A Novel β-Hairpin Peptide Z-d14CFR Enhances Multidrug-Resistant Bacterial Clearance in a Murine Model of Mastitis
Abstract
:1. Introduction
2. Results
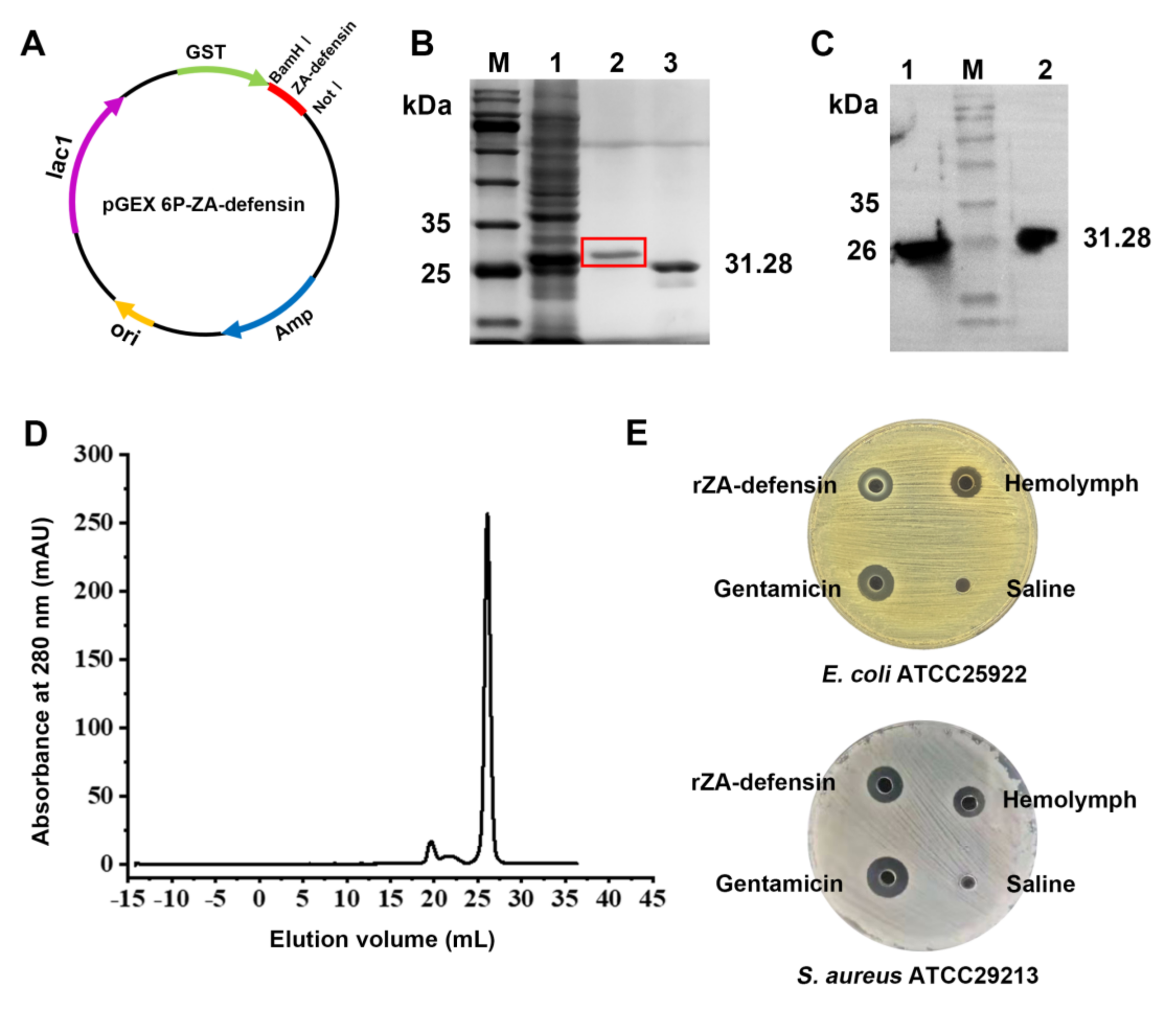
2.1. Recombinant Z. atratus Defensin Expression
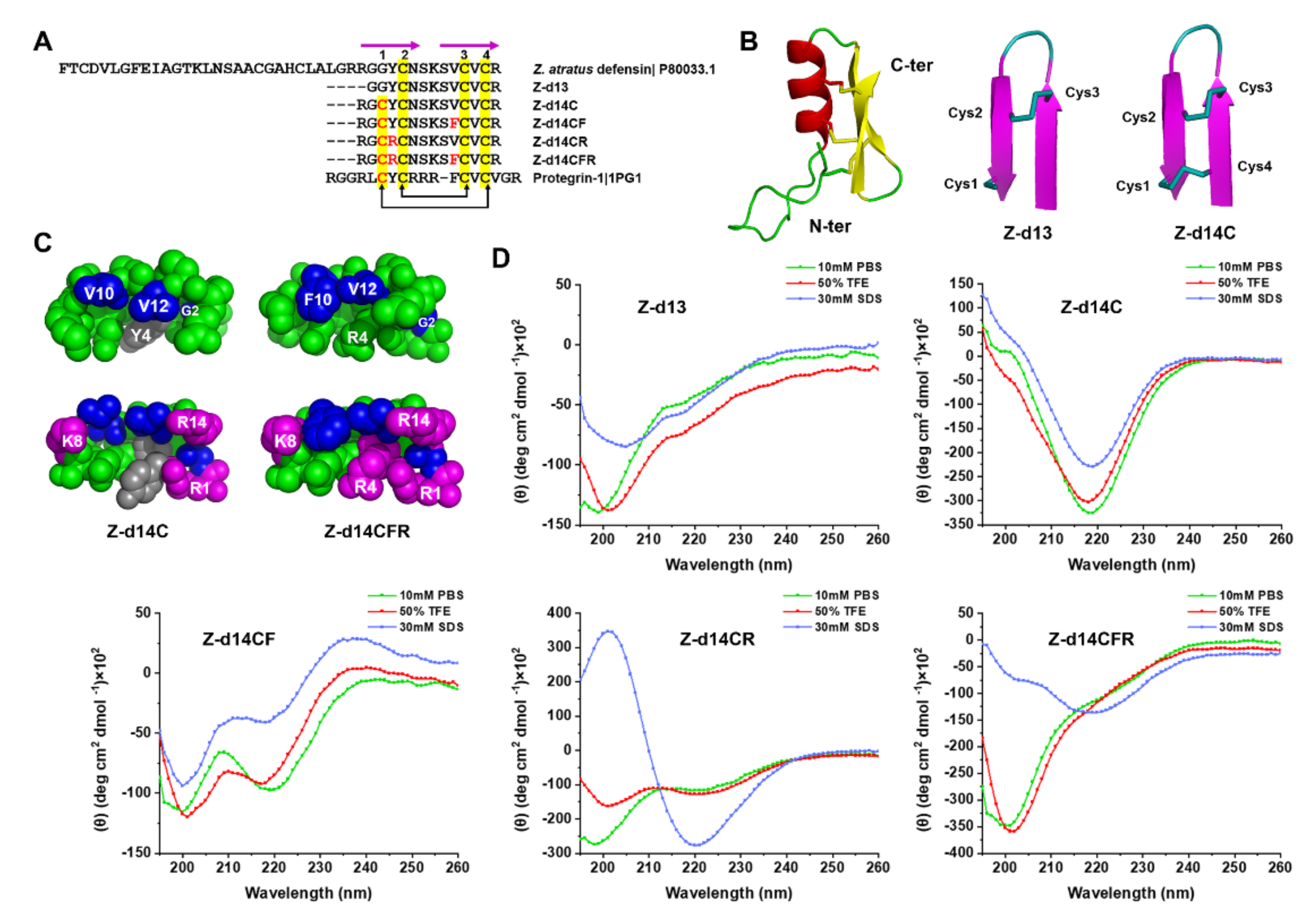
2.2. Peptide Design and Structural Analysis
2.3. Functional Screening of ZA-Defensin Analogues
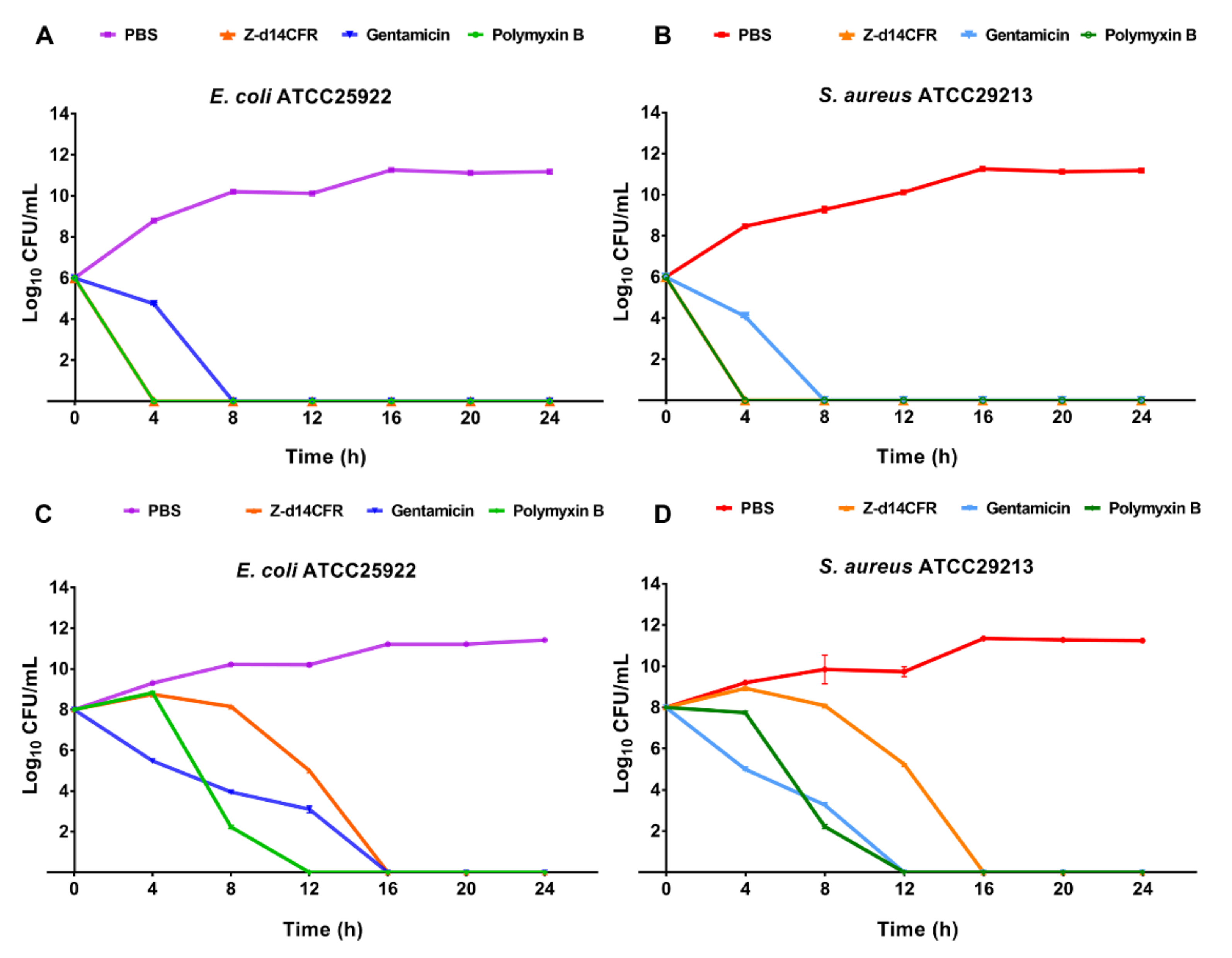
2.4. Time-Killing Curves of Z-d14CFR
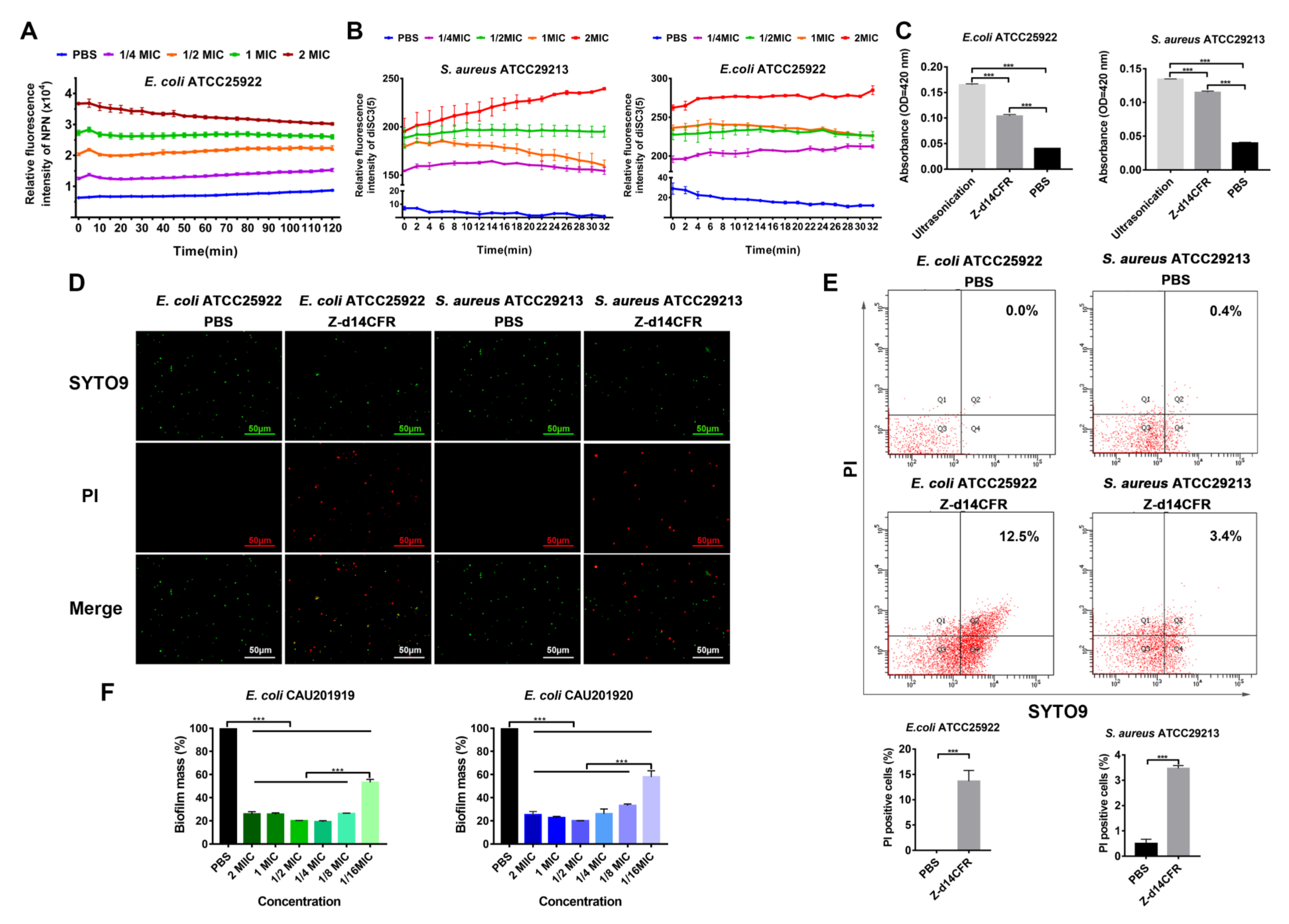
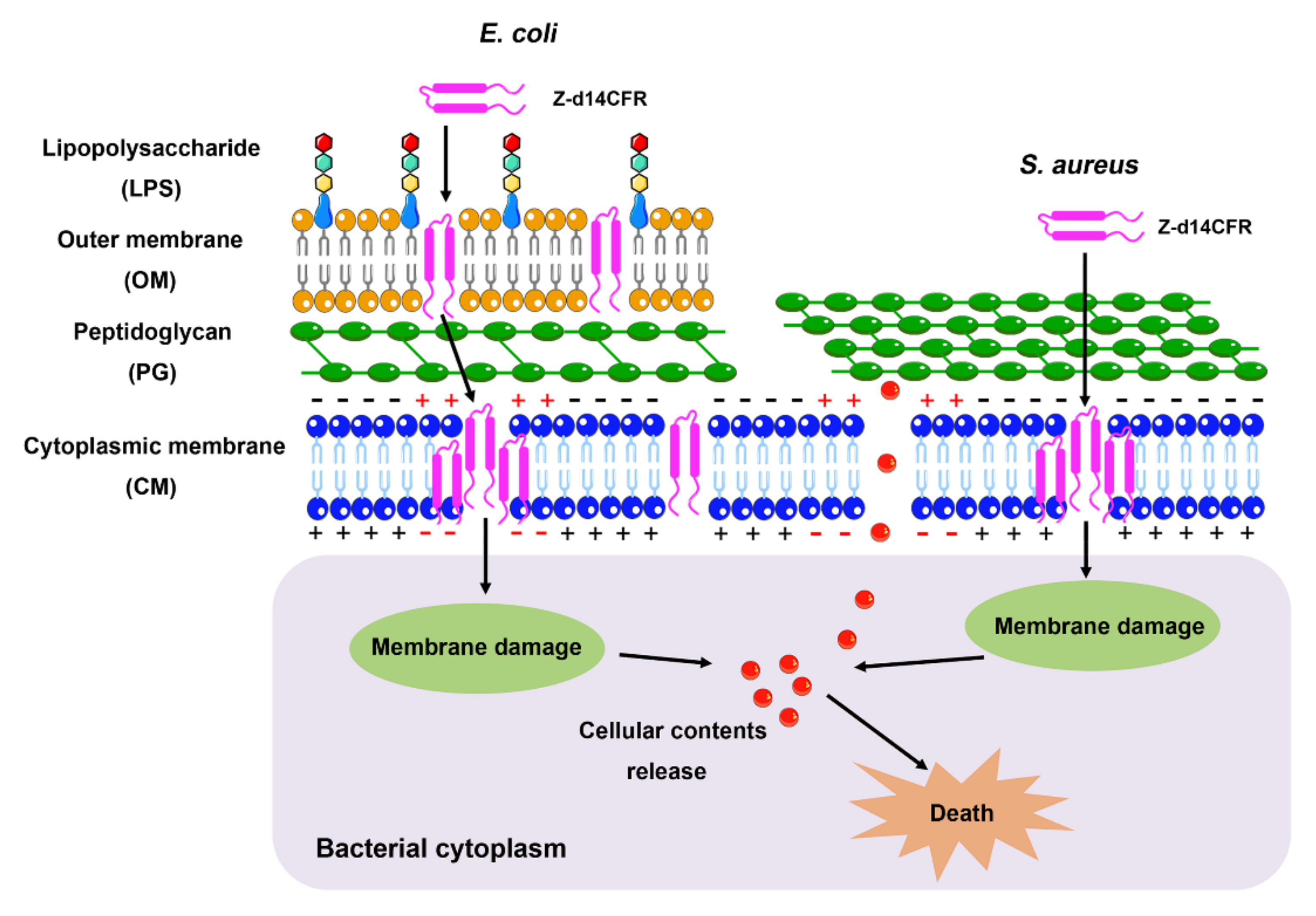
2.5. Z-d14CFR Destroyed Bacterial Membrane Integrity and Inhibited Biofilm Formation
2.6. Stability of Z-d14CFR
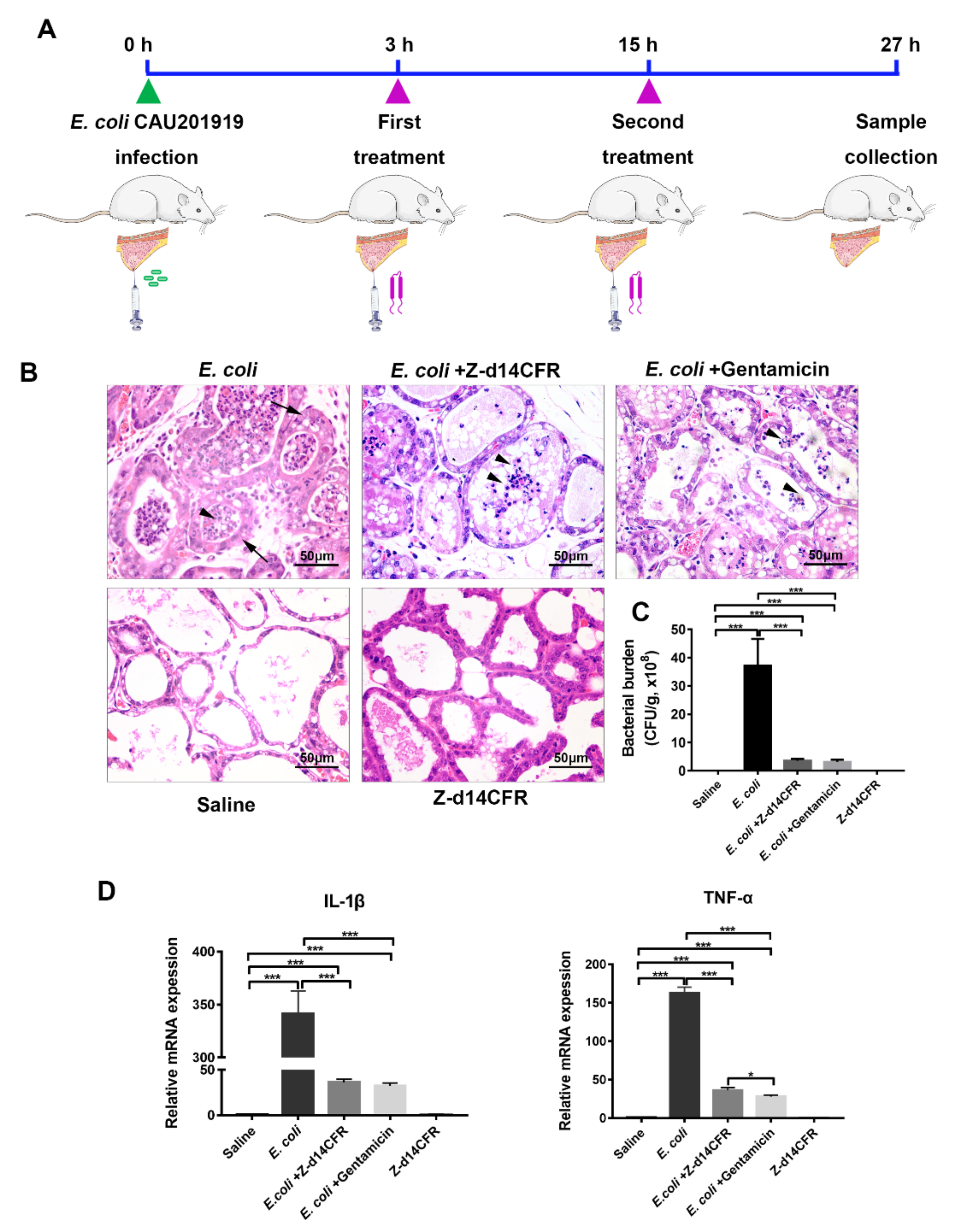
2.7. Z-d14CFR Enhanced Bacterial Clearance in a Murine Model of Mastitis
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Bacterial Strains and Cell Lines
4.3. Gene Amplification and Recombinant Protein Expression
4.4. Peptide Design and Synthesis
4.5. Circular Dichroism (CD) Spectroscopy
4.6. Antimicrobial Activity
4.7. Hemolysis
4.8. Cytotoxicity
4.9. Time-Killing Assays
4.10. Stability Assessment
4.11. Scanning Electron Microscopy (SEM)
4.12. Biofilm Inhibition Assays
4.13. Outer Membrane Permeability
4.14. Cytoplasmic Membrane Depolarization
4.15. Cytoplasmic Membrane Integrity
4.16. Extracellular β-Galactosidase Assay
4.17. Mouse Infection Models
4.18. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferri, M.; Ranucci, E.; Romagnoli, P.; Giaccone, V. Antimicrobial resistance: A global emerging threat to public health systems. Crit. Rev. Food Sci. Nutr. 2017, 57, 2857–2876. [Google Scholar] [CrossRef] [PubMed]
- Bush, K.; Courvalin, P.; Dantas, G.; Davies, J.; Eisenstein, B.; Huovinen, P.; Jacoby, G.A.; Kishony, R.; Kreiswirth, B.N.; Kutter, E.; et al. Tackling antibiotic resistance. Nat. Rev. Microbiol. 2011, 9, 894–896. [Google Scholar] [CrossRef] [PubMed]
- Tagliabue, A.; Rappuoli, R. Changing Priorities in Vaccinology: Antibiotic Resistance Moving to the Top. Front. Immunol. 2018, 9, 1068. [Google Scholar] [CrossRef] [PubMed]
- Roca, I.; Akova, M.; Baquero, F.; Carlet, J.; Cavaleri, M.; Coenen, S.; Cohen, J.; Findlay, D.; Gyssens, I.; Heuer, O.E.; et al. The global threat of antimicrobial resistance: Science for intervention. New Microbes New Infect. 2015, 6, 22–29. [Google Scholar] [CrossRef] [Green Version]
- Chernov, V.M.; Chernova, O.A.; Mouzykantov, A.A.; Lopukhov, L.L.; Aminov, R.I. Omics of antimicrobials and antimicrobial resistance. Expert Opin. Drug Discov. 2019, 14, 455–468. [Google Scholar] [CrossRef]
- Hanna, C.C.; Hermant, Y.O.; Harris, P.W.R. Discovery, Synthesis, and Optimization of Peptide-Based Antibiotics. Acc. Chem. Res. 2021, 54, 1878–1890. [Google Scholar] [CrossRef]
- Carratalá, J.V.; Serna, N.; Villaverde, A.; Vázquez, E.; Ferrer-Miralles, N. Nanostructured antimicrobial peptides: The last push towards clinics. Biotechnol. Adv. 2020, 44, 107603. [Google Scholar] [CrossRef]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef]
- Bechinger, B.; Gorr, S.U. Antimicrobial Peptides: Mechanisms of Action and Resistance. J. Dent. Res. 2017, 96, 254–260. [Google Scholar] [CrossRef] [Green Version]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility in vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, A. The big and small of drug discovery. Biotech versus pharma: Advantages and drawbacks in drug development. EMBO Rep. 2003, 4, 114–117. [Google Scholar] [CrossRef] [Green Version]
- El-Sayed Ahmed, M.A.E.; Zhong, L.L.; Shen, C. Colistin and its role in the Era of antibiotic resistance: An extended review (2000–2019). Emerg. Microbes Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazzaro, B.P.; Zasloff, M.; Rolff, J. Antimicrobial peptides: Application informed by evolution. Science 2020, 368, e5480. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, J.A.; Hetru, C. Insect defensins: Inducible antibacterial peptides. Immunol. Today 1992, 13, 411–415. [Google Scholar] [CrossRef]
- Wu, Q.; Patočka, J.; Kuča, K. Insect Antimicrobial Peptides, a Mini Review. Toxins 2018, 10, 461. [Google Scholar] [CrossRef]
- Gao, B.; Zhu, S. An insect defensin-derived β-hairpin peptide with enhanced antibacterial activity. ACS Chem. Biol. 2014, 9, 405–413. [Google Scholar] [CrossRef]
- Yamada, K.; Natori, S. Characterization of the antimicrobial peptide derived from sapecin B, an antibacterial protein of Sarcophaga peregrina (flesh fly). Biochem. J. 1994, 298, 623–628. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.H.; Hong, S.Y.; Oh, J.E.; Kwon, M.; Yoon, J.H.; Lee, J.; Lee, B.L.; Moon, H.M. Identification and characterization of the antimicrobial peptide corresponding to C-terminal beta-sheet domain of tenecin 1, an antibacterial protein of larvae of Tenebrio molitor. Biochem. J. 1998, 334, 99–105. [Google Scholar] [CrossRef]
- Ceřovský, V.; Slaninová, J.; Fučík, V.; Monincová, L.; Bednárová, L.; Maloň, P.; Stokrová, J. Lucifensin, a novel insect defensin of medicinal maggots: Synthesis and structural study. Chembiochem 2011, 12, 1352–1361. [Google Scholar] [CrossRef]
- Lazzaro, B.P.; Clark, A.G. Molecular population genetics of inducible antibacterial peptide genes in Drosophila melanogaster. Mol. Biol. Evol. 2003, 20, 914–923. [Google Scholar] [CrossRef] [PubMed]
- Li, S.X.; Liu, N.; Du, M.Z.; Zhu, Y.H. Therapeutic Effect of Darkling Beetle (Zophobas morio) Hemolymph on Skin Thermal Injury in Mice Infected by Staphylococcus haemolyticus. Vet. Sci. 2021, 8, 319. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Liu, X.; Xu, J. Antimicrobial Effect of Zophobas morio Hemolymph against Bovine Mastitis Pathogens. Microorganisms 2020, 8, 1488. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, D.A.; Hurst, M.A.; Fujii, C.A.; Kung, A.H.; Ho, J.F.; Cheng, F.C.; Loury, D.J.; Fiddes, J.C. Protegrin-1: A broad-spectrum, rapidly microbicidal peptide with in vivo activity. Antimicrob. Agents Chemother. 1997, 41, 1738–1742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, Y.; Hammer, J.; Pate, M.; Zhang, Y.; Zhu, F.; Zmuda, E.; Blazyk, J. Antimicrobial activities and structures of two linear cationic peptide families with various amphipathic beta-sheet and alpha-helical potentials. Antimicrob. Agents Chemother. 2005, 49, 4957–4964. [Google Scholar] [CrossRef] [Green Version]
- Laederach, A.; Andreotti, A.H.; Fulton, D.B. Solution and micelle-bound structures of tachyplesin I and its active aromatic linear derivatives. Biochemistry 2002, 41, 12359–12368. [Google Scholar] [CrossRef] [Green Version]
- Lynch, A.S.; Robertson, G.T. Bacterial and fungal biofilm infections. Annu. Rev. Med. 2008, 59, 415–428. [Google Scholar] [CrossRef]
- Rabin, N.; Zheng, Y.; Opoku-Temeng, C.; Du, Y.; Bonsu, E.; Sintim, H.O. Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Med. Chem. 2015, 7, 493–512. [Google Scholar] [CrossRef]
- Mandard, N.; Bulet, P.; Caille, A.; Daffre, S.; Vovelle, F. The solution structure of gomesin, an antimicrobial cysteine-rich peptide from the spider. Eur. J. Biochem. 2002, 269, 1190–1198. [Google Scholar] [CrossRef]
- Fahrner, R.L.; Dieckmann, T.; Harwig, S.S.; Lehrer, R.I.; Eisenberg, D.; Feigon, J. Solution structure of protegrin-1, a broad-spectrum antimicrobial peptide from porcine leukocytes. Chem. Biol. 1996, 3, 543–550. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Guarnieri, M.T.; Vasil, A.I.; Vasil, M.L.; Mant, C.T.; Hodges, R.S. Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides. Antimicrob. Agents Chemother. 2007, 51, 1398–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, L.M.; Edwards, M.A.; Li, J.; Yip, C.M.; Deber, C.M. Roles of hydrophobicity and charge distribution of cationic antimicrobial peptides in peptide-membrane interactions. J. Biol. Chem. 2012, 287, 7738–7745. [Google Scholar] [CrossRef] [Green Version]
- Oñate-Garzón, J.; Manrique-Moreno, M.; Trier, S.; Leidy, C.; Torres, R.; Patiño, E. Antimicrobial activity and interactions of cationic peptides derived from Galleria mellonella cecropin D-like peptide with model membranes. J. Antibiot. 2017, 70, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Frederiksen, N.; Hansen, P.R. Alternating Cationic-Hydrophobic Peptide/Peptoid Hybrids: Influence of Hydrophobicity on Antibacterial Activity and Cell Selectivity. ChemMedChem 2020, 15, 2544–2561. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Vasil, A.I.; Hale, J.D.; Hancock, R.E.; Vasil, M.L.; Hodges, R.S. Effects of net charge and the number of positively charged residues on the biological activity of amphipathic alpha-helical cationic antimicrobial peptides. Biopolymers 2008, 90, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.; Tian, H.; Wang, T.; Wang, Z.; Chou, S.; Shan, A.; Cheng, B. Central β-turn increases the cell selectivity of imperfectly amphipathic α-helical peptides. Acta Biomater. 2018, 69, 243–255. [Google Scholar] [CrossRef]
- Porter, E.A.; Weisblum, B.; Gellman, S.H. Mimicry of host-defense peptides by unnatural oligomers: Antimicrobial beta-peptides. J. Am. Chem. Soc. 2002, 124, 7324–7330. [Google Scholar] [CrossRef]
- Yan, J.; Bassler, B.L. Surviving as a Community: Antibiotic Tolerance and Persistence in Bacterial Biofilms. Cell Host Microbe 2019, 26, 15–21. [Google Scholar] [CrossRef]
- Tan, P.; Fu, H.; Ma, X. Design, optimization, and nanotechnology of antimicrobial peptides: From exploration to applications. Nano Today 2021, 39, 101229. [Google Scholar] [CrossRef]
- Malmsten, M. Interactions of Antimicrobial Peptides with Bacterial Membranes and Membrane Components. Curr. Top. Med. Chem. 2016, 16, 16–24. [Google Scholar] [CrossRef]
- Malanovic, N.; Lohner, K. Antimicrobial Peptides Targeting Gram-Positive Bacteria. Pharmaceuticals 2016, 9, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlieghe, P.; Lisowski, V.; Martinez, J.; Khrestchatisky, M. Synthetic therapeutic peptides: Science and market. Drug Discov. Today 2010, 15, 40–56. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Liu, Y.; Huang, X.; Ding, S.; Wang, Y. A broad-spectrum antibiotic adjuvant reverses multidrug-resistant Gram-negative pathogens. Nat. Microbiol. 2020, 5, 1040–1050. [Google Scholar] [CrossRef] [PubMed]


| Peptide | Sequence | Formula | MW a | Net Charge (+) | PI | RT (min) b |
|---|---|---|---|---|---|---|
| Z-d13 | GGYCNSKSVCVCR-NH2 | C54H91N19O17S3 | 1374.6 | 2 | 8.68 | 9.077 |
| Z-d14C | RGCYCNSKSVCVCR-NH2 | C61H105N23O18S4 | 1576.88 | 3 | 8.94 | 7.809 |
| Z-d14CF | RGCYCNSKSFCVCR-NH2 | C65H105N23O18S4 | 1624.92 | 3 | 8.94 | 9.538 |
| Z-d14CR | RGCRCNSKSVCVCR-NH2 | C58H108N26O17S4 | 1569.89 | 4 | 9.38 | 10.159 |
| Z-d14CFR | RGCRCNSKSFCVCR-NH2 | C62H108N26O17S4 | 1617.94 | 4 | 9.38 | 7.820 |
| Bacteria | rZA-Defensin | Z-d13 | Z-d14C | Z-d14CF | Z-d14CR | Z-d14CFR | Ampicillin | Gentamicin |
|---|---|---|---|---|---|---|---|---|
| Escherichia coli ATCC25922 | 0.5 (0.5) | NA (NA) c | 0.8 (0.8) | 0.4 (0.4) | 0.8 (0.8) | 0.1 (0.1) | 0.004 (0.016) | 0.004 (0.004) |
| Escherichia coli CVCC1450 | 0.5 (0.5) | NA (NA) | NA (NA) | 0.4 (0.4) | 0.8 (0.8) | 0.1 (0.1) | 0.008 (0.032) | 0.002 (0.004) |
| Escherichia coli CAU 201919 | 0.5 (0.5) | NA (NA) | NA (NA) | 0.4 (0.4) | 0.8 (0.8) | 0.1 (0.1) | NA (NA) | 0.04 (0.04) |
| Escherichia coli CAU 201920 | 0.5 (0.5) | NA (NA) | 0.8 (0.8) | 0.4 (0.4) | 0.8 (0.8) | 0.1 (0.1) | NA (NA) | 0.08 (0.08) |
| Salmonella typhimurium ATCC14028 | 0.5 (0.5) | NA (NA) | NA (NA) | 0.8 (0.8) | NA (NA) | 0.1 (0.1) | 0.004 (0.008) | 0.002 (0.002) |
| Klebsiella pneumoniae CAU202084 | 0.5 (0.5) | NA (NA) | NA (NA) | NA (NA) | NA (NA) | 0.2 (0.2) | NA (NA) | 0.001 (0.004) |
| Proteus vulgaris CVCC1971 | NA (NA) | NA (NA) | NA (NA) | NA (NA) | NA (NA) | NA (NA) | NA (NA) | 0.001 (0.004) |
| Staphylococcus aureus ATCC29213 | 0.5 (0.5) | NA (NA) | NA (NA) | 0.4 (0.4) | 0.8 (0.8) | 0.1 (0.1) | 0.002 (0.004) | 0.004 (0.004) |
| Staphylococcus haemolyticus CAU202078 | 1 (1) | NA (NA) | NA (NA) | 0.4 (0.4) | NA (NA) | 0.1 (0.1) | NA (NA) | 0.004 (0.004) |
| Bacillus cereus CAU 202020 | 1 (1) | NA (NA) | 0.4 (0.4) | 0.2 (0.2) | 0.1 (0.1) | 0.2 (0.2) | NA (NA) | 0.004 (0.032) |
| MRSAb ATCC33591 | NA (NA) | NA (NA) | NA (NA) | NA (NA) | NA (NA) | NA (NA) | NA (NA) | 0.004 (0.016) |
| Streptococcus suis CVCC3307 | NA (NA) | NA (NA) | NA (NA) | NA (NA) | NA (NA) | NA (NA) | 0.001 (0.002) | 0.004 (0.008) |
| E. coli ATCC25922 | S. aureus ATCC29213 | |
|---|---|---|
| 37 °C | 0.1 | 0.1 |
| 60 °C | 0.1 | 0.1 |
| 80 °C | 0.1 | 0.1 |
| 100 °C | 0.1 | 0.1 |
| 121 °C | 0.2 | 0.2 |
| pH = 2 | 0.2 | 0.2 |
| pH = 3 | 0.2 | 0.2 |
| pH = 4 | 0.1 | 0.1 |
| pH = 5 | 0.1 | 0.1 |
| pH = 6 | 0.1 | 0.1 |
| pH = 7 | 0.1 | 0.1 |
| pH = 8 | 0.1 | 0.1 |
| pH = 9 | 0.2 | 0.2 |
| pH = 10 | 0.2 | 0.2 |
| pH = 11 | 0.2 | 0.2 |
| pH = 12 | >2 | 0.2 |
| 10% FBS a | 0.2 | 0.2 |
| UV exposure | 0.1 | 0.1 |
| Pepsin | >2 | >2 |
| Trypsin | >2 | >2 |
| Papain | >2 | >2 |
| α-chymotrypsin | >2 | >2 |
| proteinase K | >2 | >2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Li, S.; Du, M.; Liu, N.; Shan, Q.; Zou, Y.; Wang, J.; Zhu, Y. A Novel β-Hairpin Peptide Z-d14CFR Enhances Multidrug-Resistant Bacterial Clearance in a Murine Model of Mastitis. Int. J. Mol. Sci. 2022, 23, 4617. https://doi.org/10.3390/ijms23094617
Wang X, Li S, Du M, Liu N, Shan Q, Zou Y, Wang J, Zhu Y. A Novel β-Hairpin Peptide Z-d14CFR Enhances Multidrug-Resistant Bacterial Clearance in a Murine Model of Mastitis. International Journal of Molecular Sciences. 2022; 23(9):4617. https://doi.org/10.3390/ijms23094617
Chicago/Turabian StyleWang, Xue, Shuxian Li, Mengze Du, Ning Liu, Qiang Shan, Yunjing Zou, Jiufeng Wang, and Yaohong Zhu. 2022. "A Novel β-Hairpin Peptide Z-d14CFR Enhances Multidrug-Resistant Bacterial Clearance in a Murine Model of Mastitis" International Journal of Molecular Sciences 23, no. 9: 4617. https://doi.org/10.3390/ijms23094617
APA StyleWang, X., Li, S., Du, M., Liu, N., Shan, Q., Zou, Y., Wang, J., & Zhu, Y. (2022). A Novel β-Hairpin Peptide Z-d14CFR Enhances Multidrug-Resistant Bacterial Clearance in a Murine Model of Mastitis. International Journal of Molecular Sciences, 23(9), 4617. https://doi.org/10.3390/ijms23094617






