The SUV4-20H Histone Methyltransferases in Health and Disease
Abstract
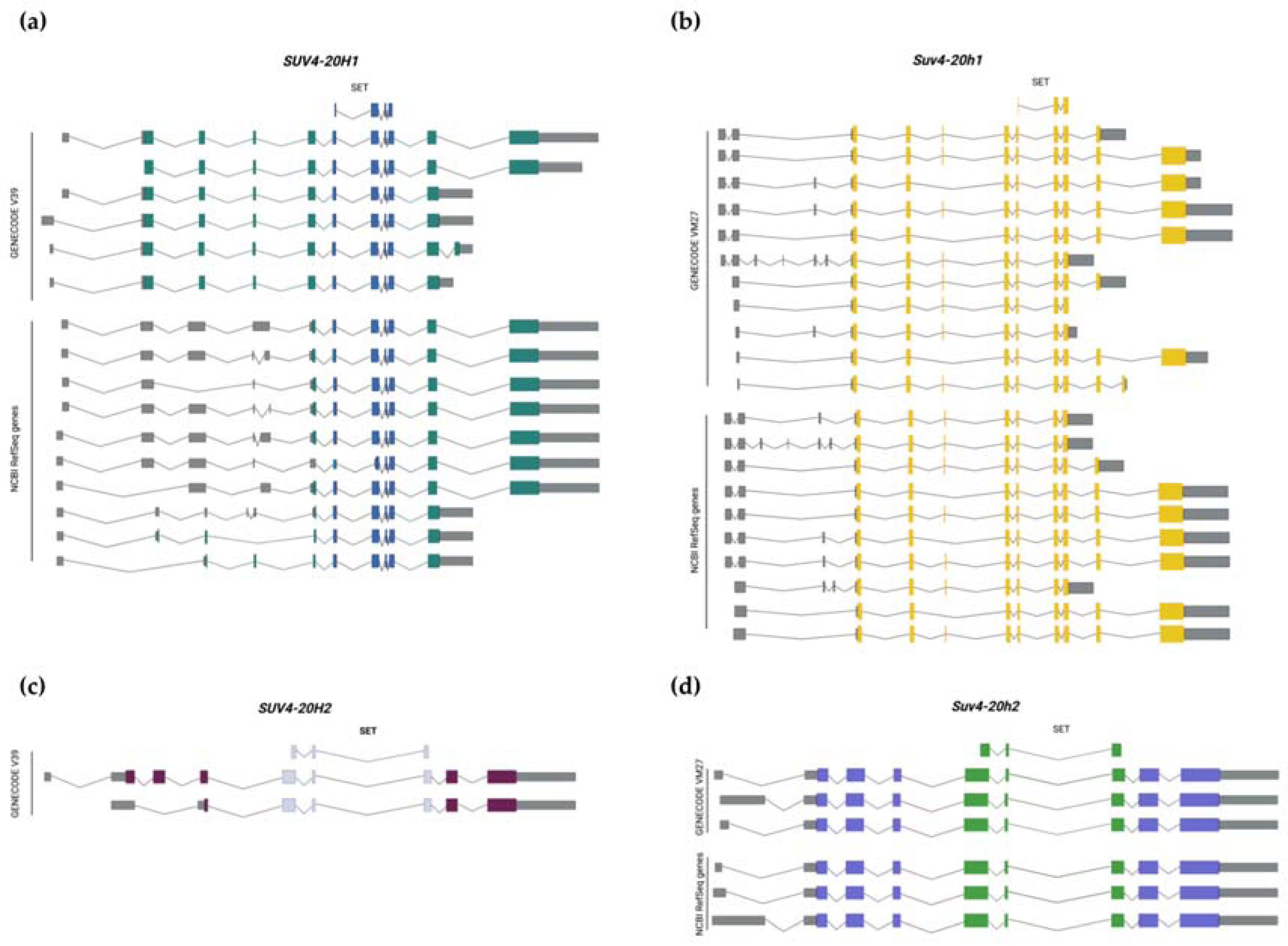
:1. Introduction
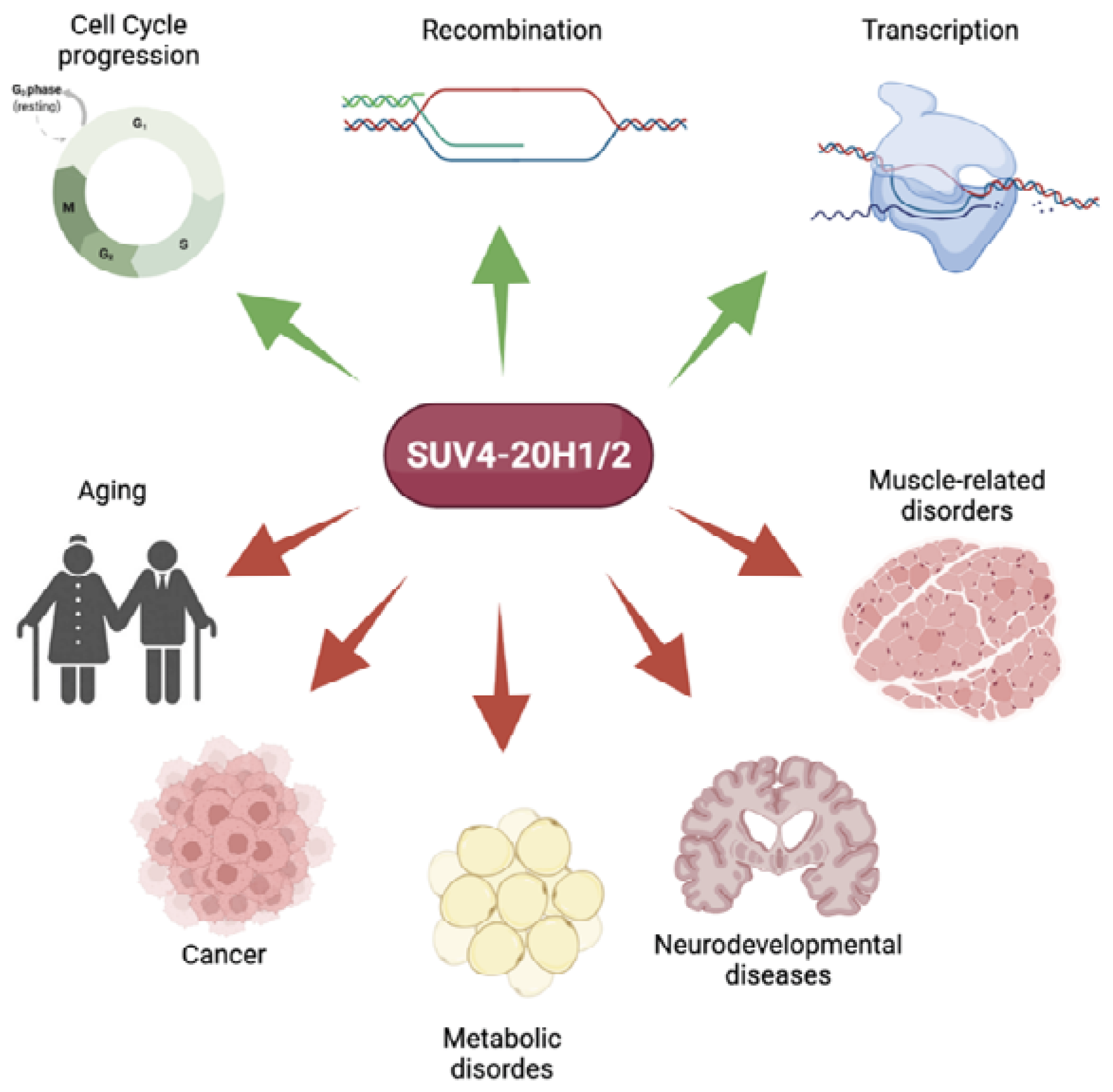
2. Role of Suv4-20h in Health
2.1. Role in Replication and Cell Cycle Progression
2.2. Role in Recombination
2.3. Role in Transcriptional Regulation
3. Suv4-20h in Disease
3.1. Aging
3.2. Cancer
3.3. Metabolic Disorders
3.4. Neurodevelopmental Disorders
3.5. Muscle-Related Disorders
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Faundes, V.; Newman, W.G.; Bernardini, L.; Canham, N.; Clayton-Smith, J.; Dallapiccola, B.; Davies, S.J.; Demos, M.K.; Goldman, A.; Gill, H.; et al. Histone Lysine Methylases and Demethylases in the Landscape of Human Developmental Disorders. Am. J. Hum. Genet. 2018, 102, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Benetti, R.; Gonzalo, S.; Jaco, I.; Schotta, G.; Klatt, P.; Jenuwein, T.; Blasco, M.A. Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J. Cell Biol. 2007, 178, 925–936. [Google Scholar] [CrossRef] [Green Version]
- Schotta, G.; Lachner, M.; Sarma, K.; Ebert, A.; Sengupta, R.; Reuter, G.; Reinberg, D.; Jenuwein, T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004, 18, 1251–1262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trojer, P.; Li, G.; Sims, R.J.; Vaquero, A.; Kalakonda, N.; Boccuni, P.; Lee, D.; Erdjument-Bromage, H.; Tempst, P.; Nimer, S.D.; et al. L3MBTL1, a Histone-Methylation-Dependent Chromatin Lock. Cell 2007, 129, 915–928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jørgensen, S.; Elvers, I.; Trelle, M.B.; Menzel, T.; Eskildsen, M.; Jensen, O.N.; Helleday, T.; Helin, K.; Sørensen, C.S. The histone methyltransferase SET8 is required for S-phase progression. J. Cell Biol. 2007, 179, 1337–1345. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Cortez, V.C.; Martínez-Redondo, P.; Català-Moll, F.; Rodríguez-Ubreva, J.; Garcia-Gomez, A.; Poorani-Subramani, G.; Ciudad, L.; Hernando, H.; Pérez-García, A.; Company, C.; et al. Activation-induced cytidine deaminase targets SUV4-20-mediated histone H4K20 trimethylation to class-switch recombination sites. Sci. Rep. 2017, 7, 7594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jørgensen, S.; Schotta, G.; Sørensen, C.S. Histone H4 Lysine 20 methylation: Key player in epigenetic regulation of genomic integrity. Nucleic Acids Res. 2013, 41, 2797–2806. [Google Scholar] [CrossRef] [PubMed]
- Brustel, J.; Kirstein, N.; Izard, F.; Grimaud, C.; Prorok, P.; Cayrou, C.; Schotta, G.; Abdelsamie, A.F.; Déjardin, J.; Méchali, M.; et al. Histone H4K20 tri-methylation at late-firing origins ensures timely heterochromatin replication. EMBO J. 2017, 36, 2726–2741. [Google Scholar] [CrossRef]
- Nishioka, K.; Rice, J.C.; Sarma, K.; Erdjument-Bromage, H.; Werner, J.; Wang, Y.; Chuikov, S.; Valenzuela, P.; Tempst, P.; Steward, R.; et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent Chromatin. Mol. Cell 2002, 9, 1201–1213. [Google Scholar] [CrossRef]
- Schotta, G.; Sengupta, R.; Kubicek, S.; Malin, S.; Kauer, M.; Callen, E.; Celeste, A.; Pagani, M.; Opravil, S.; De La Rosa-Velazquez, I.A.; et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 2008, 22, 2048–2061. [Google Scholar] [CrossRef] [Green Version]
- Fanti, L.; Pimpinelli, S. HP1: A functionally multifaceted protein. Curr. Opin. Genet. Dev. 2008, 18, 169–174. [Google Scholar] [CrossRef]
- Pannetier, M.; Julien, E.; Schotta, G.; Tardat, M.; Sardet, C.; Jenuwein, T.; Feil, R. PR-SET7 and SUV4-20H regulate H4 lysine-20 methylation at imprinting control regions in the mouse. EMBO Rep. 2008, 9, 998–1005. [Google Scholar] [CrossRef]
- Monk, D.; Arnaud, P.; Frost, J.M.; Wood, A.J.; Cowley, M.; Martin-Trujillo, A.; Guillaumet-Adkins, A.; Iglesias Platas, I.; Camprubi, C.; Bourc’His, D.; et al. Human imprinted retrogenes exhibit non-canonical imprint chromatin signatures and reside in non-imprinted host genes. Nucleic Acids Res. 2011, 39, 4577–4586. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Siarheyeva, A.; Zeng, H.; Lam, R.; Dong, A.; Wu, X.H.; Li, Y.; Schapira, M.; Vedadi, M.; Min, J. Crystal structures of the human histone H4K20 methyltransferases SUV420H1 and SUV420H2. FEBS Lett. 2013, 587, 3859–3868. [Google Scholar] [CrossRef]
- BioRender. Available online: https://app.biorender.com/ (accessed on 6 April 2022).
- Liu, W.; Tanasa, B.; Tyurina, O.V.; Yuan Zhou, T.; Gassmann, R.; Liu, W.T.; Ohgi, K.A.; Benner, C.; Garcia-Bassets, I.; Aggarwal, A.K.; et al. LETTERS PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 2010, 466, 508–512. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Telese, F.; Tan, Y.; Li, W.; Jin, C.; He, X.; Basnet, H.; Ma, Q.; Merkurjev, D.; Zhu, X.; et al. LSD1n is an H4K20 demethylase regulating memory formation via transcriptional elongation control. Nature 2015, 18, 1256–1264. [Google Scholar] [CrossRef]
- Cao, X.; Chen, Y.; Wu, B.; Wang, X.; Xue, H.; Yu, L.; Li, J.; Wang, Y.; Wang, W.; Xu, Q.; et al. Histone H4K20 Demethylation by Two hHR23 Proteins. Cell Rep. 2020, 30, 4152–4164. [Google Scholar] [CrossRef]
- Allshire, R.C.; Madhani, H.D. Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 2018, 19, 229–244. [Google Scholar] [CrossRef]
- Beck, D.B.; Burton, A.; Oda, H.; Ziegler-Birling, C.; Torres-Padilla, M.E.; Reinberg, D. The role of PR-Set7 in replication licensing depends on Suv4-20h. Genes Dev. 2012, 26, 2580–2589. [Google Scholar] [CrossRef] [Green Version]
- Bartke, T.; Vermeulen, M.; Xhemalce, B.; Robson, S.C.; Mann, M.; Kouzarides, T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 2010, 143, 470–484. [Google Scholar] [CrossRef] [Green Version]
- Vermeulen, M.; Eberl, H.C.; Matarese, F.; Marks, H.; Denissov, S.; Butter, F.; Lee, K.K.; Olsen, J.V.; Hyman, A.A.; Stunnenberg, H.G.; et al. Quantitative Interaction Proteomics and Genome-wide Profiling of Epigenetic Histone Marks and Their Readers. Cell 2010, 142, 967–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, H.; Zhang, L.; Lv, M.; Wen, Z.; Zhang, W.; Chen, X.; Zhang, P.; Li, T.; Chang, L.; Jin, C.; et al. H2A.Z facilitates licensing and activation of early replication origins. Nature 2020, 577, 576–581. [Google Scholar] [CrossRef] [PubMed]
- Eid, A.; Rodriguez-Terrones, D.; Burton, A.; Torres-Padilla, M.E. SUV4-20 activity in the preimplantation mouse embryo controls timely replication. Genes Dev. 2016, 30, 2513–2526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wongtawan, T.; Taylor, J.E.; Lawson, K.A.; Wilmut, I.; Pennings, S. Histone H4K20me3 and HP1α are late heterochromatin markers in development, but present in undifferentiated embryonic stem cells. J. Cell Sci. 2011, 124, 1878–1890. [Google Scholar] [CrossRef] [Green Version]
- Kurup, J.T.; Han, Z.; Jin, W.; Kidder, B.L. H4K20me3 methyltransferase SUV420H2 shapes the chromatin landscape of pluripotent embryonic stem cells. Development 2020, 147, dev188516. [Google Scholar] [CrossRef]
- Shue, Y.T.; Lee, K.T.; Walters, B.W.; Ong, H.B.; Silvaraju, S.; Lam, W.J.; Lim, C.Y. Dynamic shifts in chromatin states differentially mark the proliferative basal cells and terminally differentiated cells of the developing epidermis. Epigenetics 2020, 15, 932–948. [Google Scholar] [CrossRef]
- Blasco, M.A. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 2007, 8, 299–309. [Google Scholar] [CrossRef]
- Laffleur, B.; Denis-Lagache, N.; Péron, S.; Sirac, C.; Moreau, J.; Cogné, M.; Laffleur, B.; Denis-Lagache, N.; Péron, S.; Sirac, C.; et al. AID-induced remodeling of immunoglobulin genes and B cell fate. Oncotarget 2013, 5, 1118–1131. [Google Scholar] [CrossRef] [Green Version]
- Kurup, J.T.; Kidder, B.L. Identification of H4K20me3- and H3K4me3-associated RNAs using CARIP-Seq expands the transcriptional and epigenetic networks of embryonic stem cells. J. Biol. Chem. 2018, 293, 15120–15135. [Google Scholar] [CrossRef] [Green Version]
- Ji, X.; Dadon, D.B.; Abraham, B.J.; Lee, T.I.; Jaenisch, R.; Bradner, J.E.; Young, R.A. Chromatin proteomic profiling reveals novel proteins associated with histone-marked genomic regions. Proc. Natl. Acad. Sci. USA 2015, 112, 3841–3846. [Google Scholar] [CrossRef] [Green Version]
- Nicetto, D.; Hahn, M.; Jung, J.; Schneider, T.D.; Straub, T.; David, R.; Schotta, G.; Rupp, R.A.W. Suv4-20h Histone Methyltransferases Promote Neuroectodermal Differentiation by Silencing the Pluripotency-Associated Oct-25 Gene. PLoS Genet. 2013, 9, e1003188. [Google Scholar] [CrossRef] [Green Version]
- Kapoor-Vazirani, P.; Kagey, J.D.; Vertino, P.M. SUV420H2-Mediated H4K20 Trimethylation Enforces RNA Polymerase II Promoter-Proximal Pausing by Blocking hMOF-Dependent H4K16 Acetylation. Mol. Cell. Biol. 2011, 31, 1594–1609. [Google Scholar] [CrossRef] [Green Version]
- Chinenov, Y.; Sacta, M.A.; Cruz, A.R.; Rogatsky, I. GRIP1-associated SET-domain methyltransferase in glucocorticoid receptor target gene expression. Proc. Natl. Acad. Sci. USA 2008, 105, 20185–20190. [Google Scholar] [CrossRef] [Green Version]
- Bierhoff, H.; Dammert, M.A.; Brocks, D.; Dambacher, S.; Schotta, G.; Grummt, I. Quiescence-Induced LncRNAs Trigger H4K20 Trimethylation and Transcriptional Silencing. Mol. Cell 2014, 54, 675–682. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Zhang, X.; Zong, L.; Gao, Q.; Zhang, C.; Wei, R.; Guan, Y.; Huang, L.; Zhang, L.; Lyu, G.; et al. Gene body methylation safeguards ribosomal DNA transcription by preventing PHF6-mediated enrichment of repressive histone mark H4K20me3. J. Biol. Chem. 2021, 279, 101195. [Google Scholar] [CrossRef]
- Evertts, A.G.; Manning, A.L.; Wang, X.; Dyson, N.J.; Garcia, B.A.; Coller, H.A. H4K20 methylation regulates quiescence and chromatin compaction. Mol. Biol. Cell 2013, 24, 3025–3037. [Google Scholar] [CrossRef]
- Xu, J.; Kidder, B.L. H4K20me3 co-localizes with activating histone modifications at transcriptionally dynamic regions in embryonic stem cells. BMC Genom. 2018, 19, 514. [Google Scholar] [CrossRef] [Green Version]
- Lyu, G.; Guan, Y.; Zhang, C.; Zong, L.; Sun, L.; Huang, X.; Huang, L.; Zhang, L.; Tian, X.L.; Zhou, Z.; et al. TGF-β signaling alters H4K20me3 status via miR-29 and contributes to cellular senescence and cardiac aging. Nat. Commun. 2018, 9, 2560. [Google Scholar] [CrossRef]
- Kuilman, T.; Michaloglou, C.; Mooi, W.J.; Peeper, D.S. The essence of senescence. Genes Dev. 2010, 24, 2463–2479. [Google Scholar] [CrossRef] [Green Version]
- Chicas, A.; Kapoor, A.; Wang, X.; Aksoy, O.; Evertts, A.G.; Zhang, M.Q.; Garcia, B.A.; Bernstein, E.; Lowe, S.W. H3K4 demethylation by Jarid1a and Jarid1b contributes to retinoblastoma-mediated gene silencing during cellular senescence. Proc. Natl. Acad. Sci. USA 2012, 109, 8971–8976. [Google Scholar] [CrossRef] [Green Version]
- Nelson, D.M.; Jaber-Hijazi, F.; Cole, J.J.; Robertson, N.A.; Pawlikowski, J.S.; Norris, K.T.; Criscione, S.W.; Pchelintsev, N.A.; Piscitello, D.; Stong, N.; et al. Mapping H4K20me3 onto the chromatin landscape of senescent cells indicates a function in control of cell senescence and tumor suppression through preservation of genetic and epigenetic stability. Genome Biol. 2016, 17, 158. [Google Scholar] [CrossRef] [Green Version]
- Shumaker, D.K.; Dechat, T.; Kohlmaier, A.; Adam, S.A.; Bozovsky, M.R.; Erdos, M.R.; Eriksson, M.; Goldman, A.E.; Khuon, S.; Collins, F.S.; et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. USA 2006, 103, 8703–8708. [Google Scholar] [CrossRef] [Green Version]
- Sarg, B.; Koutzamani, E.; Helliger, W.; Rundquist, I.; Lindner, H.H. Postsynthetic trimethylation of histone H4 at lysine 20 in mammalian tissues is associated with aging. J. Biol. Chem. 2002, 277, 39195–39201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, M.; Li, J.; Yan, H.; Luo, T.; Huang, J.; Yuan, Y.; Hu, L.; Zheng, L. Physiological Ovarian Aging Is Associated with Altered Expression of Post-Translational Modifications in Mice. Int. J. Mol. Sci. 2021, 23, 2. [Google Scholar] [CrossRef]
- Roy, S.; Khanna, S.; Azad, A.; Schnitt, R.; He, G.; Weigert, C.; Ichijo, H.; Sen, C.K. Fra-2 mediates oxygen-sensitive induction of transforming growth factor β in cardiac fibroblasts. Cardiovasc. Res. 2010, 87, 647–655. [Google Scholar] [CrossRef] [Green Version]
- Muñoz-Espín, D.; Cañamero, M.; Maraver, A.; Gómez-López, G.; Contreras, J.; Murillo-Cuesta, S.; Rodríguez-Baeza, A.; Varela-Nieto, I.; Ruberte, J.; Collado, M.; et al. Programmed cell senescence during mammalian embryonic development. Cell 2013, 155, 1104. [Google Scholar] [CrossRef] [Green Version]
- Fraga, M.F.; Ballestar, E.; Villar-Garea, A.; Boix-Chornet, M.; Espada, J.; Schotta, G.; Bonaldi, T.; Haydon, C.; Ropero, S.; Petrie, K.; et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 2005, 37, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Marión, R.M.; Schotta, G.; Ortega, S.; Blasco, M.A. Suv4-20h abrogation enhances telomere elongation during reprogramming and confers a higher tumorigenic potential to iPs cells. PLoS ONE 2011, 6, e25680. [Google Scholar] [CrossRef] [Green Version]
- Van Den Broeck, A.; Brambilla, E.; Moro-Sibilot, D.; Lantuejoul, S.; Brambilla, C.; Eymin, B.; Khochbin, S.; Gazzeri, S. Loss of histone H4K20 trimethylation occurs in preneoplasia and influences prognosis of non-small cell lung cancer. Clin. Cancer Res. 2008, 14, 7237–7245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bröhm, A.; Elsawy, H.; Rathert, P.; Kudithipudi, S.; Schoch, T.; Schuhmacher, M.K.; Weirich, S.; Jeltsch, A. Somatic Cancer Mutations in the SUV420H1 Protein Lysine Methyltransferase Modulate Its Catalytic Activity. J. Mol. Biol. 2019, 431, 3068–3080. [Google Scholar] [CrossRef]
- Lin, W.-Y.; Fordham, S.E.; Hungate, E.; Sunter, N.J.; Elstob, C.; Xu, Y.; Park, C.; Quante, A.; Strauch, K.; Gieger, C.; et al. Genome-wide association study identifies susceptibility loci for acute myeloid leukemia. Nat. Commun. 2021, 12, 6233. [Google Scholar] [CrossRef]
- Vinci, M.; Burford, A.; Molinari, V.; Kessler, K.; Popov, S.; Clarke, M.; Taylor, K.R.; Pemberton, H.N.; Lord, C.J.; Gutteridge, A.; et al. Functional diversity and cooperativity between subclonal populations of pediatric glioblastoma and diffuse intrinsic pontine glioma cells. Nat. Med. 2018, 24, 1204–1215. [Google Scholar] [CrossRef]
- Li, Y.; Katz, R.A.; Hou, Y.; Fraga, M.F.; Es, M.; Fernández, A.F.; López, V.; Ramón Tejedor, J.; Carella, A.; García, M.G.; et al. Epigenetic Deregulation of the Histone Methyltransferase KMT5B Contributes to Malignant Transformation in Glioblastoma. Front. Cell Dev. Biol. 2021, 9, 671838. [Google Scholar] [CrossRef]
- Tryndyak, V.P.; Kovalchuk, O.; Pogribny, I.P. Loss of DNA methylation and histone H4 lysine 20 trimethylation in human breast cancer cells is associated with aberrant expression of DNA methyltransferase 1, Suv4-20h2 histone methyltransferase and methyl-binding proteins. Cancer Biol. Ther. 2006, 5, 65–70. [Google Scholar] [CrossRef] [Green Version]
- Yokoyama, Y.; Matsumoto, A.; Hieda, M.; Shinchi, Y.; Ogihara, E.; Hamada, M.; Nishioka, Y.; Kimura, H.; Yoshidome, K.; Tsujimoto, M.; et al. Loss of histone H4K20 trimethylation predicts poor prognosis in breast cancer and is associated with invasive activity. Breast Cancer Res. 2014, 16, R66. [Google Scholar] [CrossRef] [Green Version]
- Shinchi, Y.; Hieda, M.; Nishioka, Y.; Matsumoto, A.; Yokoyama, Y.; Kimura, H.; Matsuura, S.; Matsuura, N. SUV420H2 suppresses breast cancer cell invasion through down regulation of the SH2 domain-containing focal adhesion protein tensin-3. Exp. Cell Res. 2015, 334, 90–99. [Google Scholar] [CrossRef]
- Qian, X.; Li, G.; Vass, W.C.; Papageorge, A.; Walker, R.C.; Asnaghi, L.; Steinbach, P.J.; Tosato, G.; Hunter, K.; Lowy, D.R. The Tensin-3 Protein, Including its SH2 Domain, Is Phosphorylated by Src and Contributes to Tumorigenesis and Metastasis. Cancer Cell 2009, 16, 246–258. [Google Scholar] [CrossRef] [Green Version]
- Schwarzenbacher, D.; Balic, M.; Pichler, M. The Role of MicroRNAs in Breast Cancer Stem Cells. Int. J. Mol. Sci. 2013, 14, 14712–14723. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Shi, W.; Tang, T.; Wang, Y.; Yin, X.; Chen, Y.; Zhang, Y.; Xing, Y.; Shen, Y.; Xia, T.; et al. miR-29a contributes to breast cancer cells epithelial-mesenchymal transition, migration, and invasion via down-regulating histone H4K20 trimethylation through directly targeting SUV420H2. Cell Death Dis. 2019, 10, 176. [Google Scholar] [CrossRef]
- Wang, J.Y.; Zhang, Q.; Wang, D.D.; Yan, W.; Sha, H.H.; Zhao, J.H.; Yang, S.J.; Zhang, H.D.; Hou, J.C.; Xu, H.Z.; et al. MiR-29a: A potential therapeutic target and promising biomarker in tumors. Biosci. Rep. 2018, 38, BSR20171265. [Google Scholar] [CrossRef] [Green Version]
- Pal, A.S.; Agredo, A.; Lanman, N.A.; Son, J.; Sohal, I.S.; Bains, M.; Li, C.; Clingerman, J.; Gates, K.; Kasinski, A.L. Loss of KMT5C Promotes EGFR Inhibitor Resistance in NSCLC via LINC01510-Mediated Upregulation of MET. Cancer Res. 2022, 82, 1534–1547. [Google Scholar] [CrossRef] [PubMed]
- Chou, R.H.; Wang, Y.N.; Hsieh, Y.H.; Li, L.Y.; Xia, W.; Chang, W.C.; Chang, L.C.; Cheng, C.C.; Lai, C.C.; Hsu, J.L.; et al. EGFR Modulates DNA Synthesis and Repair through Tyr Phosphorylation of Histone H4. Dev. Cell 2014, 30, 224–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Choi, P.S.; Casey, S.C.; Dill, D.L.; Felsher, D.W. MYC through miR-17-92 suppresses specific target genes to maintain survival, autonomous proliferation, and a Neoplastic state. Cancer Cell 2014, 26, 262–272. [Google Scholar] [CrossRef] [Green Version]
- Elsheikh, S.E.; Green, A.R.; Rakha, E.A.; Powe, D.G.; Ahmed, R.A.; Collins, H.M.; Soria, D.; Garibaldi, J.M.; Paish, C.E.; Ammar, A.A.; et al. Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res. 2009, 69, 3802–3809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Kimball, S.; Liu, H.; Holowatyj, A.; Yang, Z.Q. Genetic alterations of histone lysine methyltransferases and their significance in breast cancer. Oncotarget 2015, 6, 2466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vougiouklakis, T.; Sone, K.; Saloura, V.; Cho, H.S.; Suzuki, T.; Dohmae, N.; Alachkar, H.; Nakamura, Y.; Hamamoto, R. SUV420H1 enhances the phosphorylation and transcription of ERK1 in cancer cells. Oncotarget 2015, 6, 43162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viotti, M.; Wilson, C.; McCleland, M.; Koeppen, H.; Haley, B.; Jhunjhunwala, S.; Klijn, C.; Modrusan, Z.; Arnott, D.; Classon, M.; et al. SUV420H2 is an epigenetic regulator of epithelial/ mesenchymal states in pancreatic cancer. J. Cell Biol. 2018, 217, 763–777. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, Y.; Liu, M.; Nie, M.; Wang, Y.; Deng, Y.; Yao, B.; Gui, T.; Li, X.; Ma, L.; et al. Suv4-20h1 promotes G1 to S phase transition by downregulating p21WAF1/CIP1 expression in chronic myeloid leukemia K562 cells. Oncol. Lett. 2018, 15, 6123–6130. [Google Scholar] [CrossRef] [Green Version]
- Isin, H.; Özgür, E.; Talu, C.K.; Trabulus, D.C.; Karaçetin, D.; Gezer, U. Impact of histone methyltransferase suv420h2 in breast cancer. Biomed. Rep. 2020, 13, 29. [Google Scholar] [CrossRef]
- Weirich, S.; Kudithipudi, S.; Jeltsch, A. Specificity of the SUV4-20H1 and SUV4-20H2 protein lysine methyltransferases and methylation of novel substrates. J. Mol. Biol. 2016, 428, 2344–2358. [Google Scholar] [CrossRef]
- Gillberg, L.; Hjort, L. Epigenetics of metabolic diseases. In Handbook of Epigenetics: The New Molecular and Medical Genetics, 2nd ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 569–580. [Google Scholar] [CrossRef]
- Yang, Y.; Luan, Y.; Feng, Q.; Chen, X.; Qin, B.; Ren, K.-D.; Luan, Y. Epigenetics and Beyond: Targeting Histone Methylation to Treat Type 2 Diabetes Mellitus. Front. Pharmacol. 2021, 12, 807413. [Google Scholar] [CrossRef]
- Ringel, A.E.; Drijvers, J.M.; Baker, G.J.; Catozzi, A.; García-Cañaveras, J.C.; Gassaway, B.M.; Miller, B.C.; Juneja, V.R.; Nguyen, T.H.; Joshi, S.; et al. Obesity Shapes Metabolism in the Tumor Microenvironment to Suppress Anti-Tumor Immunity. Cell 2020, 183, 1848–1866.e26. [Google Scholar] [CrossRef]
- Gao, W.; Liu, J.-L.; Lu, X.; Yang, Q. Epigenetic regulation of energy metabolism in obesity. J. Mol. Cell Biol. 2021, 13, 480–499. [Google Scholar] [CrossRef]
- Burgio, E.; Lopomo, A.; Migliore, L. Obesity and diabetes: From genetics to epigenetics. Mol. Biol. Rep. 2015, 42, 799–818. [Google Scholar] [CrossRef]
- Pedrotti, S.; Caccia, R.; Neguembor, M.V.; Garcia-Manteiga, J.M.; Ferri, G.; De Palma, C.; Canu, T.; Giovarelli, M.; Marra, P.; Fiocchi, A.; et al. The Suv420h histone methyltransferases regulate PPAR-γ and energy expenditure in response to environmental stimuli. Sci. Adv. 2019, 5, eaav1472. [Google Scholar] [CrossRef] [Green Version]
- Twells, R.C.; Metzker, M.L.; Brown, S.D.; Cox, R.; Garey, C.; Hammond, H.; Hey, P.J.; Levy, E.; Nakagawa, Y.; Philips, M.S.; et al. The sequence and gene characterization of a 400-kb candidate region for IDDM4 on chromosome 11q13. Genomics 2001, 72, 231–242. [Google Scholar] [CrossRef]
- Zhong, Q.; Kowluru, R.A. Epigenetic changes in mitochondrial superoxide dismutase in the retina and the development of diabetic retinopathy. Diabetes 2011, 60, 1304–1313. [Google Scholar] [CrossRef] [Green Version]
- Cassidy, S.B.; Schwartz, S.; Miller, J.L.; Driscoll, D.J. Prader-Willi syndrome. Genet. Med. 2012, 14, 10–26. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.Y.; Tsai, T.F.; Beaudet, A.L. Deficiency of Rbbp1/Arid4a and Rbbp1l1/Arid4b alters epigenetic modifications and suppresses an imprinting defect in the PWS/AS domain. Genes Dev. 2006, 20, 2859–2870. [Google Scholar] [CrossRef] [Green Version]
- Miltenberger, R.J.; Mynatt, R.L.; Wilkinson, J.E.; Woychik, R.P. The role of the agouti gene in the yellow obese syndrome. J. Nutr. 1997, 127, 1902S–1907S. [Google Scholar] [CrossRef]
- Dolinoy, D.C.; Weidman, J.R.; Waterland, R.A.; Jirtle, R.L. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ. Health Perspect. 2006, 114, 567–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardikar, A.A.; Satoor, S.N.; Karandikar, M.S.; Joglekar, M.V.; Puranik, A.S.; Wong, W.; Kumar, S.; Limaye, A.; Bhat, D.S.; Januszewski, A.S.; et al. Multigenerational Undernutrition Increases Susceptibility to Obesity and Diabetes that Is Not Reversed after Dietary Recuperation. Cell Metab 2015, 22, 312–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Q.; Zhang, Z.; Rong, W.; Jin, W.; Yan, L.; Jin, W.; Xu, Y.; Cui, X.; Tang, Q.-Q.; Pan, D. KMT5c modulates adipocyte thermogenesis by regulating Trp53 expression. Proc. Natl. Acad. Sci. USA 2020, 117, 22413–22422. [Google Scholar] [CrossRef]
- Stessman, H.A.; Xiong, B.O.; Coe, B.P.; Wang, T.; Hoekzema, K.; Fenckova, M.; Kvarnung, M.; Gerdts, J.; Trinh, S.; Cosemans, N.; et al. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat. Genet. 2017, 49, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Wickramasekara, R.N.; Robertson, B.; Hulen, J.; Hallgren, J.; Stessman, H.A.F. Differential effects by sex with Kmt5b loss. Autism Res. 2021, 14, 1554–1571. [Google Scholar] [CrossRef]
- Wang, Z.-J.; Rein, B.; Zhong, P.; Williams, J.; Cao, Q.; Yang, F.; Zhang, F.; Ma, K.; Yan, Z. Autism risk gene KMT5B deficiency in prefrontal cortex induces synaptic dysfunction and social deficits via alterations of DNA repair and gene transcription. Neuropsychopharmacology 2021, 46, 1617–1626. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, B.; Velasco, S.; Kedaigle, A.J.; Pigoni, M.; Quadrato, G.; Deo, A.J.; Adiconis, X.; Uzquiano, A.; Sartore, R.; Yang, S.M.; et al. Autism genes converge on asynchronous development of shared neuron classes. Nature 2022, 602, 268–273. [Google Scholar] [CrossRef]
- Rhodes, C.T.; Sandstrom, R.S.; Huang, S.W.A.; Wang, Y.; Schotta, G.; Berger, M.S.; Lin, C.H.A. Cross-species Analyses Unravel the Complexity of H3K27me3 and H4K20me3 in the Context of Neural Stem Progenitor Cells. Neuroepigenetics 2016, 6, 10–25. [Google Scholar] [CrossRef] [Green Version]
- Bouton, M.E. Learning and Behavior: A Contemporary Synthesis; Sinauer Associates: Sunderland, MA, USA, 2007; ISBN 978-0-87893-063-0. [Google Scholar]
- Vilema-Enríquez, G.; Quinlan, R.; Kilfeather, P.; Mazzone, R.; Saqlain, S.; del Molino del Barrio, I.; Donato, A.; Corda, G.; Li, F.; Vedadi, M.; et al. Inhibition of the SUV4-20 H1 histone methyltransferase increases frataxin expression in friedreich’s ataxia patient cells. J. Biol. Chem. 2020, 295, 17973–17985. [Google Scholar] [CrossRef]
- Boonsanay, V.; Zhang, T.; Georgieva, A.; Kostin, S.; Qi, H.; Yuan, X.; Zhou, Y.; Braun, T. Regulation of Skeletal Muscle Stem Cell Quiescence by Suv4-20h1-Dependent Facultative Heterochromatin Formation. Cell Stem Cell 2016, 18, 229–242. [Google Scholar] [CrossRef] [Green Version]
- Tsang, L.W.K.; Hu, N.; Underhill, D.A. Comparative Analyses of SUV420H1 Isoforms and SUV420H2 Reveal Differences in Their Cellular Localization and Effects on Myogenic Differentiation. PLoS ONE 2010, 5, e14447. [Google Scholar] [CrossRef] [Green Version]
- Neguembor, M.V.; Xynos, A.; Onorati, M.C.; Caccia, R.; Bortolanza, S.; Godio, C.; Pistoni, M.; Corona, D.F.; Schotta, G.; Gabellini, D. FSHD muscular dystrophy region gene 1 binds Suv4-20h1 histone methyltransferase and impairs myogenesis. J. Mol. Cell Biol. 2013, 5, 294–307. [Google Scholar] [CrossRef] [Green Version]
- Bromberg, K.D.; Mitchell, T.R.H.; Upadhyay, A.K.; Jakob, C.G.; Jhala, M.A.; Comess, K.M.; Lasko, L.M.; Li, C.; Tuzon, C.T.; Dai, Y.; et al. The SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity. Nat. Chem. Biol. 2017, 13, 317–324. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gabellini, D.; Pedrotti, S. The SUV4-20H Histone Methyltransferases in Health and Disease. Int. J. Mol. Sci. 2022, 23, 4736. https://doi.org/10.3390/ijms23094736
Gabellini D, Pedrotti S. The SUV4-20H Histone Methyltransferases in Health and Disease. International Journal of Molecular Sciences. 2022; 23(9):4736. https://doi.org/10.3390/ijms23094736
Chicago/Turabian StyleGabellini, Davide, and Simona Pedrotti. 2022. "The SUV4-20H Histone Methyltransferases in Health and Disease" International Journal of Molecular Sciences 23, no. 9: 4736. https://doi.org/10.3390/ijms23094736
APA StyleGabellini, D., & Pedrotti, S. (2022). The SUV4-20H Histone Methyltransferases in Health and Disease. International Journal of Molecular Sciences, 23(9), 4736. https://doi.org/10.3390/ijms23094736





