Profiling of Transcriptome-Wide N6-Methyladenosine (m6A) Modifications and Identifying m6A Associated Regulation in Sperm Tail Formation in Anopheles sinensis
Abstract
:1. Introduction
2. Results
2.1. Conserved mRNA m6A Toolkit in Anopheles sinensis and Other Anopheles Species
2.2. Mapping of m6A Modification on mRNA in Male and Female Anopheles sinensis
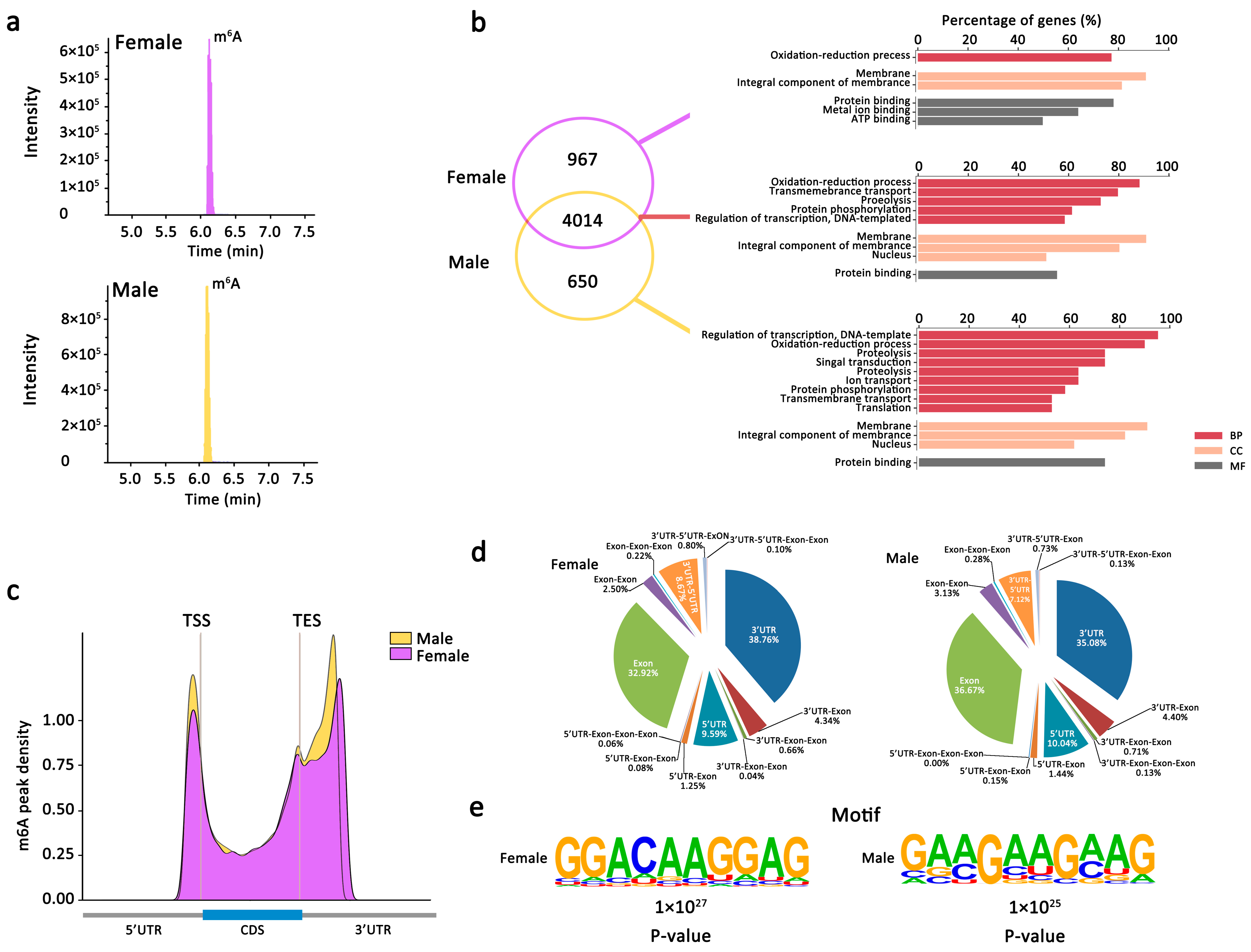
2.2.1. m6A Is Abundant and Conserved in Both Female and Male An. sinensis
2.2.2. m6A Peaks Positions in Female and Male An. sinensis
2.2.3. m6A Peaks Motifs in Female and Male An. sinensis
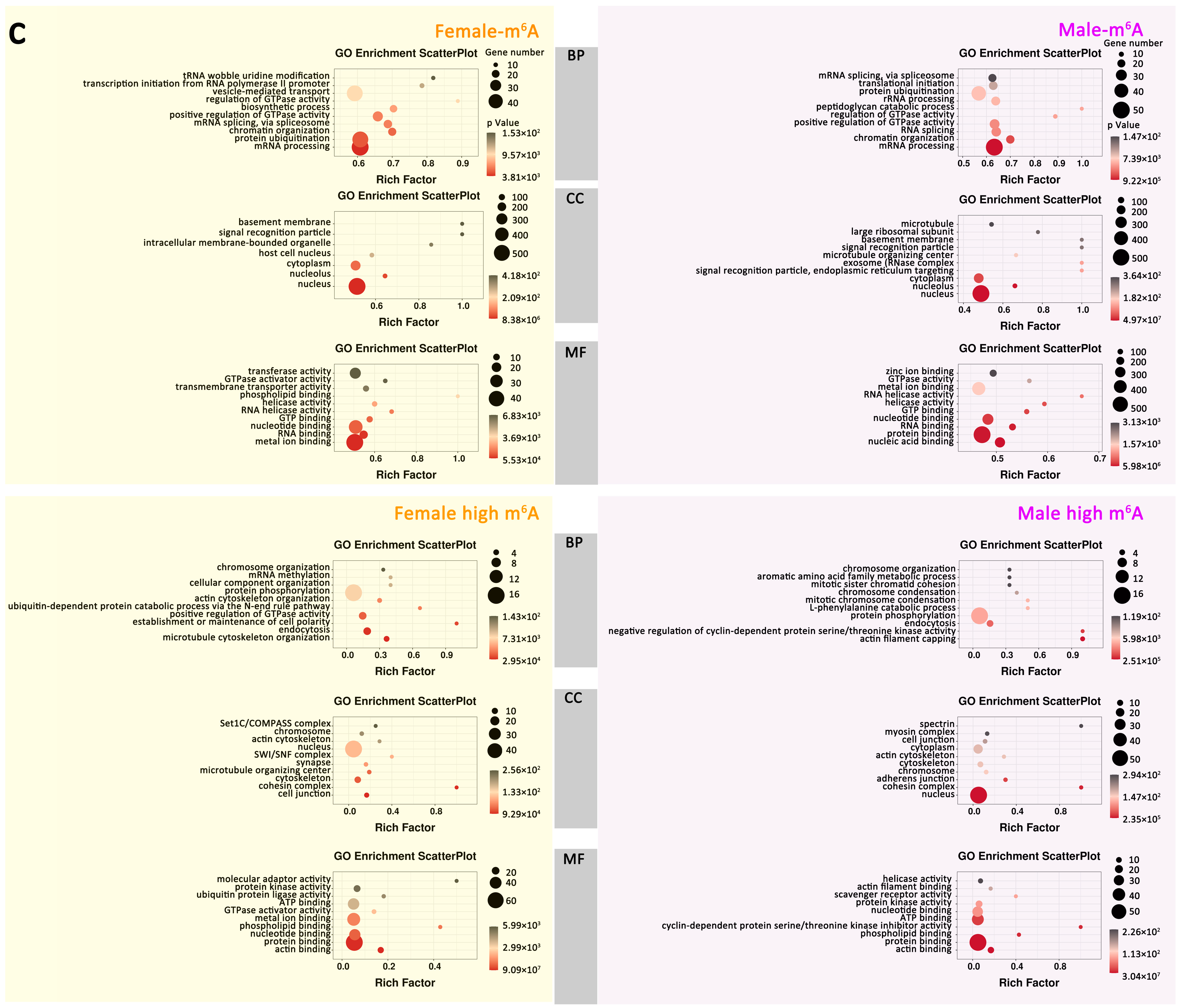
2.2.4. Gene Expression and Important Biological Pathways of m6A-Containing mRNAs Genes
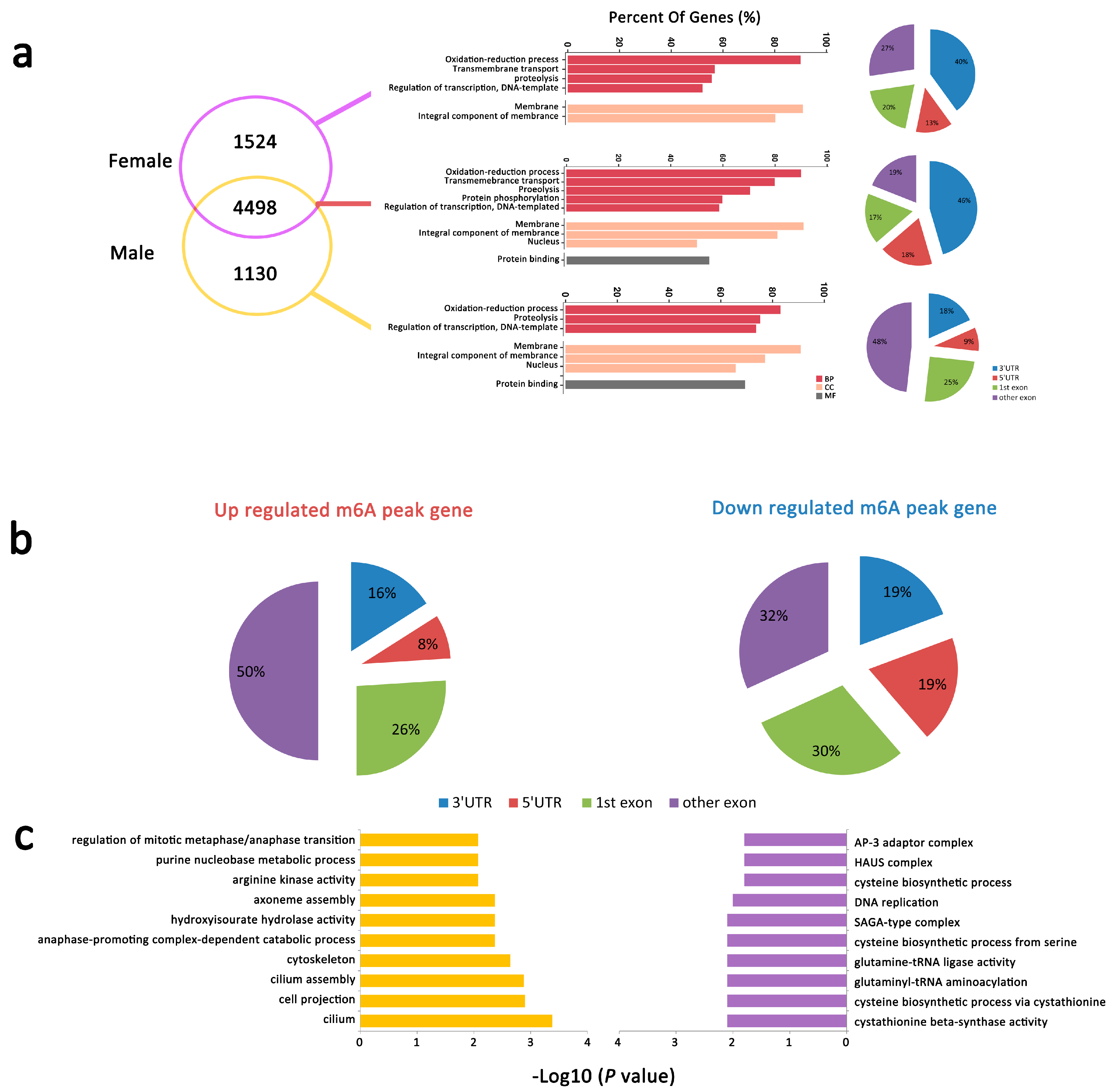
2.2.5. mRNA m6A Peaks with Different Abundance between Male and Female An. sinensis
2.3. Different Gene Expression in Male and Female An. sinensis
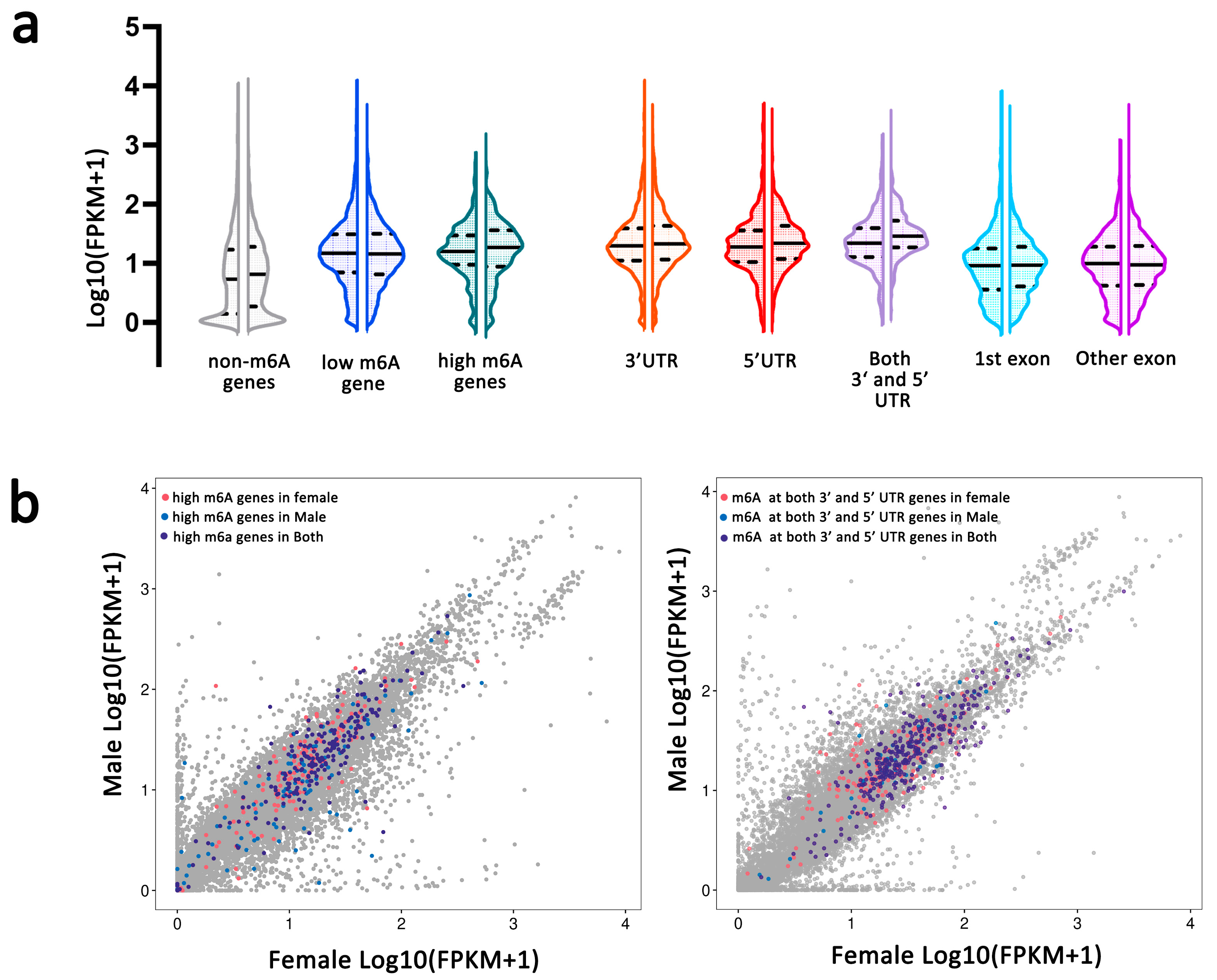
2.4. m6A Modification and Gene Expression in Female and Male An. sinensis
2.4.1. Correlation of m6A Modification and Gene Expression
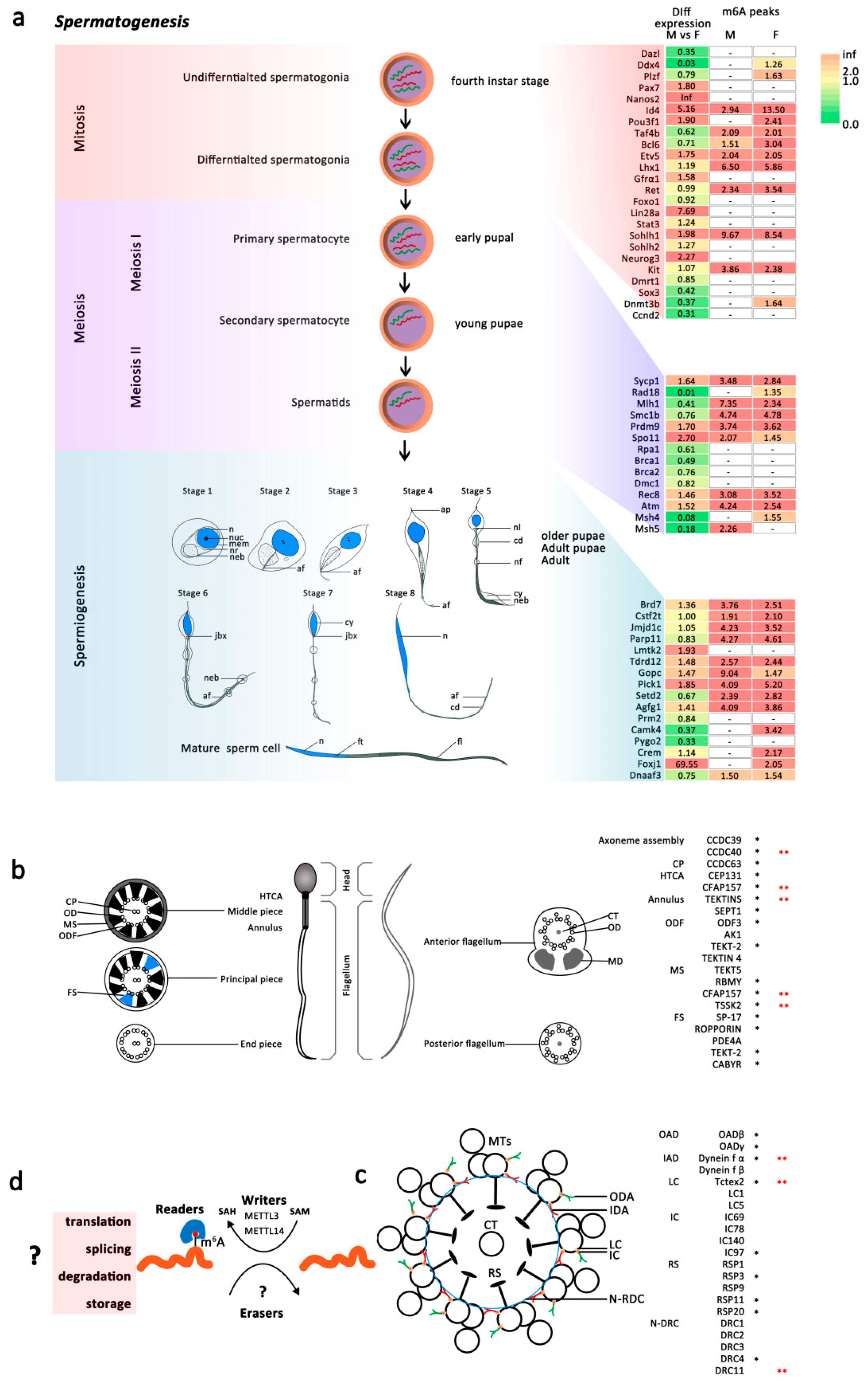
2.4.2. m6A Play Roles in Spermatogenesis Associated Genes in Male An. sinensis
m6A Associated with Spermatogenesis of Male An. sinensis
m6A in Mature Sperm of Male An. sinensis
3. Discussion
4. Materials and Methods
4.1. Mosquito Rearing and Sample Preparation
4.2. Phylogenetic Analysis of m6A Associated Proteins in Anopheles spp.
4.3. RNA Extraction and qPCR
4.4. Analysis of RNA Modification by Quantitative Mass Spectrometry
4.5. Mapping m6A Modifications in the Transcriptome
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Feng, J.; Tu, H.; Zhang, L.; Xia, Z.; Zhou, S. Imported Malaria Cases—China, 2012–2018. China CDC Wkly. 2020, 2, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Zhang, S.; Huang, F.; Zhang, L.; Feng, J.; Xia, Z.; Zhou, H.; Hu, W.; Zhou, S. Biology, Bionomics and Molecular Biology of Anopheles sinensis Wiedemann 1828 (Diptera: Culicidae), Main Malaria Vector in China. Front. Microbiol. 2017, 8, 1473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Guo, S.; Feng, X.; Afelt, A.; Frutos, R.; Zhou, S.; Manguin, S. Anopheles Vectors in Mainland China While Approaching Malaria Elimination. Trends Parasitol. 2017, 33, 889–900. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.F.; Qian, H.L.; Gu, Z.C.; Chang, X.; Wu, Z.Y.; Chen, F.Q.; Chen, Y.Z. The role of Anopheles lesteri anthropothagus in malaria transmission in Jianghuai region, Anhui. J. Parasitol. Parasit. Dis. 1984, 2, 216–219. [Google Scholar]
- Sun, Y.; Yu, D.; Chen, J.; Li, X.; Wang, B.; Wang, Z.; Mao, L.; Yao, W. Two individual incidences of vivax malaria in Dandong municipality of Liaoning province. Chin. J. Public Health 2017, 33, 314–316. [Google Scholar]
- Zhang, L.; Feng, J.; Xia, Z.; Zhou, S. Epidemiological characteristics of malaria and progress on its elimination in China in 2019. Chin. J. Parasitol. Parasit. Dis. 2020, 38, 133–138. [Google Scholar]
- Wang, D.; Li, S.; Cheng, Z.; Xiao, N.; Cotter, C.; Hwang, J.; Li, X.; Yin, S.; Wang, J.; Bai, L.; et al. Transmission Risk from Imported Plasmodium vivax Malaria in the China-Myanmar Border Region. Emerg. Infect. Dis. 2015, 21, 1861–1864. [Google Scholar] [CrossRef]
- Zhang, S.; Cheng, F.; Webber, R. A successful control programme for lymphatic filariasis in Hubei, China. Trans. R. Soc. Trop. Med. Hyg. 1994, 88, 510–512. [Google Scholar] [CrossRef]
- Manguin, S.; Bangs, M.J.; Pothikasikorn, J.; Chareonviriyaphap, T. Review on global co-transmission of human Plasmodium species and Wuchereria bancrofti by Anopheles mosquitoes. Infect. Genet. Evol. 2010, 10, 159–177. [Google Scholar] [CrossRef]
- Oliveira, A.; Strathe, E.; Etcheverry, L.; Cohnstaedt, L.W.; McVey, D.S.; Piaggio, J.; Cernicchiaro, N. Assessment of data on vector and host competence for Japanese encephalitis virus: A systematic review of the literature. Prev. Vet. Med. 2018, 154, 71–89. [Google Scholar] [CrossRef]
- Barua, S.; Hoque, M.M.; Kelly, P.J.; Poudel, A.; Adekanmbi, F.; Kalalah, A.; Yang, Y.; Wang, C. First report of Rickettsia felis in mosquitoes, USA. Emerg. Microbes. Infect. 2020, 9, 1008–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Dos-Santos, A.; Huang, W.; Liu, K.C.; Oshaghi, M.A.; Wei, G.; Agre, P.; Jacobs-Lorena, M. Driving mosquito refractoriness to Plasmodium falciparum with engineered symbiotic bacteria. Science 2017, 357, 1399–1402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glastad, K.M.; Hunt, B.G.; Goodisman, M. Epigenetics in Insects: Genome Regulation and the Generation of Phenotypic Diversity. Annu. Rev. Entomol. 2019, 64, 185–203. [Google Scholar] [CrossRef] [Green Version]
- Haussmann, I.U.; Bodi, Z.; Sanchez-Moran, E.; Mongan, N.P.; Archer, N.; Fray, R.G.; Soller, M. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 2016, 540, 301–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kan, L.; Grozhik, A.V.; Vedanayagam, J.; Patil, D.P.; Pang, N.; Lim, K.S.; Huang, Y.C.; Joseph, B.; Lin, C.J.; Despic, V.; et al. The m6A pathway facilitates sex determination in Drosophila. Nat. Commun. 2017, 8, 15737. [Google Scholar] [CrossRef] [PubMed]
- Lence, T.; Akhtar, J.; Bayer, M.; Schmid, K.; Spindler, L.; Ho, C.H.; Kreim, N.; Andrade-Navarro, M.A.; Poeck, B.; Helm, M.; et al. m6A modulates neuronal functions and sex determination in Drosophila. Nature 2016, 540, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Worpenberg, L.; Paolantoni, C.; Longhi, S.; Mulorz, M.M.; Lence, T.; Wessels, H.H.; Dassi, E.; Aiello, G.; Sutandy, F.; Scheibe, M.; et al. Ythdf is a N6-methyladenosine reader that modulates Fmr1 target mRNA selection and restricts axonal growth in Drosophila. EMBO J. 2021, 40, e104975. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Tang, H.W.; Li, J.; Perrimon, N.; Yan, D. Xio is a component of the Drosophila sex determination pathway and RNA N6-methyladenosine methyltransferase complex. Proc. Natl. Acad. Sci. USA 2018, 115, 3674–3679. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Xiao, Y.; Li, Y.; Wang, X.; Qi, S.; Wang, Y.; Zhao, L.; Wang, K.; Peng, W.; Luo, G.Z.; et al. RNA m6A Modification Functions in Larval Development and Caste Differentiation in Honeybee (Apis mellifera). Cell Rep. 2021, 34, 108580. [Google Scholar] [CrossRef]
- Rockwell, A.L.; Hongay, C.F. Dm Ime4 depletion affects permeability barrier and Chic function in Drosophila spermatogenesis. Mech. Dev. 2020, 164, 103650. [Google Scholar] [CrossRef]
- Yang, X.; Wei, X.; Yang, J.; Du, T.; Yin, C.; Fu, B.; Huang, M.; Liang, J.; Gong, P.; Liu, S.; et al. Epitranscriptomic regulation of insecticide resistance. Sci. Adv. 2021, 7, eabe5903. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Li, J.; Qian, P.; Xue, P.; Xu, J.; Chen, Y.; Zhu, J.; Tang, S.; Zhao, Q.; Qian, H.; et al. The role of N6-methyladenosine modification on diapause in silkworm (Bombyx mori) strains that exhibit different voltinism. Mol. Reprod. Dev. 2019, 86, 1981–1992. [Google Scholar] [CrossRef] [PubMed]
- Leismann, J.; Spagnuolo, M.; Pradhan, M.; Wacheul, L.; Vu, M.A.; Musheev, M.; Mier, P.; Andrade-Navarro, M.A.; Graille, M.; Niehrs, C.; et al. The 18S ribosomal RNA m6 A methyltransferase Mettl5 is required for normal walking behavior in Drosophila. EMBO Rep. 2020, 21, e49443. [Google Scholar] [CrossRef] [PubMed]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.Z.; MacQueen, A.; Zheng, G.; Duan, H.; Dore, L.C.; Lu, Z.; Liu, J.; Chen, K.; Jia, G.; Bergelson, J.; et al. Unique features of the m6A methylome in Arabidopsis thaliana. Nat. Commun. 2014, 5, 5630. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, S.; Agarwala, S.D.; Mumbach, M.R.; Jovanovic, M.; Mertins, P.; Shishkin, A.; Tabach, Y.; Mikkelsen, T.S.; Satija, R.; Ruvkun, G.; et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 2013, 155, 1409–1421. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Liu, B.; Nie, Z.; Duan, L.; Xiong, Q.; Jin, Z.; Yang, C.; Chen, Y. The role of m6A modification in the biological functions and diseases. Signal. Transduct. Target Ther. 2021, 6, 74. [Google Scholar] [CrossRef]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef] [Green Version]
- Lence, T.; Soller, M.; Roignant, J.Y. A fly view on the roles and mechanisms of the m6A mRNA modification and its players. RNA Biol. 2017, 14, 1232–1240. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Yang, Y.; Sun, B.F.; Shi, Y.; Yang, X.; Xiao, W.; Hao, Y.J.; Ping, X.L.; Chen, Y.S.; Wang, W.J.; et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014, 24, 1403–1419. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Klukovich, R.; Peng, H.; Wang, Z.; Yu, T.; Zhang, Y.; Zheng, H.; Klungland, A.; Yan, W. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3’-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. USA 2018, 115, E325–E333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, X.L.; He, S.Y.; Zhuang, X.Y.; Fan, Y.; Li, Y.H.; Yao, Y.G. mRNA expression and DNA methylation in three key genes involved in caste differentiation in female honeybees (Apis mellifera). Zool. Res. 2014, 35, 92–98. [Google Scholar] [CrossRef]
- Krafsur, E.S.; Jones, J.C. Spermiogenesis in Aedes aegypti (L.). Cytologia 1967, 32, 450–462. [Google Scholar] [CrossRef] [Green Version]
- Gui, Y.; Yuan, S. Epigenetic regulations in mammalian spermatogenesis: RNA-m6A modification and beyond. Cell. Mol. Life Sci. 2021, 78, 4893–4905. [Google Scholar] [CrossRef]
- Zhou, Y.; Kong, Y.; Fan, W.; Tao, T.; Xiao, Q.; Li, N.; Zhu, X. Principles of RNA methylation and their implications for biology and medicine. Biomed. Pharmacother. 2020, 131, 110731. [Google Scholar] [CrossRef]
- Xu, K.; Yang, Y.; Feng, G.H.; Sun, B.F.; Chen, J.Q.; Li, Y.F.; Chen, Y.S.; Zhang, X.X.; Wang, C.X.; Jiang, L.Y.; et al. Mettl3-mediated m6A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017, 27, 1100–1114. [Google Scholar] [CrossRef] [Green Version]
- Lin, Z.; Hsu, P.J.; Xing, X.; Fang, J.; Lu, Z.; Zou, Q.; Zhang, K.J.; Zhang, X.; Zhou, Y.; Zhang, T.; et al. Mettl3-/Mettl14-mediated mRNA N6-methyladenosine modulates murine spermatogenesis. Cell Res. 2017, 27, 1216–1230. [Google Scholar] [CrossRef]
- Hsu, P.J.; Zhu, Y.; Ma, H.; Guo, Y.; Shi, X.; Liu, Y.; Qi, M.; Lu, Z.; Shi, H.; Wang, J.; et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017, 27, 1115–1127. [Google Scholar] [CrossRef]
- Clements, A.N.; Potter, S.A. The fine structure of the spermathecae and their ducts in the mosquito Aedes aegypti. J. Insect Physiol. 1967, 13, 1825–1836. [Google Scholar] [CrossRef]
- Degner, E.C.; Harrington, L.C. A mosquito sperm’s journey from male ejaculate to egg: Mechanisms, molecules, and methods for exploration. Mol. Reprod. Dev. 2016, 83, 897–911. [Google Scholar] [CrossRef] [PubMed]
- Degner, E.C.; Ahmed-Braimah, Y.H.; Borziak, K.; Wolfner, M.F.; Harrington, L.C.; Dorus, S. Proteins, Transcripts, and Genetic Architecture of Seminal Fluid and Sperm in the Mosquito Aedes aegypti. Mol. Cell. Proteom. 2019, 18 (Suppl. S1), S6–S22. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zheng, H.; Esaki, Y.; Kelly, F.; Yan, W. Cullin3 is a KLHL10-interacting protein preferentially expressed during late spermiogenesis. Biol. Reprod. 2006, 74, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Gantz, V.M.; Jasinskiene, N.; Tatarenkova, O.; Fazekas, A.; Macias, V.M.; Bier, E.; James, A.A. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl. Acad. Sci. USA 2015, 112, E6736–E6743. [Google Scholar] [CrossRef] [Green Version]
- Hammond, A.; Galizi, R.; Kyrou, K.; Simoni, A.; Siniscalchi, C.; Katsanos, D.; Gribble, M.; Baker, D.; Marois, E.; Russell, S.; et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 2016, 34, 78–83. [Google Scholar] [CrossRef] [Green Version]
- Adelman, Z.N.; Tu, Z. Control of Mosquito-Borne Infectious Diseases: Sex and Gene Drive. Trends Parasitol. 2016, 32, 219–229. [Google Scholar] [CrossRef] [Green Version]
- Simoni, A.; Hammond, A.M.; Beaghton, A.K.; Galizi, R.; Taxiarchi, C.; Kyrou, K.; Meacci, D.; Gribble, M.; Morselli, G.; Burt, A.; et al. A male-biased sex-distorter gene drive for the human malaria vector Anopheles gambiae. Nat. Biotechnol. 2020, 38, 1054–1060. [Google Scholar] [CrossRef]
- Krzywinska, E.; Dennison, N.J.; Lycett, G.J.; Krzywinski, J. A maleness gene in the malaria mosquito Anopheles gambiae. Science 2016, 353, 67–69. [Google Scholar] [CrossRef] [Green Version]
- Sinkins, S.P. SEX DETERMINATION. Yob makes mosquitoes male. Science 2016, 353, 33–34. [Google Scholar] [CrossRef]
- Cator, L.J.; Wyer, C.; Harrington, L.C. Mosquito Sexual Selection and Reproductive Control Programs. Trends Parasitol. 2021, 37, 330–339. [Google Scholar] [CrossRef]
- Burki, F.; Roger, A.J.; Brown, M.W.; Simpson, A. The New Tree of Eukaryotes. Trends Ecol. Evol. 2020, 35, 43–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Wang, X.; Li, Z.; Lu, C.; Zhang, Q.; Chang, L.; Li, W.; Cheng, T.; Xia, Q.; Zhao, P. Transcriptome-wide analysis of N6-methyladenosine uncovers its regulatory role in gene expression in the lepidopteran Bombyx mori. Insect Mol. Biol. 2019, 28, 703–715. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Fu, L.; Kuang, L.; Chen, D.; Zhang, G.; Shen, Q.; Wu, D. Transcriptome-wide m6A methylation profile reveals regulatory networks in roots of barley under cadmium stress. J. Hazard. Mater. 2022, 423 Pt A, 127140. [Google Scholar] [CrossRef]
- Yang, Y.; Huang, W.; Huang, J.T.; Shen, F.; Xiong, J.; Yuan, E.F.; Qin, S.S.; Zhang, M.; Feng, Y.Q.; Yuan, B.F.; et al. Increased N6-methyladenosine in Human Sperm RNA as a Risk Factor for Asthenozoospermia. Sci. Rep. 2016, 6, 24345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, H.; Zhong, C.; Wu, X.; Chen, J.; Tao, B.; Xia, X.; Shi, M.; Zhu, Z.; Trudeau, V.L.; Hu, W. Mettl3 Mutation Disrupts Gamete Maturation and Reduces Fertility in Zebrafish. Genetics 2018, 208, 729–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [Green Version]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fu, Y.; Klungland, A.; Yang, Y.G.; He, C. Sprouts of RNA epigenetics: The discovery of mammalian RNA demethylases. RNA Biol. 2013, 10, 915–918. [Google Scholar] [CrossRef] [Green Version]
- Jain, D.; Puno, M.R.; Meydan, C.; Lailler, N.; Mason, C.E.; Lima, C.D.; Anderson, K.V.; Keeney, S. Ketu mutant mice uncover an essential meiotic function for the ancient RNA helicase YTHDC2. eLife 2018, 7, e30919. [Google Scholar] [CrossRef]
- Huang, T.; Liu, Z.; Zheng, Y.; Feng, T.; Gao, Q.; Zeng, W. YTHDF2 promotes spermagonial adhesion through modulating MMPs decay via m6A/mRNA pathway. Cell Death Dis. 2020, 11, 37. [Google Scholar] [CrossRef] [Green Version]
- Lin, Z.; Tong, M.H. m6A mRNA modification regulates mammalian spermatogenesis. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 403–411. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [Green Version]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [Green Version]
- Stöver, B.C.; Müller, K.F. TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformat. 2010, 11, 7. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [Green Version]
- Zhu, G.; Zhong, D.; Cao, J.; Zhou, H.; Li, J.; Liu, Y.; Bai, L.; Xu, S.; Wang, M.H.; Zhou, G.; et al. Transcriptome profiling of pyrethroid resistant and susceptible mosquitoes in the malaria vector, Anopheles sinensis. BMC Genom. 2014, 15, 448. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Meng, J.; Lu, Z.; Liu, H.; Zhang, L.; Zhang, S.; Chen, Y.; Rao, M.K.; Huang, Y. A protocol for RNA methylation differential analysis with MeRIP-Seq data and exomePeak R/Bioconductor package. Methods 2014, 69, 274–281. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.G.; He, Q.Y. ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31, 2382–2383. [Google Scholar] [CrossRef] [Green Version]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]


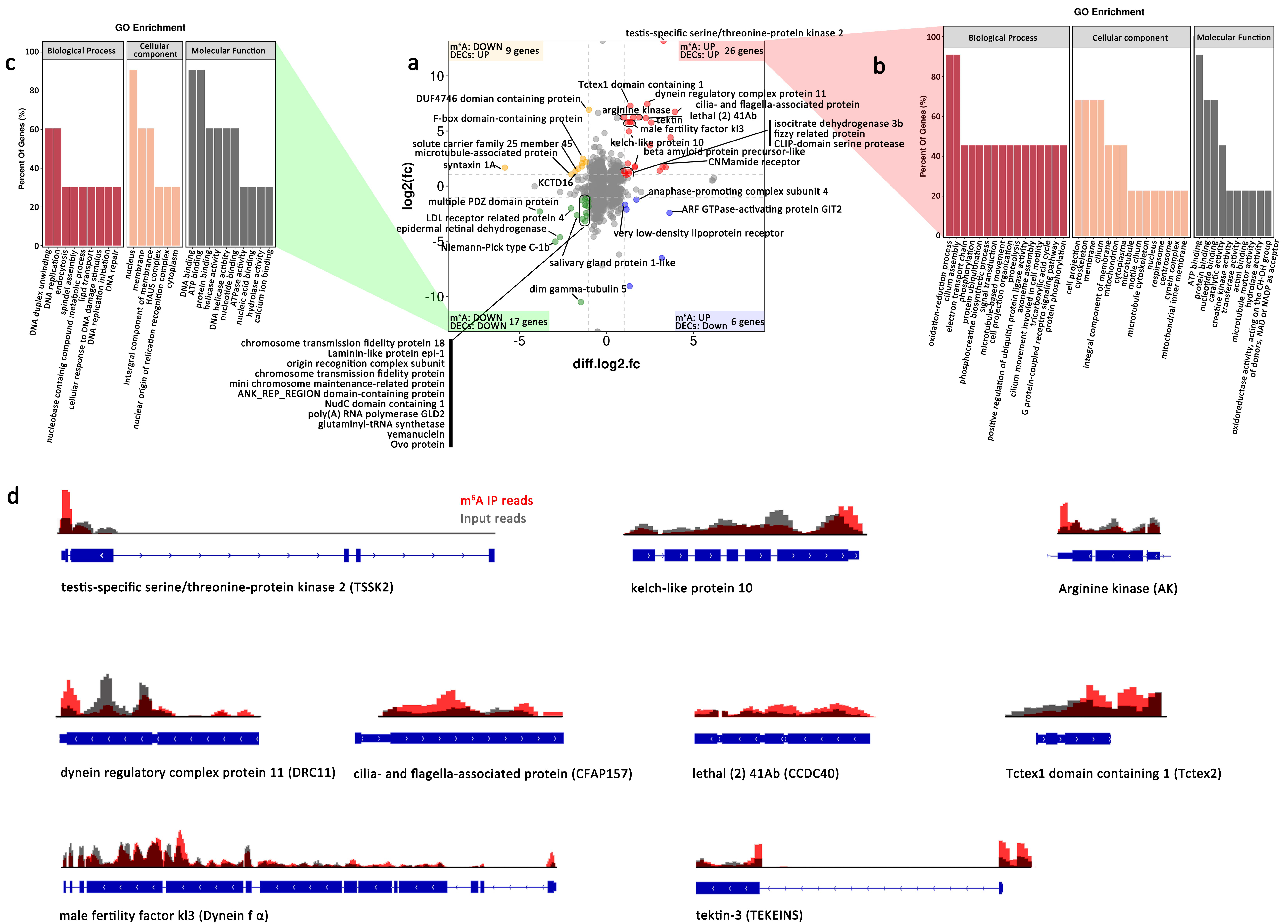
| Gene_ID | Description | m6A | Gene Expression | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Position | Diff. log2 (fc) | Diff. p_Value | Regulation | Position | log2 (fc) | p_Value | Regulation | Validated by qPCR | ||
| ASIS004369 | testis-specific serine/threonine-protein kinase 2 | 5′ UTR | 3.28 | 0.00 | up | 5′ UTR | inf | 0.00 | up | up |
| ASIS019512 | dynein regulatory complex protein 11 | 3′ UTR | 2.35 | 0.00 | up | 3′ UTR | 7.45 | 0.00 | up | |
| ASIS020833 | Tctex1 domain containing 1 | other Exon | 1.37 | 0.04 | up | other Exon | 7.28 | 0.00 | up | up |
| ASIS001560 | cilia- and flagella-associated protein | other Exon | 3.93 | 0.00 | up | other Exon | 6.73 | 0.00 | up | up |
| ASIS008463 | arginine kinase | other Exon | 1.89 | 0.01 | up | other Exon | 6.25 | 0.00 | up | up |
| other Exon | 1.54 | 0.03 | up | other Exon | 6.25 | 0.00 | up | up | ||
| 3′ UTR | 1.02 | 0.00 | up | 3′ UTR | 6.25 | 0.00 | up | up | ||
| ASIS009175 | lethal (2) 41Ab | 1st Exon | 2.27 | 0.00 | up | 1st Exon | 6.16 | 0.00 | up | up |
| ASIS021651 | male fertility factor kl3 | other Exon | 1.44 | 0.03 | up | other Exon | 5.79 | 0.00 | up | |
| other Exon | 1.35 | 0.04 | up | other Exon | 5.79 | 0.00 | up | |||
| other Exon | 1.21 | 0.01 | up | other Exon | 5.79 | 0.00 | up | |||
| ASIS021865 | tektin-3 | 5′ UTR | 2.58 | 0.00 | up | 5′ UTR | 5.76 | 0.00 | up | up |
| ASIS012597 | kelch-like protein 10 | 3′ UTR | 1.29 | 0.05 | up | 3′ UTR | 4.96 | 0.00 | up | up |
| ASIS012445 | unkown | 5′ UTR | 3.68 | 0.00 | up | 5′ UTR | 4.40 | 0.00 | up | |
| ASIS023324 | unkown | other Exon | 2.51 | 0.04 | up | other Exon | 3.70 | 0.00 | up | up |
| ASIS015071 | unkown | 3′ UTR | 1.23 | 0.00 | up | 3′ UTR | 2.05 | 0.00 | up | |
| ASIS016262 | beta amyloid protein precursor-like | 3′ UTR | 1.64 | 0.00 | up | 3′ UTR | 1.80 | 0.00 | up | |
| ASIS018187 | CNMamide receptor | 1st Exon | 3.22 | 0.01 | up | 1st Exon | 1.72 | 0.00 | up | |
| ASIS023203 | unkown | other Exon | 3.40 | 0.03 | up | other Exon | 1.72 | 0.01 | up | |
| ASIS007683 | unkown | other Exon | 1.63 | 0.00 | up | other Exon | 1.71 | 0.00 | up | |
| ASIS004699 | unkown | 1st Exon | 3.06 | 0.03 | up | 1st Exon | 1.39 | 0.00 | up | |
| ASIS000944 | Isocitte dehydrogenase [NAD] subunit, mitochondrial | 3′ UTR | 1.01 | 0.00 | up | 3′ UTR | 1.33 | 0.00 | up | |
| ASIS019305 | fizzy related protein | 1st Exon | 1.31 | 0.01 | up | 1st Exon | 1.28 | 0.00 | up | |
| ASIS013151 | unkown | 1st Exon | 1.05 | 0.01 | up | 1st Exon | 1.17 | 0.00 | up | |
| ASIS022379 | CLIP-domain serine protease | other Exon | 1.29 | 0.03 | up | other Exon | 1.13 | 0.01 | up | |
| ASIS014174 | unkown | 1st Exon | 1.13 | 0.01 | up | 1st Exon | 1.00 | 0.04 | up | |
| ASIS015991 | DUF4746 domain containing protein | other Exon | −1.01 | 0.05 | down | other Exon | 6.93 | 0.00 | up | up |
| ASIS017518 | F-box domain-containing protein | 1st Exon | −1.36 | 0.00 | down | 1st Exon | 2.48 | 0.00 | up | |
| ASIS022721 | unkown | other Exon | −1.18 | 0.00 | down | other Exon | 2.17 | 0.00 | up | |
| ASIS020661 | unkown | other Exon | −1.39 | 0.01 | down | other Exon | 2.06 | 0.00 | up | |
| ASIS022861 | unkown | 1st Exon | −1.40 | 0.00 | down | 1st Exon | 1.79 | 0.00 | up | |
| ASIS002904 | syntaxin 1A | 1st Exon | −5.84 | 0.00 | down | 1st Exon | 1.68 | 0.00 | up | up |
| ASIS003221 | solute carrier family 25 member 45 | 3′ UTR | −1.62 | 0.02 | down | 3′ UTR | 1.58 | 0.00 | up | |
| ASIS022100 | BTB/POZ domain-containing protein KCTD16 | 1st Exon | −1.78 | 0.00 | down | 1st Exon | 1.35 | 0.00 | up | |
| ASIS009765 | microtubule-associated protein | other Exon | −2.02 | 0.02 | down | other Exon | 1.06 | 0.00 | up | |
| ASIS007070 | anaphase-promoting complex subunit 4 | other Exon | 1.72 | 0.00 | up | other Exon | −1.23 | 0.00 | down | |
| ASIS004428 | very low-density lipoprotein receptor | 1st Exon | 1.07 | 0.00 | up | 1st Exon | −1.70 | 0.02 | down | |
| ASIS011700 | unkown | 3′ UTR | 1.15 | 0.00 | up | 3′ UTR | −2.12 | 0.00 | down | |
| ASIS022740 | ARF GTPase-activating protein GIT2 | 1st Exon | 3.62 | 0.00 | up | 1st Exon | −2.43 | 0.00 | down | |
| ASIS001538 | unkown | other Exon | 3.18 | 0.01 | up | other Exon | −6.53 | 0.00 | down | down |
| ASIS019082 | unkown | 3′ UTR | 1.33 | 0.01 | up | 3′ UTR | −9.07 | 0.00 | down | down |
| ASIS001385 | multiple PDZ domain protein | 1st Exon | −3.83 | 0.02 | down | 1st Exon | −2.32 | 0.00 | down | down |
| ASIS006653 | Niemann-Pick C1 protein | 1st Exon | −2.94 | 0.00 | down | 1st Exon | −5.05 | 0.00 | down | |
| ASIS000668 | epidermal retinal dehydrogenase | 3′ UTR | −2.67 | 0.02 | down | 3′ UTR | −4.65 | 0.00 | down | down |
| ASIS017710 | LDL receptor related protein 4, | 5′ UTR | −2.04 | 0.00 | down | 5′ UTR | −2.03 | 0.00 | down | |
| ASIS002714 | dim gamma-tubulin 5 | 1st Exon | −1.70 | 0.03 | down | 1st Exon | −2.64 | 0.00 | down | |
| ASIS004639 | salivary gland protein 1-like | 1st Exon | −1.47 | 0.00 | down | 1st Exon | −10.5 | 0.00 | down | down |
| ASIS008153 | chromosome transmission fidelity protein 18 | other Exon | −1.41 | 0.04 | down | other Exon | −1.26 | 0.00 | down | |
| ASIS010595 | Laminin-like protein epi-1 | 1st Exon | −1.37 | 0.01 | down | 1st Exon | −2.99 | 0.00 | down | |
| ASIS019764 | origin recognition complex subunit | 5′ UTR | −1.25 | 0.01 | down | 5′ UTR | −1.72 | 0.00 | down | |
| ASIS007143 | chromosome transmission fidelity protein | 1st Exon | −1.23 | 0.00 | down | 1st Exon | −3.20 | 0.00 | down | |
| ASIS001584 | mini chromosome maintenance-related protein | other Exon | −1.23 | 0.03 | down | other Exon | −1.16 | 0.00 | down | |
| ASIS005004 | ANK_REP_REGION domain-containing protein | 1st Exon | −1.18 | 0.04 | down | 1st Exon | −2.46 | 0.00 | down | |
| ASIS013445 | NudC domain containing 1 | 1st Exon | −1.16 | 0.00 | down | 1st Exon | −1.77 | 0.00 | down | |
| ASIS018863 | poly(A) RNA polymerase GLD2 | other Exon | −1.13 | 0.00 | down | other Exon | −2.70 | 0.00 | down | |
| ASIS020303 | glutaminyl-tRNA synthetase | 3′ UTR | −1.07 | 0.00 | down | 3′ UTR | −1.11 | 0.00 | down | |
| ASIS019128 | yemanuclein | 3′ UTR | −1.04 | 0.00 | down | 3′ UTR | −1.38 | 0.00 | down | |
| ASIS014406 | Ovo protein | 3′ UTR | −1.03 | 0.00 | down | 3′ UTR | −1.87 | 0.00 | down | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Cao, J.; Zhang, H.; Wu, J.; Yin, J. Profiling of Transcriptome-Wide N6-Methyladenosine (m6A) Modifications and Identifying m6A Associated Regulation in Sperm Tail Formation in Anopheles sinensis. Int. J. Mol. Sci. 2022, 23, 4630. https://doi.org/10.3390/ijms23094630
Liu C, Cao J, Zhang H, Wu J, Yin J. Profiling of Transcriptome-Wide N6-Methyladenosine (m6A) Modifications and Identifying m6A Associated Regulation in Sperm Tail Formation in Anopheles sinensis. International Journal of Molecular Sciences. 2022; 23(9):4630. https://doi.org/10.3390/ijms23094630
Chicago/Turabian StyleLiu, Congshan, Jianping Cao, Haobing Zhang, Jiatong Wu, and Jianhai Yin. 2022. "Profiling of Transcriptome-Wide N6-Methyladenosine (m6A) Modifications and Identifying m6A Associated Regulation in Sperm Tail Formation in Anopheles sinensis" International Journal of Molecular Sciences 23, no. 9: 4630. https://doi.org/10.3390/ijms23094630
APA StyleLiu, C., Cao, J., Zhang, H., Wu, J., & Yin, J. (2022). Profiling of Transcriptome-Wide N6-Methyladenosine (m6A) Modifications and Identifying m6A Associated Regulation in Sperm Tail Formation in Anopheles sinensis. International Journal of Molecular Sciences, 23(9), 4630. https://doi.org/10.3390/ijms23094630








