The Regulatory Circuit Underlying Downregulation of a Type III Secretion System in Yersinia enterocolitica by Transcription Factor OmpR
Abstract
1. Introduction
2. Results
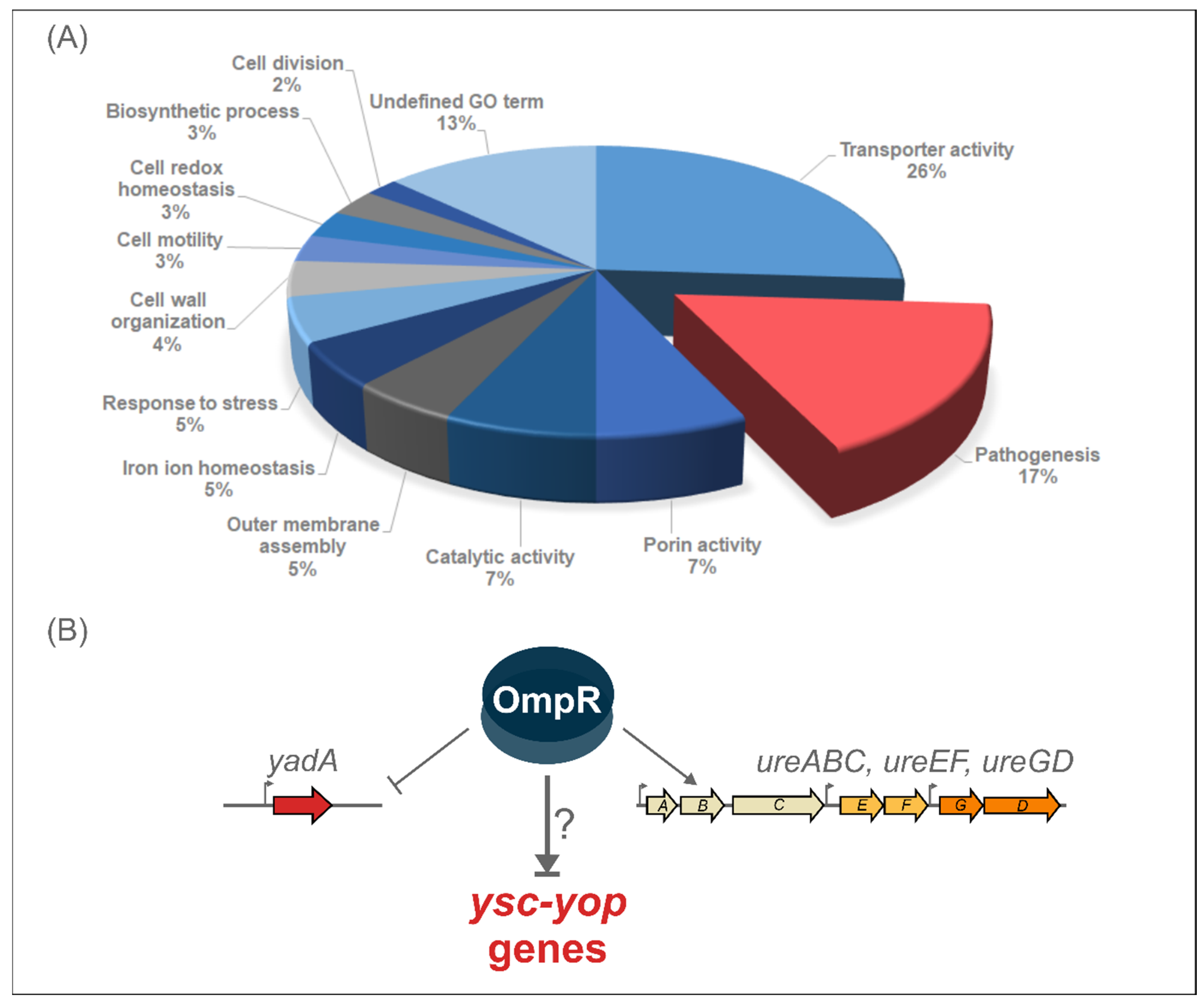
2.1. Proteomic Analysis Reveals the Impact of OmpR on the Abundance of Y. enterocolitica Pathogenicity Factors
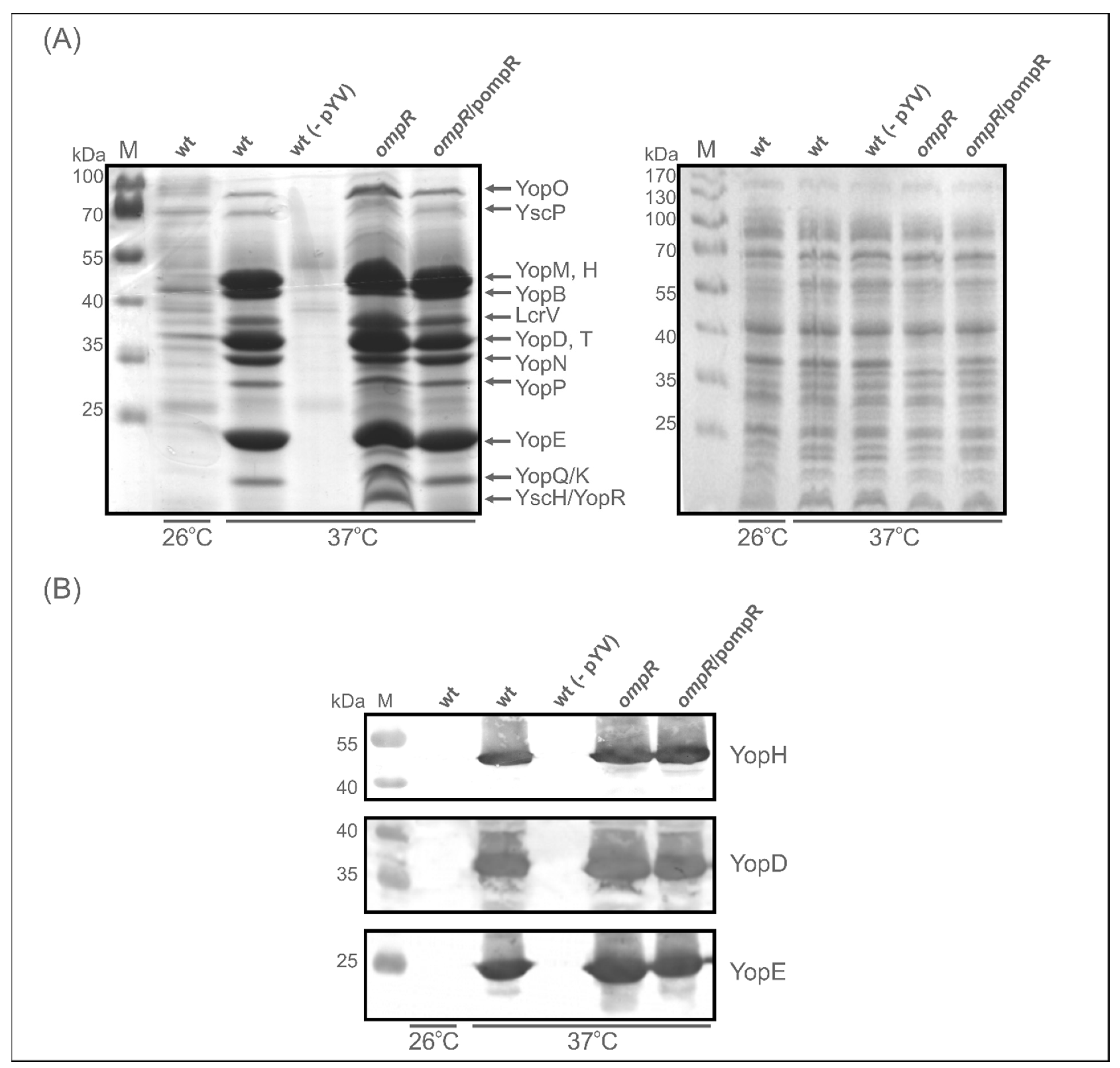
2.2. Impact of OmpR on the T3SS Proteome under Different Environmental Conditions
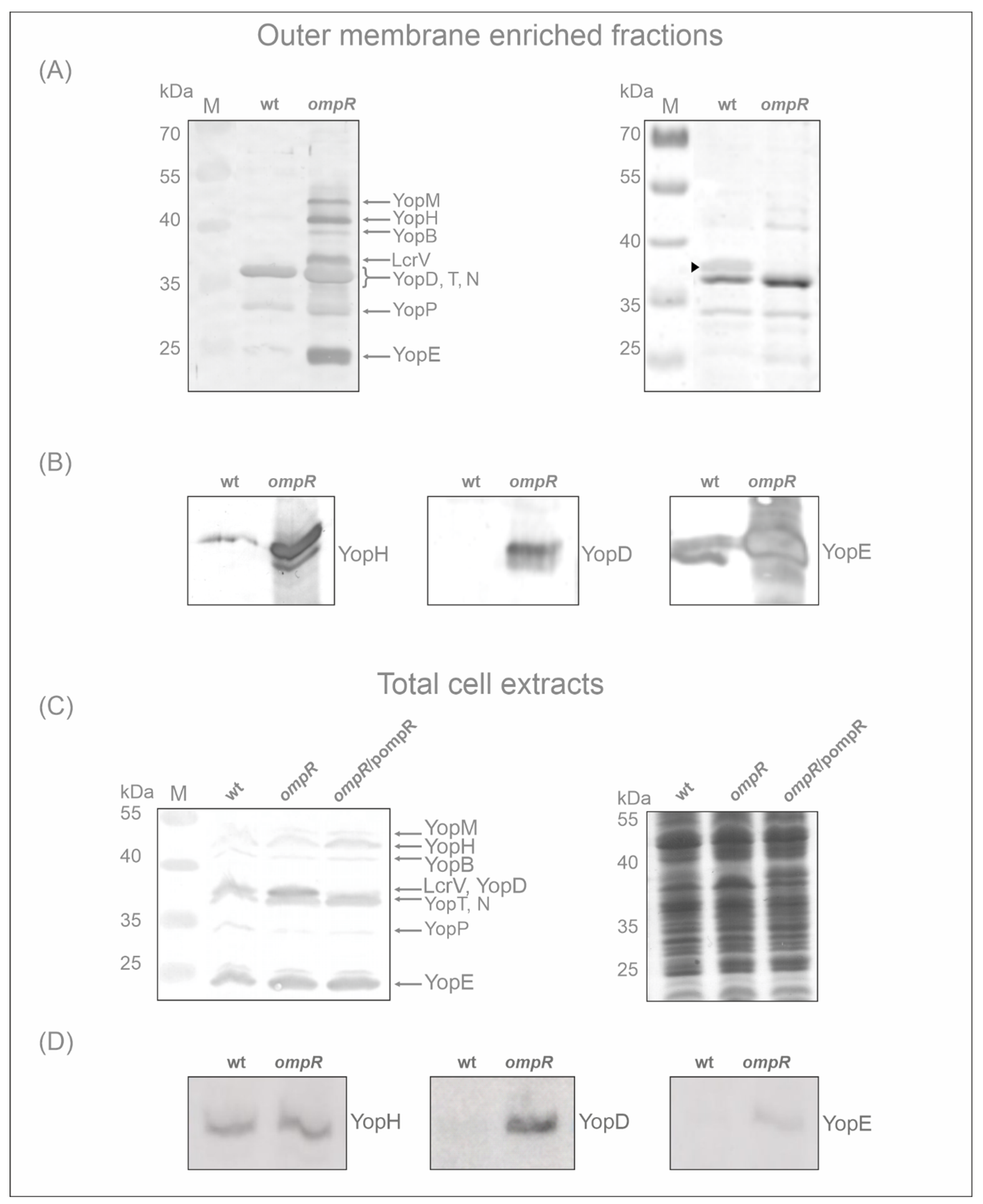
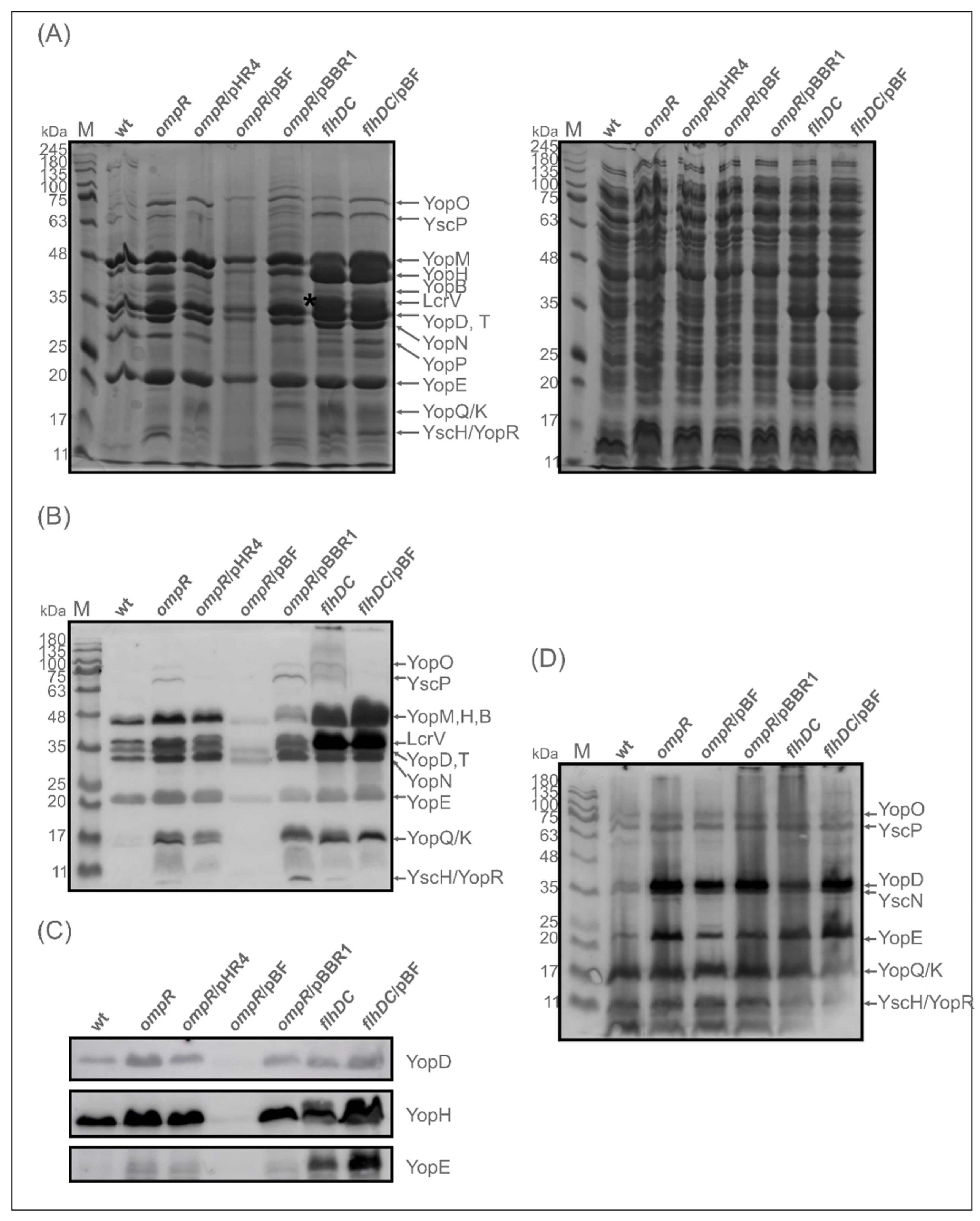
2.3. Evaluation of the Abundance of Yops in the OmpR-Deficient Strain
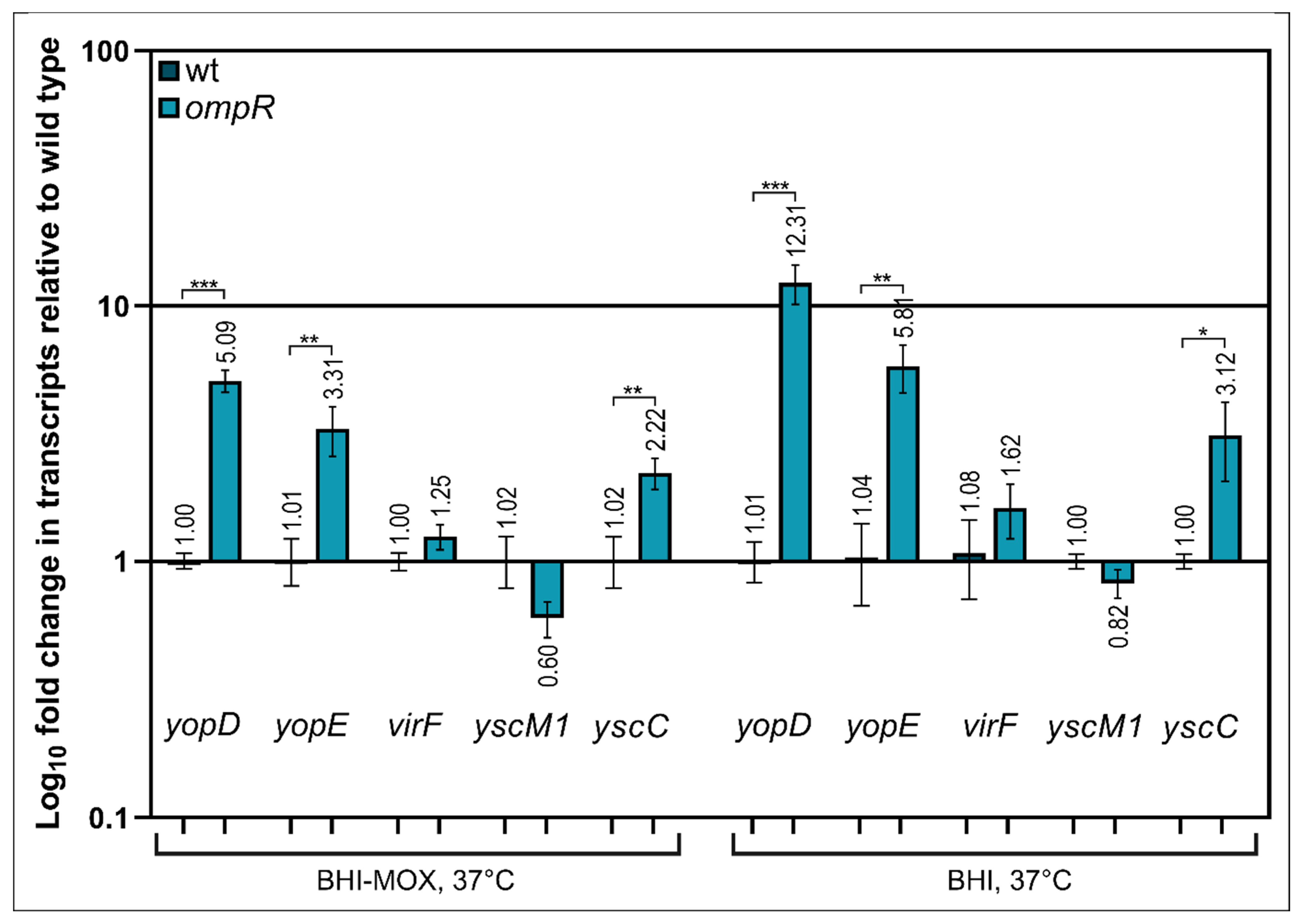
2.4. Alteration of Ysc-Yop T3SS Gene Transcription by OmpR
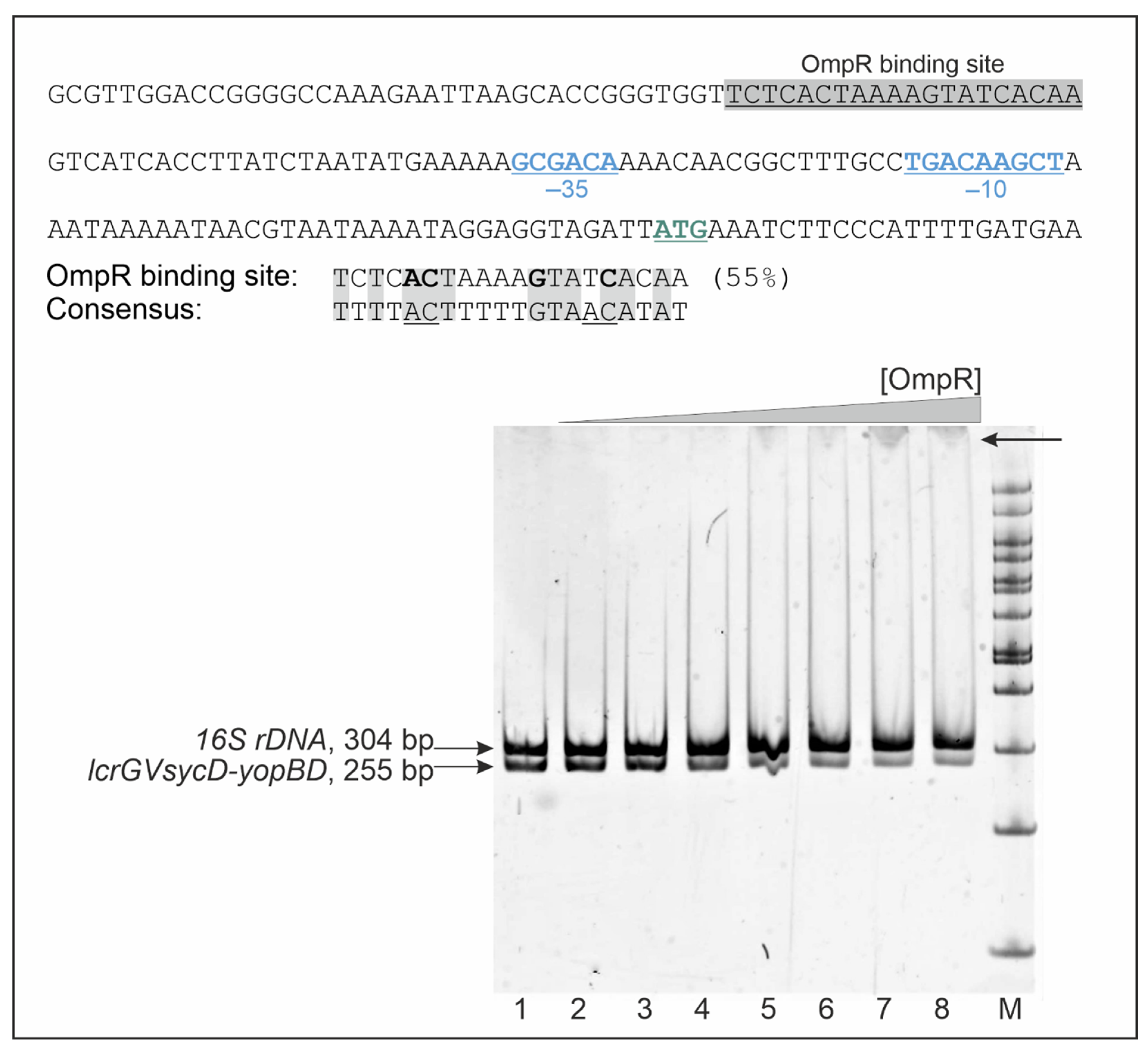
2.5. OmpR Binds to the lcrGVsycD-yopBD Operon Promoter Region
2.6. The Expression/Secretion of Yop Proteins in the ompR Mutant Is Correlated with the Amount of Regulatory Protein FlhDC
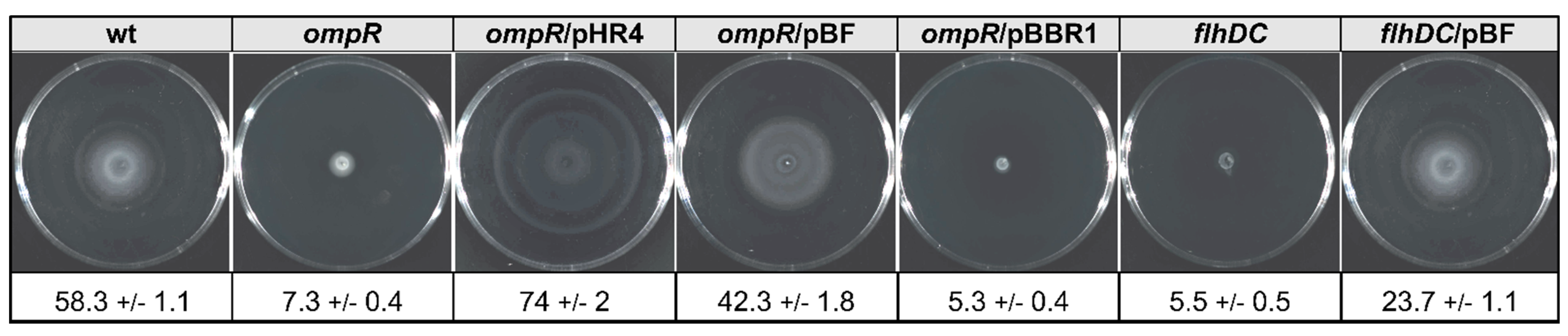
2.7. The Relevance of FlhDC Expressed in Trans in the OmpR- and FlhDC-Deficient Strains on Y. enterocolitica Motility
3. Discussion
4. Materials and Methods
4.1. Bacterial Growth Conditions
4.2. Isolation of Outer Membrane-Enriched Sarkosyl-Insoluble Fractions and Proteomic Analysis
4.3. Induction and Analysis of Secreted Yop Proteins
4.4. Western Blotting
4.5. RT-qPCR
4.6. Electrophoretic Mobility Shift Assay (EMSA)
4.7. Motility Assay
4.8. Tandem Mass Spectrometry (MS/MS) Analysis
4.9. Bioinformatic Data and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Bottone, E.J. Yersinia enterocolitica: A panoramic view of a charismatic microorganism. CRC Crit. Rev. Microbiol. 1977, 5, 211–241. [Google Scholar] [CrossRef] [PubMed]
- Bottone, E.J. Yersinia enterocolitica: Revisitation of an Enduring Human Pathogen. Clin. Microbiol. Newsl. 2015, 37, 1–8. [Google Scholar] [CrossRef]
- Wren, B.W. The yersiniae—A model genus to study the rapid evolution of bacterial pathogens. Nat. Rev. Microbiol. 2003, 1, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Bialas, N.; Kasperkiewicz, K.; Radziejewska-Lebrecht, J.; Skurnik, M. Bacterial cell surface structures in Yersinia enterocolitica. Arch. Immunol. Ther. Exp. 2012, 60, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, G.R.; Boland, A.; Boyd, A.P.; Geuijen, C.; Iriarte, M.; Neyt, C.; Sory, M.; Stainier, I. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 1998, 62, 1315–1352. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, G.R.; Biot, T.; Lambert de Rouvroit, C.; Michiels, T.; Mulder, B.; Sluiters, C.; Sory, M.-P.; Van, M. The Yersinia yop regulon. Mol. Microbiol. 1989, 3, 1455–1459. [Google Scholar] [CrossRef]
- Cornelis, G.R. Yersinia type III secretion: Send in the effectors. J. Cell. Biol. 2002, 158, 401–408. [Google Scholar] [CrossRef]
- Straley, S.C.; Perry, R.D. Environmental modulation of gene expression and pathogenesis in Yersinia. Trends Microbiol. 1995, 3, 310–317. [Google Scholar] [CrossRef]
- Rosqvist, R.; Håkansson, S.; Forsberg, A.; Wolf-Watz, H. Functional conservation of the secretion and translocation machinery for virulence proteins of yersiniae, salmonellae and shigellae. EMBO J. 1995, 14, 4187–4195. [Google Scholar] [CrossRef]
- Cornelis, G.R. The type III secretion injectosome. Nat. Rev. Microbiol. 2006, 4, 811–825. [Google Scholar] [CrossRef]
- Deng, W.; Marshall, N.C.; Rowland, J.L.; McCo, J.M.; Worrall, L.J.; Santos, A.S.; Strynadka, N.C.J.; Finlay, B.B. Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 2017, 15, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Mota, L.J.; Cornelis, G.R. The bacterial injection kit: Type III secretion systems. Ann. Med. 2005, 37, 234–249. [Google Scholar] [CrossRef] [PubMed]
- Viboud, G.I.; Bliska, J.B. Yersinia outer proteins: Role in modulation of host cell signaling responses and pathogenesis. Annu. Rev. Microbiol. 2005, 59, 69–89. [Google Scholar] [CrossRef] [PubMed]
- Parsot, C.; Hamiaux, C.; Page, A.L. The various and varying roles of specific chaperones in type III secretion systems. Curr. Opin. Microbiol. 2003, 6, 7–14. [Google Scholar] [CrossRef]
- Dewoody, R.S.; Merritt, P.M.; Marketon, M.M. Regulation of the Yersinia type III secretion system: Traffic control. Front. Cell. Infect. Microbiol. 2013, 3, 4. [Google Scholar] [CrossRef]
- Akopyan, K.; Edgren, T.; Wang-Edgren, H.; Rosqvist, R.; Fahlgren, A.; Wolf-Watz, H.; Fallman, M. Translocation of surface-localized effectors in type III secretion. Proc. Natl. Acad. Sci. USA 2011, 108, 1639–1644. [Google Scholar] [CrossRef]
- Edgren, T.; Forsberg, Å.; Rosqvist, R.; Wolf-Watz, H. Type III Secretion in Yersinia: Injectosome or Not? PLoS Pathog. 2012, 8, e1002669. [Google Scholar] [CrossRef]
- Plano, G.V.; Barve, S.S.; Straley, S.C. LcrD, a membrane-bound regulator of the Yersinia pestis low-calcium response. J. Bacteriol. 1991, 173, 7293–7303. [Google Scholar] [CrossRef]
- Pettersson, J.; Nordfelth, R.; Dubinina, E.; Bergman, T.; Gustafsson, M.; Magnusson, K.E.; Wolf-Watz, H. Modulation of virulence factor expression by pathogen target cell contact. Science 1996, 273, 1231–1233. [Google Scholar] [CrossRef]
- Williams, A.W.; Straley, S.C. YopD of Yersinia pestis plays a role in negative regulation of the low-calcium response in addition to its role in translocation of Yops. J. Bacteriol. 1998, 180, 350–358. [Google Scholar] [CrossRef]
- Wulff-Strobel, C.R.; Williams, A.W.; Straley, S.C. LcrQ and SycH function together at the Ysc type III secretion system in Yersinia pestis to impose a hierarchy of secretion. Mol. Microbiol. 2002, 43, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.S.; Plano, G.V. Identification of TyeA residues required to interact with YopN and to regulate Yop secretion. Adv. Exp. Med. Biol. 2007, 603, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, G.; Vanootegem, J.C.; Sluiters, C. Transcription of the yop regulon from Y. enterocolitica requires trans acting pYV and chromosomal genes. Microb. Pathog. 1987, 2, 367–379. [Google Scholar] [CrossRef]
- Cornelis, G.R.; Sluiters, C.; Delor, I.; Geib, D.; Kaniga, K.; Lambert de Rouvroit, C.; Sory, M.P.; Vanooteghem, J.C.; Michiels, T. ymoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol. Microbiol. 1991, 5, 1023–1034. [Google Scholar] [CrossRef]
- Lambert de Rouvroit, C.; Sluiters, C.; Cornelis, G.R. Role of the transcriptional activator, VirF, and temperature in the expression of the pYV plasmid genes of Yersinia enterocolitica. Mol. Microbiol. 1992, 6, 395–409. [Google Scholar] [CrossRef]
- Schwiesow, L.; Lam, H.; Dersch, P.; Auerbuch, V. Yersinia Type III Secretion System Master Regulator LcrF. J. Bacteriol. 2015, 198, 604–614. [Google Scholar] [CrossRef]
- Cornelis, G.R.; Sluiters, C.; Lambert de Rouvroit, C.; Michiels, T. Homology between VirF, the transcriptional activator of the Yersinia virulence region, and the Escherichia coli arabinose operon regulator. J. Bacteriol. 1989, 171, 254–262. [Google Scholar] [CrossRef]
- Hoe, N.P.; Minion, F.C.; Goguen, J.D. Temperature sensing in Yersinia pestis: Regulation of yopE transcription by lcrF. J. Bacteriol. 1992, 174, 4275–4286. [Google Scholar] [CrossRef]
- Wattiau, P.; Cornelis, G.R. Identification of DNA sequences recognized by VirF, the transcriptional activator of the Yersinia yop regulon. J. Bacteriol. 1994, 176, 3878–3884. [Google Scholar] [CrossRef][Green Version]
- Cambronne, E.D.; Cheng, L.W.; Schneewind, O. LcrQ/YscM1, regulators of the Yersinia yop virulon, are injected into host cells by a chaperone-dependent mechanism. Mol. Microbiol. 2000, 37, 263–273. [Google Scholar] [CrossRef]
- Li, Y.; Li, L.; Huang, L.; Francis, M.S.; Hu, Y.; Chen, S. Yersinia Ysc-Yop type III secretion feedback inhibition is relieved through YscV-dependent recognition and secretion of LcrQ. Mol. Microbiol. 2014, 91, 494–507. [Google Scholar] [CrossRef]
- Li, L.; Yan, H.; Feng, L.; Li, Y.; Lu, P.; Hu, Y.; Chen, S. LcrQ blocks the role of LcrF in regulating the Ysc-Yop type III secretion genes in Yersinia pseudotuberculosis. PLoS ONE 2014, 9, e92243. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M.; Ramamurthi, K.S.; Tam, C.; Schneewind, O. YopD and LcrH regulate expression of Yersinia enterocolitica YopQ by a posttranscriptional mechanism and bind to yopQ RNA. J. Bacteriol. 2002, 184, 1287–1295. [Google Scholar] [CrossRef] [PubMed]
- Fei, K.; Yan, H.; Zeng, X.; Huang, S.; Tang, W.; Francis, M.S.; Chen, S.; Hu, Y. LcrQ Coordinates with the YopD-LcrH Complex To Repress lcrF Expression and Control Type III Secretion by Yersinia pseudotuberculosis. mBio 2021, 12, e0145721. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, K.E.; Liu, J.; Edqvist, P.J.; Francis, M.S. Extracytoplasmic-stress-responsive pathways modulate type III secretion in Yersinia pseudotuberculosis. Infect. Immun. 2007, 75, 3913–3924. [Google Scholar] [CrossRef]
- Liu, J.; Thanikkal, E.J.; Obi, I.R.; Francis, M.S. Elevated CpxR~P levels repress the Ysc-Yop type III secretion system of Yersinia pseudotuberculosis. Res. Microbiol. 2012, 163, 518–530. [Google Scholar] [CrossRef]
- Li, Y.; Hu, Y.; Francis, M.S.; Chen, S. RcsB positively regulates the Yersinia/Ysc-Yop type III secretion system by activating expression of the master transcriptional regulator LcrF. Environ. Microbiol. 2015, 17, 1219–1233. [Google Scholar] [CrossRef]
- Stock, J.B.; Ninfa, A.J.; Stock, A.M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 1989, 53, 450–490. [Google Scholar] [CrossRef]
- Chakraborty, S.; Kenney, L.J. A New Role of OmpR in Acid and Osmotic Stress in Salmonella and E. coli. Front. Microbiol. 2018, 9, 2656. [Google Scholar] [CrossRef]
- Kenney, L.J.; Anand, G.S. EnvZ/OmpR Two-Component Signaling: An Archetype System That Can Function Noncanonically. EcoSal Plus. 2020, 9. [Google Scholar] [CrossRef]
- Russo, F.D.; Silhavy, T.J. EnvZ controls the concentration of phosphoryled OmpR to mediate osmoregulation of the porin genes. J. Mol. Biol. 1990, 222, 567–580. [Google Scholar] [CrossRef]
- Head, C.G.; Tardy, A.; Kenney, L.J. Relative binding affinities of OmpR and OmpR-phosphate at the ompF and ompC regulatory sites. J. Mol. Biol. 1998, 281, 857–870. [Google Scholar] [CrossRef] [PubMed]
- Kenney, L.J. Structure/function relationships in OmpR and other winged-helix transcription factors. Curr. Opin. Microbiol. 2002, 5, 135–141. [Google Scholar] [CrossRef]
- Bernardini, M.J.; Fontaine, A.; Sansonetti, P.J. The two-component regulatory system OmpR-EnvZ controls the virulence of Shigella flexneri. J. Bacteriol. 1990, 172, 6274–6281. [Google Scholar] [CrossRef] [PubMed]
- Bang, I.S.; Kim, B.H.; Foster, J.W.; Park, Y.K. OmpR regulates the stationary-phase acid tolerance response of Salmonella enterica serovar typhimurium. J. Bacteriol. 2000, 182, 2245–2252. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.K.; Detweiler, C.S.; Falkow, S. OmpR regulates the two-component system SsrA-SsrB in Salmonella pathogenicity island 2. J. Bacteriol. 2000, 182, 771–781. [Google Scholar] [CrossRef]
- Bang, I.S.; Audia, J.P.; Park, Y.K.; Foster, J.W. Autoinduction of the ompR response regulator by acid shock and control of the Salmonella typhimurium acid tolerance response. Mol. Microbiol. 2002, 44, 1235–1250. [Google Scholar] [CrossRef] [PubMed]
- Jubelin, G.; Vianney, A.; Beloin, C.; Ghigo, J.M.; Lazzaroni, J.C.; Lejeune, P.; Dorel, C. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol. 2005, 187, 2038–2049. [Google Scholar] [CrossRef]
- Cameron, A.D.; Dorman, C.J. A fundamental regulatory mechanism operating through OmpR and DNA topology controls expression of Salmonella pathogenicity islands SPI-1 and SPI-2. PLoS Genet. 2012, 8, e1002615. [Google Scholar] [CrossRef]
- Rentschler, A.E.; Lovrich, S.D.; Fitton, R.; Enos-Berlage, J.; Schwan, W.R. OmpR regulation of the uropathogenic Escherichia coli fimB gene in an acidic/high osmolality environment. Microbiology 2013, 159 Pt 2, 316–327. [Google Scholar] [CrossRef]
- Gerken, H.; Vuong, P.; Soparkar, K.; Misra, R. Roles of the EnvZ/OmpR Two-Component System and Porins in Iron Acquisition in Escherichia coli. mBio 2020, 11, e01192-20. [Google Scholar] [CrossRef]
- Perkins, T.T.; Davies, M.R.; Klemm, E.J.; Rowley, G.; Wileman, T.; James, K.; Keane, T.; Maskell, D.; Hinton, J.C.D.; Dougan, G.; et al. ChIP-seq and transcriptome analysis of the OmpR regulon of Salmonella enterica serovars Typhi and Typhimurium reveals accessory genes implicated in host colonization. Mol. Microbiol. 2013, 87, 526–538. [Google Scholar] [CrossRef] [PubMed]
- Quinn, H.J.; Cameron, A.D.; Dorman, C.J. Bacterial regulon evolution: Distinct responses and roles for the identical OmpR proteins of Salmonella Typhimurium and Escherichia coli in the acid stress response. PLoS Genet. 2014, 10, e1004215. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Lu, P.; Wang, Y.; Ding, L.; Atkinson, S.; Chen, S. OmpR positively regulates urease expression to enhance acid survival of Yersinia pseudotuberculosis. Microbiology 2009, 155 Pt 8, 2522–2531. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Wang, Y.; Ding, L.; Lu, P.; Atkinson, S.; Chen, S. Positive regulation of flhDC expression by OmpR in Yersinia pseudotuberculosis. Microbiology 2009, 155, 3622–3631. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gao, H.; Zhang, Y.; Han, Y.; Yang, L.; Liu, X.; Guo, Z.; Tan, Y.; Huang, X.; Yang, R.; Zhou, D. Phenotypic and transcriptional analysis of the osmotic regulator OmpR in Yersinia pestis. BMC Microbiol. 2011, 11, 39. [Google Scholar] [CrossRef]
- Gueguen, E.; Durand, E.; Zhang, X.Y.; d’Amalric, Q.; Journet, L.; Cascales, E. Expression of a Yersinia pseudotuberculosis type VI secretion system is responsive to envelope stresses through the OmpR transcriptional activator. PLoS ONE 2013, 8, e66615. [Google Scholar] [CrossRef]
- Dorrell, N.; Li, S.R.; Everest, P.H.; Dougan, G.; Wren, B.W. Construction and characterisation of a Yersinia enterocolitica O:8 ompR mutant. FEMS Microbiol. Lett. 1998, 165, 145–151. [Google Scholar] [CrossRef][Green Version]
- Brzostek, K.; Raczkowska, A.; Zasada, A. The osmotic regulator OmpR is involved in the response of Yersinia enterocolitica O:9 to environmental stresses and survival within macrophages. FEMS Microbiol. Lett. 2003, 228, 265–271. [Google Scholar] [CrossRef][Green Version]
- Raczkowska, A.; Skorek, K.; Brzóstkowska, M.; Lasińska, A.; Brzostek, K. Pleiotropic effects of a Yersinia enterocolitica ompR mutation on adherent-invasive abilities and biofilm formation. FEMS Microbiol. Lett. 2011, 321, 43–49. [Google Scholar] [CrossRef]
- Raczkowska, A.; Skorek, K.; Bielecki, J.; Brzostek, K. OmpR controls Yersinia enterocolitica motility by positive regulation of flhDC expression. Anton. Van Leeuwenhoek 2011, 99, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Skorek, K.; Raczkowska, A.; Dudek, B.; Miętka, K.; Guz-Regner, K.; Pawlak, A.; Klausa, E.; Bugla-Płoskońska, G.; Brzostek, K. Regulatory protein OmpR influences the serum resistance of Yersinia enterocolitica O:9 by modifying the structure of the outer membrane. PLoS ONE 2013, 8, e79525. [Google Scholar] [CrossRef] [PubMed]
- Jaworska, K.; Nieckarz, M.; Ludwiczak, M.; Raczkowska, A.; Brzostek, K. OmpR-Mediated Transcriptional Regulation and Function of Two Heme Receptor Proteins of Yersinia enterocolitica Bio-Serotype 2/O:9. Front. Cell. Infect. Microbiol. 2018, 8, 333. [Google Scholar] [CrossRef] [PubMed]
- Jaworska, K.; Ludwiczak, M.; Murawska, E.; Raczkowska, A.; Brzostek, K. The Regulator OmpR in Yersinia enterocolitica Participates in Iron Homeostasis by Modulating Fur Level and Affecting the Expression of Genes Involved in Iron Uptake. Int. J. Mol. Sci. 2021, 22, 1475. [Google Scholar] [CrossRef]
- Nieckarz, M.; Raczkowska, A.; Debski, J.; Kistowski, M.; Dadlez, M.; Heesemann, J.; Rossier, O.; Brzostek, K. Impact of OmpR on the membrane proteome of Yersinia enterocolitica in different environments: Repression of major adhesin YadA and heme receptor HemR. Environ. Microbiol. 2016, 18, 997–1021. [Google Scholar] [CrossRef]
- Nieckarz, M.; Kaczor, P.; Jaworska, K.; Raczkowska, A.; Brzostek, K. Urease Expression in Pathogenic Yersinia enterocolitica Strains of Bio-Serotypes 2/O:9 and 1B/O:8 Is Differentially Regulated by the OmpR Regulator. Front. Microbiol. 2020, 11, 607. [Google Scholar] [CrossRef]
- Koster, M.; Bitter, W.; de Cock, H.; Allaoui, A.; Cornelis, G.R.; Tommassen, J. The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocolitica forms a ring-shaped multimeric complex. Mol. Microbiol. 1997, 26, 789–797. [Google Scholar] [CrossRef]
- Diepold, A.; Wiesand, U.; Amstutz, M.; Cornelis, G.R. Assembly of the Yersinia injectosome: The missing pieces. Mol. Microbiol. 2012, 85, 878–892. [Google Scholar] [CrossRef]
- Blaylock, B.; Riordan, K.E.; Missiakas, D.M.; Schneewind, O. Characterization of the Yersinia enterocolitica type III secretion ATPase YscN and its regulator, YscL. J. Bacteriol. 2006, 188, 3525–3534. [Google Scholar] [CrossRef]
- Agrain, C.; Sorg, I.; Paroz, C.; Cornelis, G.R. Secretion of YscP from Yersinia enterocolitica is essential to control the length of the injectisome needle but not to change the type III secretion substrate specificity. Mol. Microbiol. 2005, 57, 1415–1427. [Google Scholar] [CrossRef]
- Forsberg, A.; Viitanen, A.M.; Skurnik, M.; Wolf-Watz, H. The surface-located YopN protein is involved in calcium signal transduction in Yersinia pseudotuberculosis. Mol. Microbiol. 1991, 5, 977–986. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Qin, L.; Egger, L.A.; Inouye, M. Transcription regulation of ompF and ompC by a single transcription factor, OmpR. J. Biol. Chem. 2006, 281, 17114–17123. [Google Scholar] [CrossRef] [PubMed]
- Cambronne, E.D.; Schneewind, O. Yersinia enterocolitica type III secretion: yscM1 and yscM2 regulate yop gene expression by a posttranscriptional mechanism that targets the 5’ untranslated region of yop mRNA. J. Bacteriol. 2002, 184, 5880–5893. [Google Scholar] [CrossRef] [PubMed]
- Maeda, S.; Takayanagi, K.; Nishimura, Y.; Maruyama, T.; Sato, K.; Mizuno, T. Activation of the osmoregulated ompC gene by the OmpR protein in Escherichia coli: A study involving synthetic OmpR-binding sequences. J. Biochem. 1991, 110, 324–327. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Harlocker, S.L.; Bergstrom, L.; Inouye, M. Tandem binding of six OmpR proteins to the ompF upstream regulatory sequence of Escherichia coli. J. Biol. Chem. 1995, 270, 26849–26856. [Google Scholar] [CrossRef]
- Sarker, M.R.; Neyt, C.; Stainier, I.; Cornelis, G.R. The Yersinia Yop virulon: LcrV is required for extrusion of the translocators YopB and YopD. J. Bacteriol. 1998, 180, 1207–1214. [Google Scholar] [CrossRef]
- Bleves, S.M.N.; Marenne, G.; Detry, G.R. Cornelis. Up-regulation of the Yersinia enterocolitica yop regulon by deletion of the flagellum master operon flhDC. J. Bacteriol. 2002, 184, 3214–3223. [Google Scholar] [CrossRef][Green Version]
- Horne, S.M.; Pruss, B.M. Global gene regulation in Yersinia enterocolitica: Effect of FliA on the expression levels of flagellar and plasmid-encoded virulence genes. Arch. Microbiol. 2006, 185, 115–126. [Google Scholar] [CrossRef]
- Kovach, M.E.; Elzer, P.H.; Hill, D.S.; Robertson, G.T.; Farris, M.A.; Roop, R.M.; Peterson, K.M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995, 166, 175–176. [Google Scholar] [CrossRef]
- Watarai, M.; Tobe, T.; Yoshikawa, M.; Sasakawa, C. Contact of Shigella with host cells triggers release of Ipa invasins and is an essential function of invasiveness. EMBO J. 1995, 14, 2461–2470. [Google Scholar] [CrossRef]
- Francis, M.S.; Lloyd, S.A.; Wolf-Watz, H. The type III secretion chaperone LcrH co-operates with YopD to establish a negative, regulatory loop for control of Yop synthesis in Yersinia pseudotuberculosis. Mol. Microbiol. 2001, 42, 1075–1093. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Q.; Anderson, D.M. Expression hierarchy in the Yersinia type III secretion system established through YopD recognition of RNA. Mol. Microbiol. 2011, 80, 966–980. [Google Scholar] [CrossRef] [PubMed]
- Kusmierek, M.; Hoßmann, J.; Witte, R.; Opitz, W.; Vollmer, I.; Volk, M.; Heroven, A.K.; Wolf-Watz, H.; Dersch, P. A bacterial secreted translocator hijacks riboregulators to control type III secretion in response to host cell contact. PLoS Pathog. 2019, 15, e1007813. [Google Scholar] [CrossRef] [PubMed]
- Young, B.M.; Young, G.M. YplA is exported by the Ysc, Ysa, and flagellar type III secretion systems of Yersinia enterocolitica. J. Bacteriol. 2002, 184, 1324–1334. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, T.M.; Brandt, K.; Starke, M.; Rattei, T. Shotgun sequencing of Yersinia enterocolitica strain W22703 (biotype 2, serotype O:9): Genomic evidence for oscillation between invertebrates and mammals. BMC Genom. 2011, 12, 168. [Google Scholar] [CrossRef]
- Young, G.M.; Smith, M.J.; Minnich, S.A.; Miller, V.L. The Yersinia enterocolitica motility master regulatory operon, flhDC, is required for flagellin production, swimming motility, and swarming motility. J. Bacteriol. 1999, 181, 2823–2833. [Google Scholar] [CrossRef]
- Kubori, T.; Matsushima, Y.; Nakamura, D.; Uralil, J.; Lara-Tejero, M.; Sukhan, A.; Galán, J.E.; Aizawa, S.I. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 1998, 280, 602–605. [Google Scholar] [CrossRef]
- Blocker, A.; Komoriya, K.; Aizawa, S. Type III secretion systems and bacterial flagella: Insights into their function from structural similarities. Proc. Natl. Acad. Sci. USA 2003, 100, 3027–3030. [Google Scholar] [CrossRef]
- Brzostek, K.; Raczkowska, A. The level of Yop proteins secreted by Yersinia enterocolitica is changed in maltose mutants. FEMS Microbiol. Lett. 2001, 204, 95–100. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
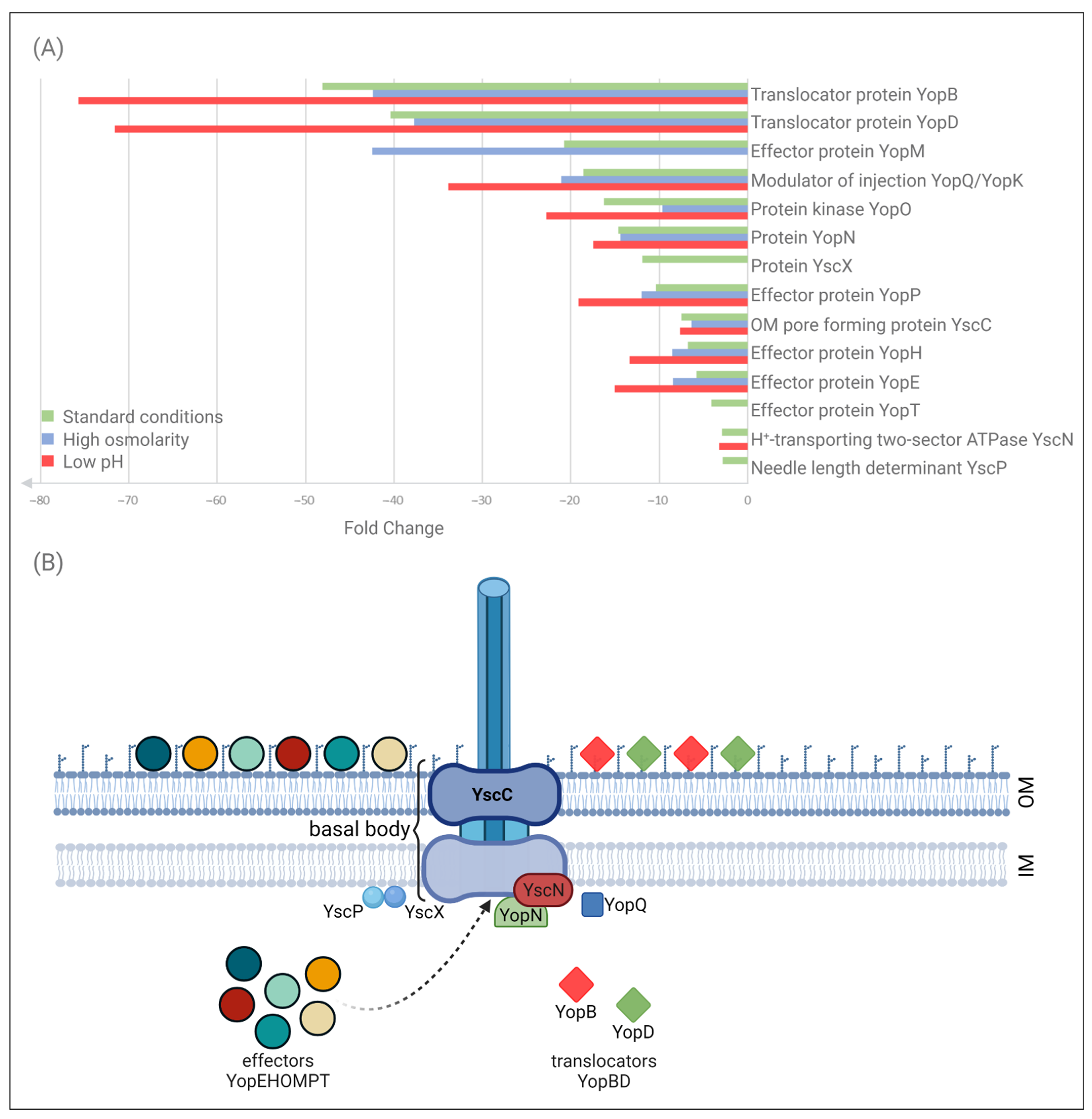
| Differentially Expressed Proteins | Regulation Ye9 vs. AR4 b | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Standard Conditions | High Osmolarity | Low pH | |||||||||||
| Accession Number (GenBank) | Protein Description a | q-Value c | Ratio d | fc e | pep f | q-Value | Ratio | fc | pep | q-Value | Ratio | fc | pep |
| ADZ44444 | Translocator protein YopB | 0.00005 | 0.02 | 48.12 | 41 | 0.00007 | 0.02 | 42.4 | 42 | 0.00005 | 0.01 | 75.73 | 41 |
| ADZ44443 | Translocator protein YopD | 0.00005 | 0.02 | 40.37 | 48 | 0.00007 | 0.03 | 37.76 | 48 | 0.00005 | 0.01 | 71.6 | 48 |
| ADZ44440 | Effector protein YopM | 0.00073 | 0.05 | 20.79 | 23 | 0.00276 | 0.02 | 42.45 | 21 | 0.07599 | 0.25 | 3.95 | 22 |
| ADZ44434 | Negative modulator of injection YopQ/YopK | 0.00005 | 0.05 | 18.54 | 18 | 0.0003 | 0.05 | 21.11 | 17 | 0.00005 | 0.03 | 33.89 | 18 |
| ADZ44516 | Protein kinase YopO | 0.00005 | 0.06 | 16.25 | 64 | 0.00007 | 0.1 | 9.64 | 57 | 0.00005 | 0.04 | 22.77 | 61 |
| ADZ44454 | OM protein YopN | 0.00147 | 0.07 | 14.62 | 12 | 0.04588 | 0.07 | 14.4 | 10 | 0.01486 | 0.06 | 17.46 | 11 |
| ADZ44451 | Protein YscX | 0.0194 | 0.08 | 11.89 | 6 | 0.34986 | 0.18 | 5.52 | 6 | 0.13906 | 0.17 | 5.83 | 6 |
| ADZ44518 | Effector protein YopP | 0.00048 | 0.1 | 10.37 | 26 | 0.00057 | 0.08 | 11.94 | 25 | 0.0019 | 0.05 | 19.12 | 25 |
| ADZ44467 | OM pore forming protein YscC | 0.00005 | 0.13 | 7.48 | 31 | 0.00007 | 0.16 | 6.37 | 31 | 0.00005 | 0.13 | 7.66 | 31 |
| ADZ44479 | Tyrosine-protein phosphatase effector protein YopH | 0.00005 | 0.15 | 6.78 | 76 | 0.00007 | 0.12 | 8.49 | 69 | 0.00005 | 0.08 | 13.32 | 72 |
| EOR65641 | Effector protein YopE | 0.00005 | 0.17 | 5.77 | 29 | 0.00007 | 0.12 | 8.41 | 27 | 0.00005 | 0.07 | 15.03 | 30 |
| ADZ44435 | Effector protein YopT | 0.00998 | 0.24 | 4.12 | 10 | 0.18349 | 0.24 | 4.08 | 9 | 0.10699 | 0.2 | 4.97 | 8 |
| ADZ44455 | H+-transporting two-sector ATPase YscN | 0.0128 | 0.35 | 2.87 | 13 | 0.21592 | 0.49 | 2.04 | 12 | 0.04486 | 0.31 | 3.23 | 12 |
| ADZ44457 | Needle length determinant YscP | 0.02605 | 0.36 | 2.78 | 13 | 0.26892 | 0.45 | 2.23 | 12 | 0.11434 | 0.42 | 2.39 | 13 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nieckarz, M.; Jaworska, K.; Raczkowska, A.; Brzostek, K. The Regulatory Circuit Underlying Downregulation of a Type III Secretion System in Yersinia enterocolitica by Transcription Factor OmpR. Int. J. Mol. Sci. 2022, 23, 4758. https://doi.org/10.3390/ijms23094758
Nieckarz M, Jaworska K, Raczkowska A, Brzostek K. The Regulatory Circuit Underlying Downregulation of a Type III Secretion System in Yersinia enterocolitica by Transcription Factor OmpR. International Journal of Molecular Sciences. 2022; 23(9):4758. https://doi.org/10.3390/ijms23094758
Chicago/Turabian StyleNieckarz, Marta, Karolina Jaworska, Adrianna Raczkowska, and Katarzyna Brzostek. 2022. "The Regulatory Circuit Underlying Downregulation of a Type III Secretion System in Yersinia enterocolitica by Transcription Factor OmpR" International Journal of Molecular Sciences 23, no. 9: 4758. https://doi.org/10.3390/ijms23094758
APA StyleNieckarz, M., Jaworska, K., Raczkowska, A., & Brzostek, K. (2022). The Regulatory Circuit Underlying Downregulation of a Type III Secretion System in Yersinia enterocolitica by Transcription Factor OmpR. International Journal of Molecular Sciences, 23(9), 4758. https://doi.org/10.3390/ijms23094758






