at the plasma membrane, which allows PDK1 to phosphorylate Thr308; this allows for the partial activation of AKT, which leads to the activation of mTORC1 and tuberous sclerosis protein 2 (TSC2) [71]. The downstream effect of this partial activation promotes cellular proliferation and protein synthesis [72]. The full activation of AKT results in phosphorylation occurring both in the cytoplasm and in the nuclei of proteins such as FOXO, which leads to attenuation of apoptosis and the promotion of the proliferation, angiogenesis, and survival of cells [73].
12. Metalloproteinase MMP1,3,9 and ADAMTS1 Pathway
ERG has been shown to be recruited by the Fos/Jun complex, bind to the promoter, and activate matrix metalloproteinase 1 (MMP1) transcription. Furthermore, ETS2 has been shown to directly activate both MMP1 and MMP3 [74,75]. Overall, ERG and ETS2 induce the loss of focal adhesion and therefore the de-differentiation of the prostate cells [21]. Additionally, MMP9 has been shown to be positively correlated with TMPRSS2-ERG, which is upregulated in VCaP cells; however, the mechanism behind this interaction has yet to be determined [76]. MMP1 and MMP3 are proteolytic enzymes that degrade bonds and connective tissue in the basement membrane; therefore, their activation is directly correlated with the ability of cells to migrate and invade [21].
Another family of proteins that share the metalloproteinase domain with MMPs are disintegrin and metalloproteinase (ADAMs). They regulate various cell functions such as migration and invasion and have been known as cancer progression regulators. ERG has been shown to upregulate ADAMTS1 genes in prostate cancer cells, which show an attraction towards fibroblasts [77].
ERG activates C-MYC to promote the de-differentiation of the PCa cells, and C-MYC has been identified as a major transcriptional factor that is involved in various cellular functions such as proliferation, differentiation, apoptosis, and cellular motility. C-MYC has also been shown to be a major oncogene that is implicated in the progression of prostate cancer [78]. It has been reported that C-MYC regulates and increases the expression of AR genes and helps to stabilize various proteins such as AR-FL and AR-V, which are observed in castration-resistant prostate cancer [79]. C-MYC knockdown in enzalutamide resistant PCa cells results in enhanced cells sensitivity to enzalutamide [79]. Previous studies conclude that C-MYC expression may be used as a predictor for biochemical recurrence in primary prostate tumors. C-MYC expression may not be directly related to ERG protein expression, but it is strongly correlated with TMPRSS2-ERG status [80]. It appears that the high expression of ERG due to TMPRSS2-ERG fusion results in the upregulation the C-MYC oncogene, which attenuates the differentiation of the prostate epithelium [38]. When ERG and C-MYC levels were reduced using siRNA, the upregulation of prostate differentiation genes such as PSA, SL34A3/prostein, and MSMB was observed [79]. Furthermore, studies have shown that when ERG is repressed, the C-MYC targets that are activated express self-renewal genes, resulting in decreased HSC differentiation and an increased HSC population [81]. Overall, it appears that ERG activates C-MYC in an effort to promote the de-differentiation of the prostate epithelial cells [21,82].
13. ERG and microRNAs
Kim et al. reported that in PCa, ERG directly binds to the ETS motif within the promoter of miR-200c and inhibits its expression [83]. miR-200c is one of the members of the miR-200 family that has been shown to be downregulated in metastatic compared to primary tumors, and its loss has been correlated with poor cell differentiation [21]. It was reported that miR-200 acts a tumor suppressor and downregulates EMT markers such as ZEB1 and Vimentin [83]. Another study indicated that miR-221 is actually downregulated in patients with more aggressive PCa with TMPRSS2-ERG gene fusion [84]. miR-221 has been known to be overexpressed in various tumors such as bladder [85], glioblastoma [86], breast [87], and chronic lymphocytic leukemia [88]. It appears that in these types of cancers, miR-221 and miR-449 suppress the expression of p27 and p21, respectively, which are tumor suppressors, and are responsible for regulating the phase transition from G0 to S phase, which further stimulates proliferation [84,89]. ERG overexpression ultimately results in the overexpression of genes in the EMT pathway and enhances the migration and invasion ability of PCa cells [83]. Studies have shown that there is a correlation between the downregulation of miR-221 and metastatic tumors, but to this date. the mechanism of action has yet to be elucidated by researchers.
14. Nitric Oxide NO-cGMP Signaling Pathway
Previous studies have shown that the alpha1 and beta1 subunits of Soluble guanylyl cylase (sGC) and cGMP synthesis are elevated by TMPRSS2-ERG in PCa cells [90]. More importantly, studies have shown that not only does sGC inhibitor treatment suppress tumor growth and proliferation in TMPRSS2-ERG-positive PCa xerographs, but that it can also act strategically with an AR antagonist such as enzalutamide [90]. sGC is a regulator of the nitric oxide (NO)-cGMP signaling pathway. Upon the binding of NO, sGC synthesizes cGMP and activates protein kinase G (PKG), which is directly related to cell proliferation and tumorigenesis in various cancers [90,91,92].
15. TMPRSS2-ERG Fusion Upregulates CXCR4 Enhances Tumor Adhesion and Aggregation
Previous studies have concluded that there are eight ERG/Ets factor binding sites near the promoter of chemokine receptor type 4 (CXCR4) (Figure 3) and that TMPRSS2-ERG expression enhances the function and expression of CXCR4 [93,94]. CXCL12 is a known CXCR4 receptor ligand, and the interactions between the two functions enhance tumor aggressiveness and increase the ability of cancer cells to adhere to the extracellular matrix [95,96]. The CXCR4/CXCL12 axis has been shown to increase MMP expression, which promotes cell migration and growth in PCa [93]. An analysis of PCa demographics revealed relatively higher CXCR4 expression during PCa progression and in metastatic bone tissue compared to benign PCa cells that were reported. In vitro, a direct correlation between TMPRSS-ERG and CXCR4 was observed when ERG was knocked down [96].
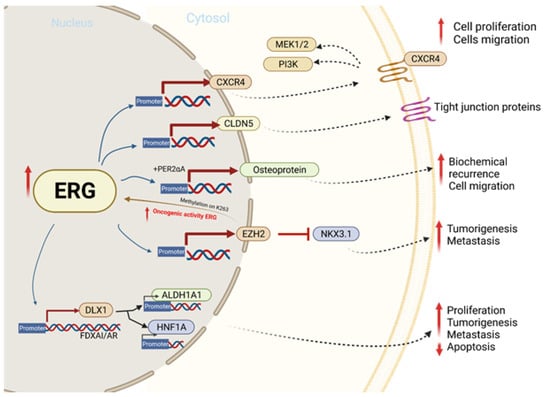
Figure 3.
The molecular mechanism involved in ERG overexpression and related outcomes in tumor cell proliferation, migration, invasion, and metastasis.
16. TMPRSS2-ERG Binds to EMT Key Regulators ZEB1/ZEB2
Prior studies show that TMPRSS2-ERG can directly bind to the Zinc finger e-box binding homeobox 1 (ZEB1) promoter and binds to ZEB2 modulators such as ILIR2 and SPINT1 and increases their overall expression [97]. ZEB1 and ZEB2 are members of the ZEB family of transcriptional factors and are key regulators in the EMT and disease progression pathways [98]. Furthermore, ZEB1 knockdown revealed a significant decline in the migration and invasion capacity of TMPRSS2-ERG-expressing cells.
17. EZH2 Enhances ERG Oncogenic Activity
It appears that the enhancer of zeste jomolog 2 (EZH2), which is a histone H3K27 methyltransferase, catalyzes the methylation of ERG at the lysine 263 residue. This interaction promotes the translocation and DNA binding of ERG in the nucleus, which results in the enhancement of the oncogenic activity of ERG [99,100]. Studies have shown that the methylation of ERG at the lysine362 residue is associated with metastatic properties and increased tumorigenic characteristics in cell lines [99]. Further, these interactions seem to enhance the progression of PCa from non-invasive lesions to invasive adenocarcinomas in Erg/Pten mice [99].
18. ERG and Tight Junction Protein CLDN5
Recent Studies have shown that ERG is positively correlated with the expression of an important tight junction protein known as Claudin 5 (CLDN5); it has been observed that there are two major ERG binding sites near the promoter of the CLDN5 gene, making ERG a direct transcriptional regulator of CLDN5. Moreover, CLDN5 is known as a member of the 24 tetraspan transmembrane protein family and one of the major components of the tight junction strands that are responsible for regulating barrier functions [101]. This protein is involved in many of the processes related to vascular homeostasis. Research has shown that the knockout of ERG expression in mice resulted in increased endothelial cell permeability due to decreased CLDN5 expression [102]. Interestingly, CLDN5 upregulation results in decreased the cell migration and invasion ability of lung cancer cells due to the decreased permeability of the cell membrane due to the enhancement of the CLDN5 tight junctions [103,104]. In addition to ERG’s function in maintaining vascular homeostasis, it also plays a critical role in maintaining the population of hematopoietic stem cells (HSC) by regulating their differentiation. A recent study demonstrated that ERG is necessary for arresting HSCs in a dormant G0 phase by analyzing the number of cells within each phase of the cell cycle. CLDN5 has been implicated as a negative regulator of many biological processes such as angiogenesis, cell migration, and vascular permeability; alternatively, it has also been shown to be a positive regulator of tight junction assembly, cell population differentiation, protein binding, and endothelial barrier development [103,105,106]. Furthermore, TNF-alpha has been classified as a direct inhibitor of ERG and CLDN5 by extension [101].
19. ERG Upregulates Distal-Less Homeobox-1(DLX1)
It is thought that ERG upregulates distal-less homeobox 1 (DLX1) by interacting with enhanced bound AR and FOXA1 [107]. To support this hypothesis, it has been observed that when ERG and therefore DLX1 transcription is inhibited via BET inhibitors, there is a significant reduction in the oncogenic effects of DLX1 [107]. DLX is a transcriptional factor and is part of the homeobox-containing family [108]. Several malignancies, such as those including the prostate, have been linked to the deregulation of the homeobox gene, and therefore, DLX1 has been validated as a potential PCa biomarker [107,109].
20. GSK3B and WEE1 Induces TMPRSS2-ERG Degradation
Hong et al. [110] revealed that the glycogen synthase kinase 3 beta (GSK3B) and WEE1 induce TMPRSS2-ERG degradation via the dual phosphorylation of ERG threonine-187 and tyrosine-190. Such phosphorylation allows for the recognition and degradation of the ERG oncoprotein by the E3 ubiquitin ligase FBW7. GSK3B and WEE1 have been found to be associated with the DNA damage that is induced proteasomal degradation in PCa [111]. The relationship between TMPRSS2-ERG and PTEN has been described previously, but it is interesting to note that this degradation pathway is eradicated in the case of PTEN loss or GSK3B inactivation. This has further been implicated with the growth of chemoresistant PCa cell lines in culture and in mice.
21. Summary
Clinical data demonstrate the incidence of ERG overexpression in approximately 50% of all patients diagnosed for prostate cancer. The most common mechanism associated with ERG overexpression arises from the gene fusion of TMPRSS2 and ERG. This fusion allows for the androgen responsive promoter TMPRSS2 to act on ERG, resulting in its transcriptional upregulation. Researchers have begun to elucidate the role of ERG in the oncogenic mechanisms associated with tumorigenesis and cancer progression in their search for potential non-invasive biomarkers and novel targeted therapeutics. Here, we reviewed multiple pathways that are affected by the upregulation of ERG in PCa. We conclude that ERG is a pivotal mediator in tumor development and is involved in many cellular processes, including cell proliferation, de-differentiation, angiogenesis, and cancer stem cell homeostasis and is implicated in the epithelial to mesenchymal transition pathway. Future research should explore the potential applications of ERG in improving diagnostic and prognostic methods in PCa as well as potential opportunities for therapeutic targeting.
Author Contributions
Conceptualization, T.A.B.; writing orginal draft preparation, E.K.K., Y.G., M.C., S.M.B. and D.D.; resources, Y.G. and M.C.; validation, E.K.K. and Y.G.; Writing-original draft, E.K.K., Y.G. and M.C.; Writing-review & editing, E.K.K., Y.G. and T.A.B. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported in part by the Prostate Cancer Foundation USA, Young Investigator Award (YI-2012) (T.A.B.). This work is also supported by Prostate Cancer Canada Movember Award (TAG 2018-2060).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Sundquist, J.; Sundquist, K.; Ji, J. Prostate cancer incidence and survival in relation to prostate cancer as second cancer in relatives. Cancer Med. 2022, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Macrini, S.; Francesconi, S.; Caprera, C.; Lancia, D.; Corsi, M.; Gunnellini, M.; Rocchi, A.; Pireddu, A.; Marziani, F.; Mosillo, C.; et al. Looking for a Simplified Diagnostic Model to Identify Potentially Lethal Cases of Prostate Cancer at Initial Diagnosis: An ImGO Pilot Study. Cancers 2022, 14, 1542. [Google Scholar] [CrossRef]
- Petrovics, G.; Liu, A.; Shaheduzzaman, S.; Furasato, B.; Sun, C.; Chen, Y.; Nau, M.; Ravindranath, L.; Chen, Y.; Dobi, A.; et al. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene 2005, 24, 3847–3852, Erratum in Oncogene 2007, 26, 6684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rezk, M.; Chandra, A.; Addis, D.; Moller, H.; Youssef, M.; Dasgupta, P.; Yamamoto, H. ETS-related gene(ERG) expression as a predictor of oncological outcomes in patients with high-grade prostate cancer treated with primary androgen deprivation therapy: A cohort study. BMJ Open 2019, 9, e025161. [Google Scholar] [CrossRef]
- Liu, F.; Patient, R. Genome-wide analysis of the zebrafish ETS family identifies three genes required for hemangioblast differentiation or angiogenesis. Circ. Res. 2008, 103, 1147–1154. [Google Scholar] [CrossRef] [Green Version]
- Testa, U.; Castelli, G.; Pelosi, E. Cellular and Molecular Mechanisms Underlying Prostate Cancer Development: Therapeutic Implications. Medicines 2019, 6, 82. [Google Scholar] [CrossRef] [Green Version]
- Tomlins, S.A.; Laxman, B.; Varambally, S.; Cao, X.; Yu, J.; Helgeson, B.E.; Cao, Q.; Prensner, J.R.; Rubin, M.A.; Shah, R.B.; et al. Role of the TMPRSS2-ERG Gene Fusion in Prostate Cancer. Neoplasia 2008, 10, 177–188. [Google Scholar] [CrossRef] [Green Version]
- Tomlins, S.A.; Mehra, R.; Rhodes, D.R.; Smith, L.R.; Roulston, D.; Helgeson, B.E.; Cao, X.; Wei, J.T.; Rubin, M.A.; Shah, R.B.; et al. TMPRSS2:ETV4 Gene Fusions Define a Third Molecular Subtype of Prostate Cancer. Cancer Res. 2006, 66, 3396–3400. [Google Scholar] [CrossRef] [Green Version]
- Tomlins, S.A.; Rhodes, D.R.; Perner, S.; Dhanasekaran, S.M.; Mehra, R.; Sun, X.-W.; Varambally, S.; Cao, X.; Tchinda, J.; Kuefer, R.; et al. Recurrent Fusion of TMPRSS2 and ETS Transcription Factor Genes in Prostate Cancer. Science 2005, 310, 644–648. [Google Scholar] [CrossRef]
- Helgeson, B.E.; Tomlins, S.; Shah, N.; Laxman, B.; Cao, Q.; Prensner, J.; Cao, X.; Singla, N.; Montie, J.E.; Varambally, S.; et al. Characterization of TMPRSS2:ETV5 and SLC45A3:ETV5 Gene Fusions in Prostate Cancer. Cancer Res. 2008, 68, 73–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, B.C.; Zhang, W. Fusion genes in solid tumors: An emerging target for cancer diagnosis and treatment. Chin. J. Cancer 2013, 32, 594–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krumbholz, M.; Agaimy, A.; Stoehr, R.; Burger, M.; Wach, S.; Taubert, H.; Wullich, B.; Hartmann, A.; Metzler, M. Molecular Composition of Genomic TMPRSS2-ERG Rearrangements in Prostate Cancer. Dis. Markers 2019, 2019, 5085373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, Q.; Sun, Y.; Dobi, A.; Srivastava, S.; Wang, W.; Srivastava, S.; Ji, Y.; Hou, J.; Zhao, G.-P.; Li, Y.; et al. Systematic analysis reveals molecular characteristics of ERG-negative prostate cancer. Sci. Rep. 2018, 8, 14336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ateeq, B.; Bhatia, V.; Goel, S. Molecular Discriminators of Racial Disparities in Prostate Cancer. Trends Cancer 2016, 2, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Vijayaraj, P.; Le Bras, A.; Mitchell, N.; Kondo, M.; Juliao, S.; Wasserman, M.; Beeler, D.; Spokes, K.; Aird, W.C.; Baldwin, H.S.; et al. Erg is a crucial regulator of endocardial-mesenchymal transformation during cardiac valve morphogenesis. Development 2012, 139, 3973–3985. [Google Scholar] [CrossRef] [Green Version]
- Birdsey, G.M.; Shah, A.V.; Dufton, N.; Reynolds, L.E.; Almagro, L.O.; Yang, Y.; Aspalter, I.M.; Khan, S.T.; Mason, J.C.; Dejana, E.; et al. The endothelial transcription factor ERG promotes vascular stability and growth through Wnt/beta-catenin signaling. Dev. Cell 2015, 32, 82–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sreenath, T.L.; Dobi, A.; Petrovics, G.; Srivastava, S. Oncogenic activation of ERG: A predominant mechanism in prostate cancer. J. Carcinog. 2011, 10, 37. [Google Scholar]
- Nauseef, J.T.; Henry, M.D. Epithelial-to-mesenchymal transition in prostate cancer: Paradigm or puzzle? Nat. Rev. Urol. 2011, 8, 428–439. [Google Scholar] [CrossRef]
- Yu, J.; Yu, J.; Mani, R.-S.; Cao, Q.; Brenner, C.J.; Brenner, X.; Wang, X.; Wu, L.; Li, J.; Hu, M.; et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell 2010, 17, 443–454. [Google Scholar] [CrossRef] [Green Version]
- Adamo, P.; Ladomery, M.R. The oncogene ERG: A key factor in prostate cancer. Oncogene 2016, 35, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Klezovitch, O.; Risk, M.; Coleman, I.; Lucas, J.M.; Null, M.; True, L.D.; Nelson, P.S.; Vasioukhin, V. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc. Natl. Acad. Sci. USA 2008, 105, 2105–2110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hägglöf, C.; Hammarsten, P.; Strömvall, K.; Egevad, L.; Josefsson, A.; Stattin, P.; Granfors, T.; Bergh, A. TMPRSS2-ERG Expression Predicts Prostate Cancer Survival and Associates with Stromal Biomarkers. PLoS ONE 2014, 9, e86824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babal, Y.; Kandemir, B.; Kurnaz, I. Gene Regulatory Network of ETS Domain Transcription Factors in Different Stages of Glioma. J. Pers. Med. 2021, 11, 138. [Google Scholar] [CrossRef] [PubMed]
- Parolia, A.; Cieslik, M.; Chu, S.-C.; Xiao, L.; Ouchi, T.; Zhang, Y.; Wang, X.; Vats, P.; Cao, X.; Pitchiaya, S.; et al. Distinct structural classes of activating FOXA1 alterations in advanced prostate cancer. Nature 2019, 571, 413–418. [Google Scholar] [CrossRef]
- Majesky, M.W. Vascular Development. Arterioscler. Thromb. Vasc. Biol. 2018, 38, e17–e24. [Google Scholar] [CrossRef] [Green Version]
- Vivekanand, P. Lessons from Drosophila Pointed, an ETS family transcription factor and key nuclear effector of the RTK signaling pathway. Genesis 2018, 56, e23257. [Google Scholar] [CrossRef]
- Pham, D.; Moseley, C.E.; Gao, M.; Savic, D.; Winstead, C.J.; Sun, M.; Kee, B.L.; Myers, R.M.; Weaver, C.T.; Hatton, R.D. Batf Pioneers the Reorganization of Chromatin in Developing Effector T Cells via Ets1-Dependent Recruitment of Ctcf. Cell Rep. 2019, 29, 1203–1220.e7. [Google Scholar] [CrossRef] [Green Version]
- Vasuri, F.; Valente, S.; Motta, I.; Degiovanni, A.; Ciavarella, C.; Pasquinelli, G. ETS-Related Gene Expression in Healthy Femoral Arteries With Focal Calcifications. Front. Cell Dev. Biol. 2021, 9, 623782. [Google Scholar] [CrossRef]
- Tharakan, B.; Hunter, F.A.; Muthusamy, S.; Randolph, S.; Byrd, C.; Rao, V.N.; Reddy, E.S.P.; Childs, E.W. ETS-Related Gene Activation Preserves Adherens Junctions and Permeability in Microvascular Endothelial Cells. Shock 2021, 57, 309–315. [Google Scholar] [CrossRef]
- Neal, A.; Nornes, S.; Louphrasitthiphol, P.; Sacilotto, N.; Preston, M.D.; Fleisinger, L.; Payne, S.; De Val, S. ETS factors are required but not sufficient for specific patterns of enhancer activity in different endothelial subtypes. Dev. Biol. 2021, 473, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Looney, A.P.; Han, R.; Stawski, L.; Marden, G.; Iwamoto, M.; Trojanowska, M. Synergistic Role of Endothelial ERG and FLI1 in Mediating Pulmonary Vascular Homeostasis. Am. J. Respir. Cell Mol. Biol. 2017, 57, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Lorenzin, F.; Demichelis, F. Past, Current, and Future Strategies to Target ERG Fusion-Positive Prostate Cancer. Cancers 2022, 14, 1118. [Google Scholar] [CrossRef] [PubMed]
- Kohaar, I.; Li, Q.; Chen, Y.; Ravindranath, L.; Young, D.; Ali, A.; Sesterhenn, I.A.; Rosner, I.L.; Cullen, J.; Srivastava, S.; et al. Association of germline genetic variants with TMPRSS2-ERG fusion status in prostate cancer. Oncotarget 2020, 11, 1321–1333. [Google Scholar] [CrossRef] [Green Version]
- Kobelyatskaya, A.; Pudova, E.; Snezhkina, A.; Fedorova, M.; Pavlov, V.; Guvatova, Z.; Savvateeva, M.; Melnikova, N.; Dmitriev, A.; Trofimov, D.; et al. Impact TMPRSS2–ERG Molecular Subtype on Prostate Cancer Recurrence. Life 2021, 11, 588. [Google Scholar] [CrossRef]
- Giunchi, F.; Massari, F.; Altimari, A.; Gruppioni, E.; Nobili, E.; Fiorentino, M.; Ardizzoni, A. Dual TMPRSS2:ERG Fusion in a Patient with Lung and Prostate Cancers. Diagnostics 2020, 10, 1109. [Google Scholar] [CrossRef]
- Yamoah, K.; Lal, P.; Awasthi, S.; Naghavi, A.O.; Rounbehler, R.J.; Gerke, T.; Berglund, A.E.; Pow-Sang, J.M.; Schaeffer, E.M.; Dhillon, J.; et al. TMPRSS2-ERG fusion impacts anterior tumor location in men with prostate cancer. Prostate 2020, 81, 109–117. [Google Scholar] [CrossRef]
- Fang, L.; Li, D.; Yin, J.; Pan, H.; Ye, H.; Bowman, J.; Capaldo, B.; Kelly, K. TMPRSS2-ERG promotes the initiation of prostate cancer by suppressing oncogene-induced senescence. Cancer Gene Ther. 2022, 1–14. [Google Scholar] [CrossRef]
- Sakamoto, K.; Endo, K.; Sakamoto, K.; Kayamori, K.; Ehata, S.; Ichikawa, J.; Ando, T.; Nakamura, R.; Kimura, Y.; Yoshizawa, K.; et al. EHF suppresses cancer progression by inhibiting ETS1-mediated ZEB expression. Oncogenesis 2021, 10, 26. [Google Scholar] [CrossRef]
- Greulich, B.M.; Plotnik, J.P.; Jerde, T.J.; Hollenhorst, P.C. Toll-like receptor 4 signaling activates ERG function in prostate cancer and provides a therapeutic target. NAR Cancer 2021, 3, zcaa046. [Google Scholar] [CrossRef]
- Tsourlakis, M.C.; Khosrawi, P.; Weigand, P.; Kluth, M.; Hube-Magg, C.; Minner, S.; Koop, C.; Graefen, M.; Heinzer, H.; Wittmer, C.; et al. VEGFR-1 Overexpression Identifies a Small Subgroup of Aggressive Prostate Cancers in Patients Treated by Prostatectomy. Int. J. Mol. Sci. 2015, 16, 8591–8606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, C.; Dobi, A.; Mohamed, A.; Li, H.; Thangapazham, R.L.; Furusato, B.; Shaheduzzaman, S.; Tan, S.H.; Vaidyanathan, G.; Whitman, E.; et al. TMPRSS2-ERG fusion, a common genomic alteration in prostate cancer activates C-MYC and abrogates prostate epithelial differentiation. Oncogene 2008, 27, 5348–5353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vestweber, D. VE-cadherin: The major endothelial adhesion molecule controlling cellular junctions and blood vessel formation. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 223–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blee, A.M.; He, Y.; Yang, Y.; Ye, Z.; Yan, Y.; Pan, Y.; Ma, T.; Dugdale, J.; Kuehn, E.; Kohli, M.; et al. TMPRSS2-ERG Controls Luminal Epithelial Lineage and Antiandrogen Sensitivity in PTEN and TP53-Mutated Prostate Cancer. Clin. Cancer Res. 2018, 24, 4551–4565. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Yuan, Q.; Di, W.; Xia, X.; Liu, Z.; Mao, N.; Li, L.; Li, C.; He, J.; Li, Y.; et al. ERG orchestrates chromatin interactions to drive prostate cell fate reprogramming. J. Clin. Investig. 2020, 130, 5924–5941. [Google Scholar] [CrossRef]
- Wasmuth, E.V.; Hoover, E.A.; Antar, A.; Klinge, S.; Chen, Y.; Sawyers, C.L. Modulation of androgen receptor DNA binding activity through direct interaction with the ETS transcription factor ERG. Proc. Natl. Acad. Sci. USA 2020, 117, 8584–8592. [Google Scholar] [CrossRef] [Green Version]
- Kohvakka, A.; Sattari, M.; Shcherban, A.; Annala, M.; Urbanucci, A.; Kesseli, J.; Tammela, T.L.J.; Kivinummi, K.; Latonen, L.; Nykter, M.; et al. AR and ERG drive the expression of prostate cancer specific long noncoding RNAs. Oncogene 2020, 39, 5241–5251. [Google Scholar] [CrossRef]
- Shah, N.; Kesten, N.; Font-Tello, A.; Chang, M.E.K.; Vadhi, R.; Lim, K.; Flory, M.R.; Cejas, P.; Mohammed, H.; Long, H.W.; et al. ERG-Mediated Coregulator Complex Formation Maintains Androgen Receptor Signaling in Prostate Cancer. Cancer Res. 2020, 80, 4612–4619. [Google Scholar] [CrossRef]
- Feng, X.; Zhou, C.K.; Clish, C.B.; Wilson, K.M.; Pernar, C.H.; Dickerman, B.A.; Loda, M.; Finn, S.P.; Penney, K.L.; Schmidt, D.R.; et al. Association of Prediagnostic Blood Metabolomics with Prostate Cancer Defined by ERG or PTEN Molecular Subtypes. Cancer Epidemiol. Biomark. Prev. 2021, 30, 1000–1008. [Google Scholar] [CrossRef]
- Mao, N.; Gao, D.; Hu, W.; Hieronymus, H.; Wang, S.; Lee, Y.S.; Lee, C.; Choi, D.; Gopalan, A.; Chen, Y.; et al. Aberrant Expression of ERG Promotes Resistance to Combined PI3K and AR Pathway Inhibition through Maintenance of AR Target Genes. Mol. Cancer Ther. 2019, 18, 1577–1586. [Google Scholar] [CrossRef] [Green Version]
- Pettersson, A.; Graff, R.E.; Bauer, S.R.; Pitt, M.J.; Lis, R.T.; Stack, E.C.; Martin, N.E.; Kunz, L.; Penney, K.L.; Ligon, A.H.; et al. The TMPRSS2:ERG rearrangement, ERG expression, and prostate cancer outcomes: A cohort study and meta-analysis. Cancer Epidemiol. Prev. Biomark. 2012, 21, 1497–1509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jillson, L.; Yette, G.; Laajala, T.; Tilley, W.; Costello, J.; Cramer, S. Androgen Receptor Signaling in Prostate Cancer Genomic Subtypes. Cancers 2021, 13, 3272. [Google Scholar] [CrossRef] [PubMed]
- Brady, L.; Carlsson, J.; Baird, A.-M.; Casey, O.; Vlajnic, T.; Murchan, P.; Cormican, D.; Costigan, D.; Gray, S.; Sheils, O.; et al. Correlation of integrated ERG/PTEN assessment with biochemical recurrence in prostate cancer. Cancer Treat. Res. Commun. 2021, 29, 100451. [Google Scholar] [CrossRef] [PubMed]
- Imada, E.L.; Sanchez, D.F.; Dinalankara, W.; Vidotto, T.; Ebot, E.M.; Tyekucheva, S.; Franco, G.R.; Mucci, L.A.; Loda, M.; Schaeffer, E.M.; et al. Transcriptional landscape of PTEN loss in primary prostate cancer. BMC Cancer 2021, 21, 856. [Google Scholar] [CrossRef]
- Abou-Ouf, H.; Assem, H.; Ghosh, S.; Karnes, R.J.; Stoletov, K.; Palanisamy, N.; Lewis, J.D.; Bismar, T.A. High Serine-arginine Protein Kinase 1 Expression with PTEN Loss Defines Aggressive Phenotype of Prostate Cancer Associated with Lethal Outcome and Decreased Overall Survival. Eur. Urol. Open Sci. 2021, 23, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Murillo-Garzón, V.; Kypta, R. WNT signalling in prostate cancer. Nat. Rev. Urol. 2017, 14, 683–696. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthi, B.V.S.K.; Chandrashekar, D.S.; Balasubramanya, S.A.H.; Robinson, A.; Carskadon, S.; Rao, U.; Gordetsky, J.; Manne, U.; Netto, G.J.; Sudarshan, S.; et al. Wnt receptor Frizzled 8 is a target of ERG in prostate cancer. Prostate 2018, 78, 1311–1320. [Google Scholar] [CrossRef]
- Kaplan, Z.; Zielske, S.P.; Ibrahim, K.G.; Cackowski, F.C. WNT and beta-Catenin Signaling in the Bone Metastasis of Prostate Cancer. Life 2021, 11, 1099. [Google Scholar] [CrossRef]
- Wu, L.; Zhao, J.C.; Kim, J.; Jin, H.; Wang, C.-Y.; Yu, J. ERG Is a Critical Regulator of Wnt/LEF1 Signaling in Prostate Cancer. Cancer Res. 2013, 73, 6068–6079. [Google Scholar] [CrossRef] [Green Version]
- Bisson, I.; Prowse, D.M. WNT signaling regulates self-renewal and differentiation of prostate cancer cells with stem cell charac-teristics. Cell Res. 2009, 19, 683–697. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Singhal, U.; Qiao, Y.; Kasputis, T.; Chung, J.-S.; Zhao, H.; Chammaa, F.; Belardo, J.A.; Roth, T.M.; Zhang, H.; et al. Wnt Signaling Drives Prostate Cancer Bone Metastatic Tropism and Invasion. Transl. Oncol. 2020, 13, 100747. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Hernández, C.D.; Cruz-Burgos, M.; Ramírez, S.A.C.; Losada-García, A.; Camacho-Arroyo, I.; García-López, P.; Langley, E.; González-Covarrubias, V.; Llaguno-Munive, M.; Albino-Sánchez, M.E.; et al. SFRP1 increases TMPRSS2-ERG expression promoting neoplastic features in prostate cancer in vitro and in vivo. Cancer Cell Int. 2020, 20, 312. [Google Scholar] [CrossRef]
- Lodygin, D.; Epanchintsev, A.; Menssen, A.; Diebold, J.; Hermeking, H. Functional Epigenomics Identifies Genes Frequently Silenced in Prostate Cancer. Cancer Res. 2005, 65, 4218–4227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Tobilla, P.; Solórzano, S.R.; Salido-Guadarrama, I.; González-Covarrubias, V.; Morales-Montor, G.; Díaz-Otañez, C.E.; Rodríguez-Dorantes, M. SFRP1 repression in prostate cancer is triggered by two different epigenetic mechanisms. Gene 2016, 593, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Becker-Santos, D.D.; Guo, Y.; Ghaffari, M.; Vickers, E.D.; Lehman, M.; Altamirano-Dimas, M.; Oloumi, A.; Furukawa, J.; Sharma, M.; Wang, Y.; et al. Integrin-linked kinase as a target for ERG-mediated invasive properties in prostate cancer models. Carcinogenesis 2012, 33, 2558–2567. [Google Scholar] [CrossRef] [Green Version]
- Kilinc, A.N.; Han, S.; Barrett, L.A.; Anandasivam, N.; Nelson, C.M. Integrin-linked kinase tunes cell-cell and cell-matrix adhesions to regulate the switch between apoptosis and EMT downstream of TGFbeta1. Mol. Biol. Cell 2021, 32, 402–412. [Google Scholar] [CrossRef]
- Zhou, W.; Su, Y.; Zhang, Y.; Han, B.; Liu, H.; Wang, X. Endothelial Cells Promote Docetaxel Resistance of Prostate Cancer Cells by Inducing ERG Expression and Activating Akt/mTOR Signaling Pathway. Front. Oncol. 2020, 10, 584505. [Google Scholar] [CrossRef]
- Squire, J. TMPRSS2-ERG and PTEN loss in prostate cancer. Nat. Genet. 2009, 41, 509–510. [Google Scholar] [CrossRef]
- Feng, J.; Dang, Y.; Zhang, W.; Zhao, X.; Zhang, C.; Hou, Z.; Jin, Y.; McNutt, M.A.; Marks, A.R.; Yin, Y. PTEN arginine methylation by PRMT6 suppresses PI3K-AKT signaling and modulates pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 2019, 116, 6868–6877. [Google Scholar] [CrossRef] [Green Version]
- Hemmings, B.A.; Restuccia, D.F. PI3K-PKB/Akt Pathway. Cold Spring Harb. Perspect. Biol. 2012, 4, a011189. [Google Scholar] [CrossRef] [Green Version]
- Amin, H.; Wani, N.A.; Farooq, S.; Nayak, D.; Chakraborty, S.; Shankar, S.; Rasool, R.u.; Koul, S.; Goswami, A.; Rai, R. Inhibition of Invasion in Pancreatic Cancer Cells by Conjugate of EPA with beta(3,3)-Pip-OH via PI3K/Akt/NF-kB Pathway. ACS Med. Chem. Lett. 2015, 6, 1071–1074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dilmaghani, N.A.; Safaroghli-Azar, A.; Pourbagheri-Sigaroodi, A.; Bashash, D. The PI3K/Akt/mTORC signaling axis in head and neck squamous cell carcinoma: Possibilities for therapeutic interventions either as single agents or in combination with conventional therapies. IUBMB Life 2021, 73, 618–642. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.Y.; Meng, X.T.; Xu, Y.N.; Tian, X.J. Role of FOXO protein’s abnormal activation through PI3K/AKT pathway in platinum resistance of ovarian cancer. J. Obstet. Gynaecol. Res. 2021, 47, 1946–1957. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Zhou, X.; Wang, R.; Zhao, J.; Tang, J.; Zhang, Q.; Du, Y.; Pang, Y. Identification of potential diagnostic biomarkers in MMPs for pancreatic carcinoma. Medicine 2021, 100, e26135. [Google Scholar] [CrossRef]
- Johnson, T.R.; Koul, S.; Kumar, B.; Khandrika, L.; Venezia, S.; Maroni, P.D.; Meacham, R.B.; Koul, H.K. Retraction Note to: Loss of PDEF, a prostate-derived Ets factor is associated with aggressive phenotype of prostate cancer: Regulation of MMP 9 by PDEF. Mol. Cancer 2021, 20, 109. [Google Scholar] [CrossRef]
- Liu, B.; Gu, X.; Huang, T.; Luan, Y.; Ding, X. Identification of TMPRSS2-ERG mechanisms in prostate cancer invasiveness: Involvement of MMP-9 and plexin B1. Oncol. Rep. 2016, 37, 201–208. [Google Scholar] [CrossRef] [Green Version]
- Scaravilli, M.; Koivukoski, S.; Latonen, L. Androgen-Driven Fusion Genes and Chimeric Transcripts in Prostate Cancer. Front. Cell Dev. Biol. 2021, 9, 623809. [Google Scholar] [CrossRef]
- Wang, X.; Yu, J.; Yan, J.; Peng, K.; Zhou, H. Single-cell sequencing reveals MYC targeting gene MAD2L1 is associated with prostate cancer bone metastasis tumor dormancy. BMC Urol. 2022, 22, 37. [Google Scholar] [CrossRef]
- Bai, S.; Cao, S.; Jin, L.; Kobelski, M.; Schouest, B.; Wang, X.; Ungerleider, N.; Baddoo, M.; Zhang, W.; Corey, E.; et al. A positive role of c-Myc in regulating androgen receptor and its splice variants in prostate cancer. Oncogene 2019, 38, 4977–4989. [Google Scholar] [CrossRef]
- Zeng, W.; Sun, H.; Meng, F.; Liu, Z.; Xiong, J.; Zhou, S.; Li, F.; Hu, J.; Hu, Z.; Liu, Z. Nuclear C-MYC expression level is associated with disease progression and potentially predictive of two year overall survival in prostate cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 1878–1888. [Google Scholar]
- Knudsen, K.J.; Rehn, M.; Hasemann, M.S.; Rapin, N.; Bagger, F.O.; Ohlsson, E.; Willer, A.; Frank, A.-K.; Søndergaard, E.; Jendholm, J.; et al. ERG promotes the maintenance of hematopoietic stem cells by restricting their differentiation. Genes Dev. 2015, 29, 1915–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boldrini, L.; Bartoletti, R.; Giordano, M.; Manassero, F.; Selli, C.; Panichi, M.; Galli, L.; Farci, F.; Faviana, P. C-MYC, HIF-1alpha, ERG, TKT, and GSTP1: An Axis in Prostate Cancer? Pathol. Oncol. Res. 2019, 25, 1423–1429. [Google Scholar] [CrossRef]
- Kim, J.; Wu, L.; Zhao, J.C.; Jin, H.; Yu, J. TMPRSS2-ERG gene fusions induce prostate tumorigenesis by modulating microRNA miR-200c. Oncogene 2013, 33, 5183–5192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordanpour, A.; Stanimirovic, A.; Nam, R.K.; Moreno, C.S.; Sherman, C.; Sugar, L.; Seth, A. miR-221 Is down-regulated in TMPRSS2:ERG fusion-positive prostate cancer. Anticancer Res. 2011, 31, 403–410. [Google Scholar] [PubMed]
- Li, J.; Li, Q.; Huang, H.; Li, Y.; Li, L.; Hou, W.; You, Z. Overexpression of miRNA-221 promotes cell proliferation by targeting the apoptotic protease activating factor-1 and indicates a poor prognosis in ovarian cancer. Int. J. Oncol. 2017, 50, 1087–1096. [Google Scholar] [CrossRef] [Green Version]
- Swellam, M.; El Arab, L.E.; Al-Posttany, A.S.; Said, S.B. Clinical impact of circulating oncogenic MiRNA-221 and MiRNA-222 in glioblastoma multiform. J. Neuro-Oncol. 2019, 144, 545–551. [Google Scholar] [CrossRef]
- Niu, X.-Y.; Zhang, Z.-Q.; Ma, P.-L. MiRNA-221-5p promotes breast cancer progression by regulating E-cadherin expression. Eur. Rev. Med. Pharm. Sci 2019, 23, 6983–6990. [Google Scholar]
- Zhang, R.; Huo, C.-H. Long Noncoding RNA SOCS2-AS Promotes Leukemogenesis in FLT3-ITD+ Acute Myeloid Leukemia Through miRNA-221. OncoTargets Ther. 2020, 13, 2925–2934. [Google Scholar] [CrossRef] [Green Version]
- Bauer, S.; Ratz, L.; Heckmann-Nötzel, D.; Kaczorowski, A.; Hohenfellner, M.; Kristiansen, G.; Duensing, S.; Altevogt, P.; Klauck, S.; Sültmann, H. miR-449a Repression Leads to Enhanced NOTCH Signaling in TMPRSS2:ERG Fusion Positive Prostate Cancer Cells. Cancers 2021, 13, 964. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Gao, S.; Han, D.; Han, W.; Chen, S.; Patalano, S.; Macoska, J.A.; He, H.H.; Cai, C.C. TMPRSS2-ERG activates NO-cGMP signaling in prostate cancer cells. Oncogene 2019, 38, 4397–4411. [Google Scholar] [CrossRef]
- Klinger, J.R.; Kadowitz, P.J. The Nitric Oxide Pathway in Pulmonary Vascular Disease. Am. J. Cardiol. 2017, 120, S71–S79. [Google Scholar] [CrossRef] [Green Version]
- Fajardo, A.M.; Piazza, G.A.; Tinsley, H.N. The Role of Cyclic Nucleotide Signaling Pathways in Cancer: Targets for Prevention and Treatment. Cancers 2014, 6, 436–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, J.; Kandagatla, P.; Singareddy, R.; Kropinski, A.; Sheng, S.; Cher, M.L.; Chinni, S.R. Androgens Induce Functional CXCR4 through ERG Factor Expression in TMPRSS2-ERG Fusion-Positive Prostate Cancer Cells. Transl. Oncol. 2010, 3, 195–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clézardin, P.; Coleman, R.; Puppo, M.; Ottewell, P.; Bonnelye, E.; Paycha, F.; Confavreux, C.B.; Holen, I. Bone metastasis: Mechanisms, therapies, and biomarkers. Physiol. Rev. 2021, 101, 797–855. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.X.; Fang, M.; Wang, J.; Cooper, C.R.; Pienta, K.J.; Taichman, R.S. Expression and activation of alpha v beta 3 integrins by SDF-1/CXC12 increases the aggressiveness of prostate cancer cells. Prostate 2007, 67, 61–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singareddy, R.; Semaan, L.; Conley-LaComb, M.K.; John, J.S.; Powell, K.; Iyer, M.; Smith, D.; Heilbrun, L.K.; Shi, N.; Sakr, W.; et al. Transcriptional Regulation of CXCR4 in Prostate Cancer: Significance of TMPRSS2-ERG Fusions. Mol. Cancer Res. 2013, 11, 1349–1361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarrabi, A.; Hashemi, F.; Hashemi, F.; Zabolian, A.; Banihashemi, S.M.; Moghadam, S.S.; Hushmandi, K.; Samarghandian, S.; Ashrafizadeh, M.; Khan, H. Role of ZEB Family Members in Proliferation, Metastasis, and Chemoresistance of Prostate Cancer Cells: Revealing Signaling Networks. Curr. Cancer Drug Targets 2021, 21, 749–767. [Google Scholar]
- Wang, J.; Farkas, C.; Benyoucef, A.; Carmichael, C.; Haigh, K.; Wong, N.; Huylebroeck, D.; Stemmler, M.P.; Brabletz, S.; Brabletz, T.; et al. Interplay between the EMT transcription factors ZEB1 and ZEB2 regulates hematopoietic stem and progenitor cell differentiation and hematopoietic lineage fidelity. PLOS Biol. 2021, 19, e3001394. [Google Scholar] [CrossRef]
- Zoma, M.; Curti, L.; Shinde, D.; Albino, D.; Mitra, A.; Sgrignani, J.; Mapelli, S.N.; Sandrini, G.; Civenni, G.; Merulla, J.; et al. EZH2-induced lysine K362 methylation enhances TMPRSS2-ERG oncogenic activity in prostate cancer. Nat. Commun. 2021, 12, 4147. [Google Scholar] [CrossRef]
- Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef] [Green Version]
- Yuan, L.; Le Bras, A.; Sacharidou, A.; Itagaki, K.; Zhan, Y.; Kondo, M.; Carman, C.V.; Davis, G.E.; Aird, W.C.; Oettgen, P. ETS-related Gene (ERG) Controls Endothelial Cell Permeability via Transcriptional Regulation of the Claudin 5 (CLDN5) Gene. J. Biol. Chem. 2012, 287, 6582–6591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Liu, Q.; Yuan, F.; Guo, S.; Liu, J.; Sun, Z.; Gao, P.; Wang, Y.; Yan, S.; Liu, J. Erg mediates downregulation of claudin-5 in the brain endothelium of a murine experimental model of cerebral malaria. FEBS Lett. 2019, 593, 2585–2595. [Google Scholar] [CrossRef]
- Ma, S.-C.; Li, Q.; Peng, J.-Y.; Zhouwen, J.-L.; Diao, J.-F.; Niu, J.-X.; Wang, X.; Guan, X.-D.; Jia, W.; Jiang, W.-G. Claudin-5 regulates blood-brain barrier permeability by modifying brain microvascular endothelial cell proliferation, migration, and adhesion to prevent lung cancer metastasis. CNS Neurosci. Ther. 2017, 23, 947–960. [Google Scholar] [CrossRef] [PubMed]
- Kind, S.; Büscheck, F.; Höflmayer, D.; Hube-Magg, C.; Kluth, M.; Tsourlakis, M.C.; Steurer, S.; Clauditz, T.S.; Luebke, A.M.; Burandt, E.; et al. Claudin-1 upregulation is associated with favorable tumor features and a reduced risk for biochemical recurrence in ERG-positive prostate cancer. World J. Urol. 2019, 38, 2185–2196. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Luo, Y.; Liu, Q.; Cai, M.; Li, J.; Sun, W.; Wang, J.; He, C.; Liu, Y.; Liu, X. The effect of claudin-5 overexpression on the interactions of claudin-1 and -2 and barrier function in retinal cells. Curr. Mol. Med. 2014, 14, 1226–1237. [Google Scholar] [CrossRef]
- Kluger, M.S.; Clark, P.R.; Tellides, G.; Gerke, V.; Pober, J.S. Claudin-5 Controls Intercellular Barriers of Human Dermal Microvascular but Not Human Umbilical Vein Endothelial Cells. Arter. Thromb. Vasc. Biol. 2013, 33, 489–500. [Google Scholar] [CrossRef] [Green Version]
- Goel, S.; Bhatia, V.; Kundu, S.; Biswas, T.; Carskadon, S.; Gupta, N.; Asim, M.; Morrissey, C.; Palanisamy, N.; Ateeq, B. Transcriptional network involving ERG and AR orchestrates Distal-less homeobox-1 mediated prostate cancer progression. Nat. Commun. 2021, 12, 5325. [Google Scholar] [CrossRef]
- Cani, A.K.; Hu, K.; Liu, C.J.; Siddiqui, J.; Zheng, Y.; Han, S.; Nallandhighal, S.; Hovelson, D.H.; Xiao, L.; Pham, T.; et al. Development of a Whole-urine, Multiplexed, Next-generation RNA-sequencing Assay for Early Detection of Aggressive Prostate Cancer. Eur. Urol. Oncol. 2021. [Google Scholar] [CrossRef]
- Van Neste, L.; Hendriks, R.J.; Dijkstra, S.; Trooskens, G.; Cornel, E.B.; Jannink, S.A.; de Jong, H.; Hessels, D.; Smit, F.P.; Melchers, W.J.G.; et al. Detection of High-grade Prostate Cancer Using a Urinary Molecular Biomarker–Based Risk Score. Eur. Urol. 2016, 70, 740–748. [Google Scholar] [CrossRef]
- Hong, Z.; Zhang, W.; Ding, D.; Huang, Z.; Yan, Y.; Cao, W.; Pan, Y.; Hou, X.; Weroha, S.J.; Karnes, R.J.; et al. DNA Damage Promotes TMPRSS2-ERG Oncoprotein Destruction and Prostate Cancer Suppression via Signaling Converged by GSK3beta and WEE1. Mol. Cell 2020, 79, 1008–1023. [Google Scholar] [CrossRef]
- Strittmatter, B.G.; Jerde, T.J.; Hollenhorst, P.C. Ras/ERK and PI3K/AKT signaling differentially regulate oncogenic ERG medi-ated transcription in prostate cells. PLoS Genet. 2021, 17, e1009708. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).