NMR Characterization of Long-Chain Fatty Acylcarnitine Binding to the Mitochondrial Carnitine/Acylcarnitine Carrier
Abstract
1. Introduction
2. Results
2.1. An NMR-Feasible Sample of CAC
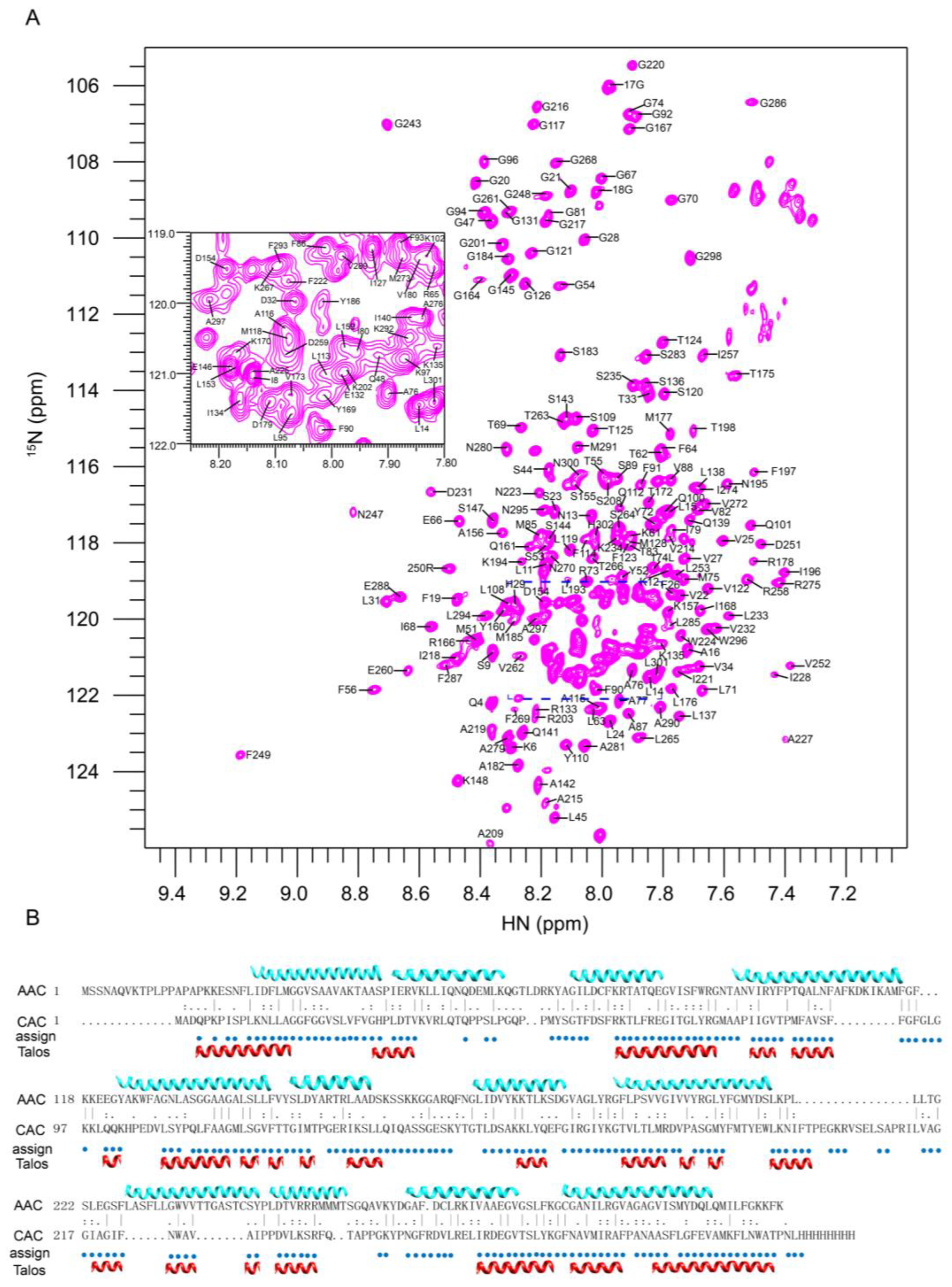
2.2. Resonance Assignment and Initial Structure Characterization of CAC
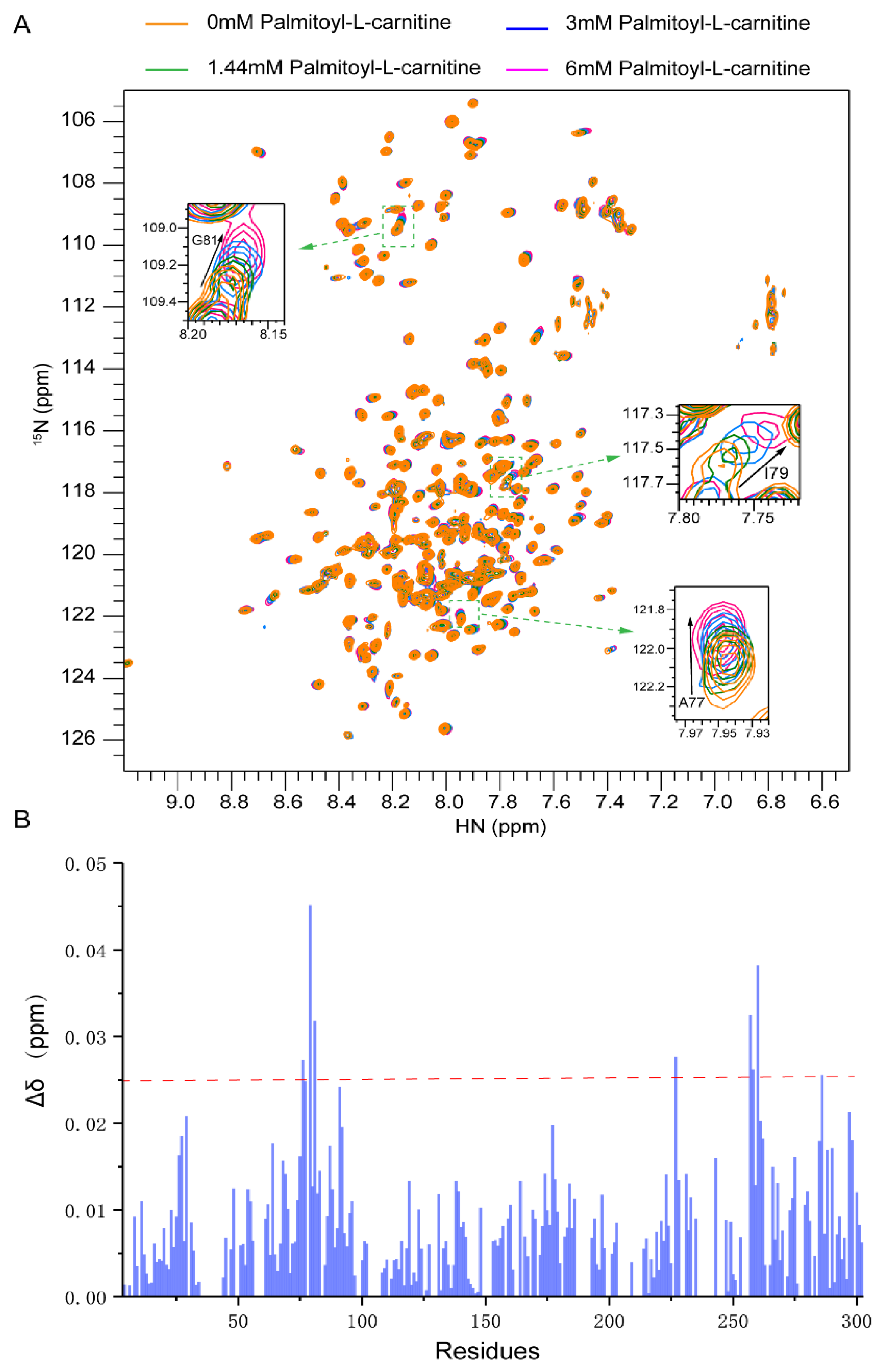
2.3. Substrate Binding to CAC from Chemical-Shift Titrations
2.4. Molecular Docking of Palmitoyl-L-Carnitine to CAC
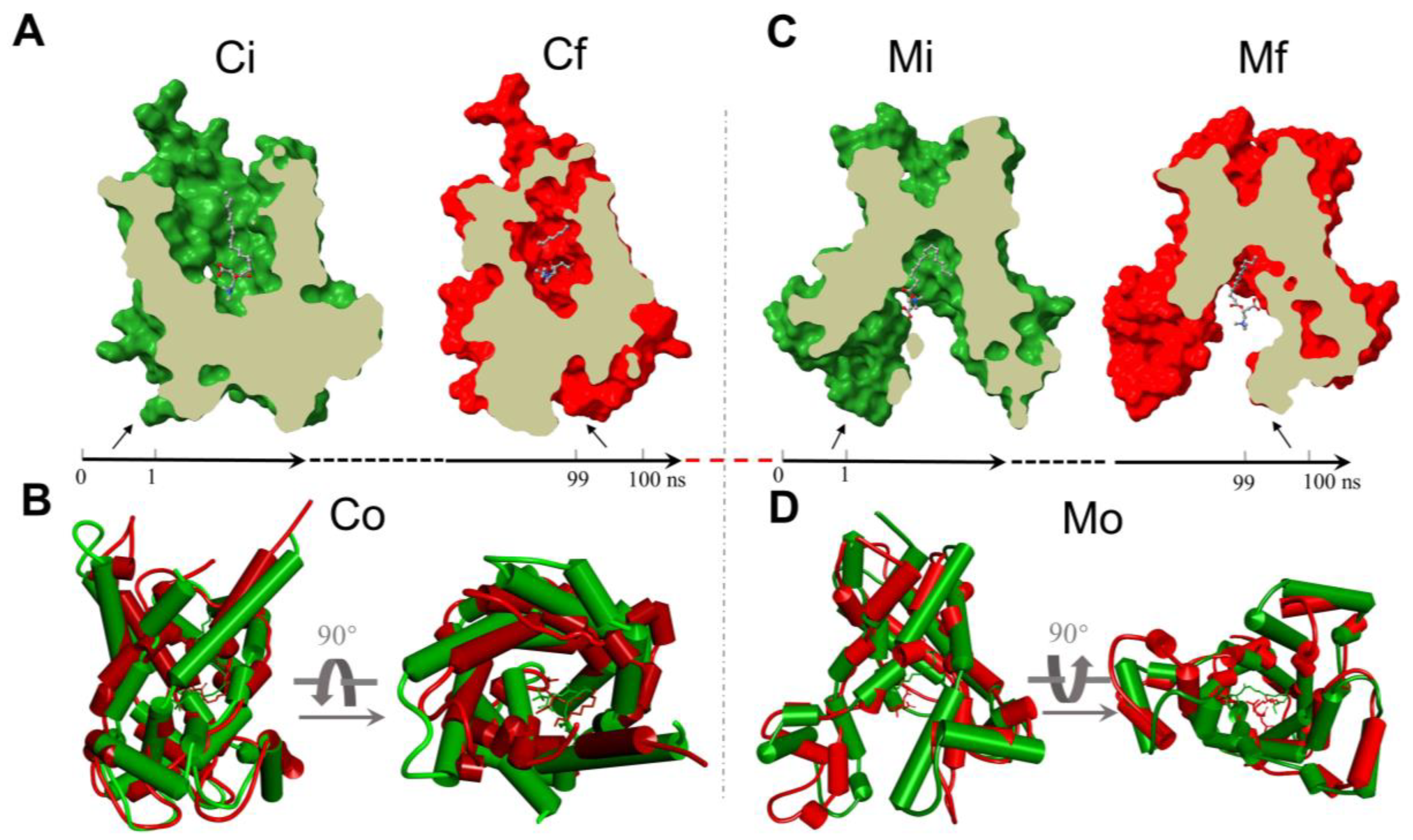
2.5. The Analysis of the MD Simulations of the Palmitoyl-L-Carnitine with CAC
3. Discussion
4. Materials and Methods
4.1. Protein Expression and Purification
4.2. NMR Resonance Assignment
4.3. NMR Titration Experiments
4.4. Protein Modeling
4.5. Molecular Docking
4.6. Molecular Dynamics Simulations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zammit, V.A.; Ramsay, R.R.; Bonomini, M.; Arduini, A. Carnitine, mitochondrial function and therapy. Adv. Drug Deliv. Rev. 2009, 61, 1353–1362. [Google Scholar] [CrossRef] [PubMed]
- Indiveri, C.; Iacobazzi, V.; Tonazzi, A.; Giangregorio, N.; Infantino, V.; Convertini, P.; Console, L.; Palmieri, F. The mitochondrial carnitine/acylcarnitine carrier: Function, structure and physiopathology. Mol. Aspects Med. 2011, 32, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Iacobazzi, V.; Convertini, P.; Infantino, V.; Scarcia, P.; Todisco, S.; Palmieri, F. Statins, fibrates and retinoic acid upregulate mitochondrial acylcarnitine carrier gene expression. Biochem. Biophys. Res. Commun. 2009, 388, 643–647. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, F.; Monne, M. Discoveries, metabolic roles and diseases of mitochondrial carriers: A review. Biochim. Biophys. Acta 2016, 1863, 2362–2378. [Google Scholar] [CrossRef]
- Kuan, J.; Saier, M.H., Jr. The mitochondrial carrier family of transport proteins: Structural, functional, and evolutionary relationships. Crit. Rev. Biochem. Mol. Biol. 1993, 28, 209–233. [Google Scholar] [CrossRef]
- Pebay-Peyroula, E.; Dahout-Gonzalez, C.; Kahn, R.; Trezeguet, V.; Lauquin, G.J.; Brandolin, G. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 2003, 426, 39–44. [Google Scholar] [CrossRef]
- Ruprecht, J.J.; Hellawell, A.M.; Harding, M.; Crichton, P.G.; McCoy, A.J.; Kunji, E.R. Structures of yeast mitochondrial ADP/ATP carriers support a domain-based alternating-access transport mechanism. Proc. Natl. Acad. Sci. USA 2014, 111, E426–E434. [Google Scholar] [CrossRef]
- Ruprecht, J.J.; King, M.S.; Zogg, T.; Aleksandrova, A.A.; Pardon, E.; Crichton, P.G.; Steyaert, J.; Kunji, E.R.S. The Molecular Mechanism of Transport by the Mitochondrial ADP/ATP Carrier. Cell 2019, 176, 435–447.e15. [Google Scholar] [CrossRef]
- Robinson, A.J.; Kunji, E.R. Mitochondrial carriers in the cytoplasmic state have a common substrate binding site. Proc. Natl. Acad. Sci. USA 2006, 103, 2617–2622. [Google Scholar] [CrossRef]
- Bruschweiler, S.; Yang, Q.; Run, C.; Chou, J.J. Substrate-modulated ADP/ATP-transporter dynamics revealed by NMR relaxation dispersion. Nat. Struct. Mol. Biol. 2015, 22, 636–641. [Google Scholar] [CrossRef]
- Tonazzi, A.; Giangregorio, N.; Indiveri, C.; Palmieri, F. Identification by site-directed mutagenesis and chemical modification of three vicinal cysteine residues in rat mitochondrial carnitine/acylcarnitine transporter. J. Biol. Chem. 2005, 280, 19607–19612. [Google Scholar] [CrossRef]
- Giangregorio, N.; Tonazzi, A.; Console, L.; Indiveri, C.; Palmieri, F. Site-directed mutagenesis of charged amino acids of the human mitochondrial carnitine/acylcarnitine carrier: Insight into the molecular mechanism of transport. Biochim. Biophys. Acta 2010, 1797, 839–845. [Google Scholar] [CrossRef]
- IJlst, L.; van Roermund, C.W.; Iacobazzi, V.; Oostheim, W.; Ruiter, J.P.; Williams, J.C.; Palmieri, F.; Wanders, R.J. Functional analysis of mutant human carnitine acylcarnitine translocases in yeast. Biochem. Biophys. Res. Commun. 2001, 280, 700–706. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, S.; Zhu, Q.; Wu, B.; Liu, Z.; OuYang, B.; Chou, J.J. Specific Interaction of the Human Mitochondrial Uncoupling Protein 1 with Free Long-Chain Fatty Acid. Structure 2017, 25, 1371–1379.e3. [Google Scholar] [CrossRef]
- Run, C.; Yang, Q.; Liu, Z.; OuYang, B.; Chou, J.J. Molecular Basis of MgATP Selectivity of the Mitochondrial SCaMC Carrier. Structure 2015, 23, 1394–1403. [Google Scholar] [CrossRef][Green Version]
- Shen, Y.; Delaglio, F.; Cornilescu, G.; Bax, A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 2009, 44, 213–223. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Dallakyan, S.; Olson, A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar] [CrossRef]
- Yeagle, P.L.; Albert, A.D. G-protein coupled receptor structure. Biochim. Biophys. Acta 2007, 1768, 808–824. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, S.; Run, C.; OuYang, B.; Chou, J.J. Specific Lipid Binding of Membrane Proteins in Detergent Micelles Characterized by NMR and Molecular Dynamics. Biochemistry 2016, 55, 5317–5320. [Google Scholar] [CrossRef]
- Tonazzi, A.; Console, L.; Giangregorio, N.; Indiveri, C.; Palmieri, F. Identification by site-directed mutagenesis of a hydrophobic binding site of the mitochondrial carnitine/acylcarnitine carrier involved in the interaction with acyl groups. Biochim. Biophys. Acta 2012, 1817, 697–704. [Google Scholar] [CrossRef]
- Wang, Y.; Tajkhorshid, E. Electrostatic funneling of substrate in mitochondrial inner membrane carriers. Proc. Natl. Acad. Sci. USA 2008, 105, 9598–9603. [Google Scholar] [CrossRef]
- Yao, S.; Ma, B.; Yi, Q.; Guan, M.X.; Cang, X. Investigating the Broad Matrix-Gate Network in the Mitochondrial ADP/ATP Carrier through Molecular Dynamics Simulations. Molecules 2022, 27, 1071. [Google Scholar] [CrossRef]
- Indiveri, C.; Iacobazzi, V.; Giangregorio, N.; Palmieri, F. Bacterial overexpression, purification, and reconstitution of the carnitine/acylcarnitine carrier from rat lliver mitochondria. Biochem. Biophys. Res. Commun. 1998, 249, 589–594. [Google Scholar] [CrossRef]
- Kay, L.E.; Torchia, D.A.; Bax, A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Biochemistry 1989, 28, 8972–8979. [Google Scholar] [CrossRef]
- Pervushin, K.; Riek, R.; Wider, G.; Wuthrich, K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. USA 1997, 94, 12366–12371. [Google Scholar] [CrossRef]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef]
- Bartels, C.; Xia, T.H.; Billeter, M.; Guntert, P.; Wuthrich, K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR 1995, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Vranken, W.F.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, M.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 2005, 59, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Gurbuz, B.B.; Yilmaz, D.Y.; Ozgul, R.K.; Kosukcu, C.; Dursun, A.; Sivri, H.S.; Coskun, T.; Tokatli, A. Clinical and molecular characteristics of carnitineacylcarnitine translocase deficiency with c.270delC and a novel c.408C>A variant. Turk. J. Pediatrics 2021, 63, 691–696. [Google Scholar] [CrossRef] [PubMed]
- Jason, B.D.S.B. Discovery Studio Modeling Environment, Release 4.0, San Diego: Dassault Systèmes. Available online: https://www.3dsbiovia.com/ (accessed on 15 October 2016).
- Shaw, D.E. A fast, scalable method for the parallel evaluation of distance-limited pairwise particle interactions. J. Comput. Chem. 2005, 26, 1318–1328. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Tirado-Rives, J. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J. Am. Chem. Soc. 1988, 110, 1657–1666. [Google Scholar] [CrossRef]
- Hoover, W.G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 1985, 31, 1695–1697. [Google Scholar] [CrossRef]
- Lippert, R.A.; Predescu, C.; Ierardi, D.J.; Mackenzie, K.M.; Eastwood, M.P.; Dror, R.O.; Shaw, D.E. Accurate and efficient integration for molecular dynamics simulations at constant temperature and pressure. J. Chem. Phys. 2013, 139, 164106. [Google Scholar] [CrossRef]
- Deng, Z.; Martyna, G.J.; Klein, M.L. Structure and dynamics of bipolarons in liquid ammonia. Phys. Rev. Lett. 1992, 68, 2496–2499. [Google Scholar] [CrossRef]
- Wang, H.; Dommert, F.; Holm, C. Optimizing working parameters of the smooth particle mesh Ewald algorithm in terms of accuracy and efficiency. J. Chem. Phys. 2010, 133, 034117. [Google Scholar] [CrossRef]
- Petersen, H.G. Accuracy and efficiency of the particle mesh Ewald method. J. Chem. Phys. 1995, 103. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, N.; Jia, X.; Fan, S.; Wu, B.; Wang, S.; OuYang, B. NMR Characterization of Long-Chain Fatty Acylcarnitine Binding to the Mitochondrial Carnitine/Acylcarnitine Carrier. Int. J. Mol. Sci. 2022, 23, 4608. https://doi.org/10.3390/ijms23094608
Zhang N, Jia X, Fan S, Wu B, Wang S, OuYang B. NMR Characterization of Long-Chain Fatty Acylcarnitine Binding to the Mitochondrial Carnitine/Acylcarnitine Carrier. International Journal of Molecular Sciences. 2022; 23(9):4608. https://doi.org/10.3390/ijms23094608
Chicago/Turabian StyleZhang, Ningning, Xiaopu Jia, Shuai Fan, Bin Wu, Shuqing Wang, and Bo OuYang. 2022. "NMR Characterization of Long-Chain Fatty Acylcarnitine Binding to the Mitochondrial Carnitine/Acylcarnitine Carrier" International Journal of Molecular Sciences 23, no. 9: 4608. https://doi.org/10.3390/ijms23094608
APA StyleZhang, N., Jia, X., Fan, S., Wu, B., Wang, S., & OuYang, B. (2022). NMR Characterization of Long-Chain Fatty Acylcarnitine Binding to the Mitochondrial Carnitine/Acylcarnitine Carrier. International Journal of Molecular Sciences, 23(9), 4608. https://doi.org/10.3390/ijms23094608





