Abstract
Glioblastoma stem cells (GSCs) are cells with a self-renewal ability and capacity to initiate tumors upon serial transplantation that have been linked to tumor cell heterogeneity. Most standard treatments fail to completely eradicate GSCs, causing the recurrence of the disease. GSCs could represent one reason for the low efficacy of cancer therapy and for the short relapse time. Nonetheless, experimental data suggest that the presence of therapy-resistant GSCs could explain tumor recurrence. Therefore, to effectively target GSCs, a comprehensive understanding of their biology and the survival and developing mechanisms during treatment is mandatory. This review provides an overview of the molecular features, microenvironment, detection, and targeting strategies of GSCs, an essential information required for an efficient therapy. Despite the outstanding results in oncology, researchers are still developing novel strategies, of which one could be targeting the GSCs present in the hypoxic regions and invasive edge of the glioblastoma.
1. Introduction
Glioblastoma (GBM) is the most common type of primary malignant brain tumor, accounting for 55% of primary brain tumors. GBM has a poor prognosis, with a mean survival rate between 14 to 16 months for patients treated with the standard treatment strategies (maximal safe surgical resection, followed by radiotherapy together with concomitant and adjuvant chemotherapy using temozolomide (TMZ)). Temozolomide is the first-line chemotherapy drug in GBM, an available orally alkylating agent that causes apoptosis by generating single-strand and double-strand breaks in DNA [1]. The 2-year survival rate was still less than 30%, even in the most recent reports.
GBM is a heterogeneous tumor resistant to all therapeutic approaches because of the multiple subclonal driver mutations [2,3]. Almost all tumors consist of various cell populations and heterogeneity that make them difficult to treat. The causes of recurrence are complex and include the absence of a clear tumor margin useful for a complete resection, the presence of migrating cancer cells into the surrounding normal tissue, high proliferative index, chemotherapy and radiotherapy resistance of the cancer stem cells (CSCs), and cerebrospinal fluid (CSF) dissemination. CSCs, also known as tumor-initiating cells (TICs), are suggested to be responsible for cancer relapse and drug resistance due to their ability to self-renew and to differentiate into a heterogeneous population of cancer cells [4,5].
Therefore, the surgical removal of the entire tumor mass is impossible. The presence of tumor cells at 2–3 cm from the original site of the tumor has led to even more aggressive tumoral recurrences after the removal of the tumor mass [6,7].
TICs and GSCs represent a subpopulation of self-renewing cells involved in the process of tumor initiation (for TICs) and tumor maintenance (for GSCs). Both types have distinct markers and biological functions and can be approached using different molecular and cellular methods. GSCs may be derived from TICs [8].
TICs (referred to as the cell of origin) are normal cells that acquire the first mutation that results in cancer promotion. The cells that promote GBM regrowth after surgical resection are supposed to be the undifferentiated GSCs, which produce differentiated progeny, creating a rapidly dividing tumor. They are also responsible for tumor heterogeneity and therapy resistance [9,10,11,12]. These cells demonstrate the main characteristics of stem cells: proliferation, differentiation, and self-renewal [9,13,14].
The GSCs’ resistance to radio- and chemotherapy could be explained by their high capacity for extensive DNA repair, quiescence, higher mitochondrial reserve, and localization in the hypoxic niche [9]. Mitochondrial activity is modulated by hypoxic conditions, controlling cell apoptosis and necroptosis, and reactive oxygen species (ROS) generation, therefore reducing susceptibility to chemo- and radio-induced apoptosis [15,16]. Moreover, anticancer therapies induce cell death by direct or indirect DNA damage. Targeting the DNA repairing pathways could increase the sensitivity of tumor to various cancer therapies [17]. Quiescence is a mechanism that turns the stem cells into a low metabolic state. The quiescence is correlated with the tumor microenvironment, including the extracellular matrix, immune cells, signaling molecules, and surrounding blood vessels. In GSCs, low ROS levels are associated with the quiescence/dormancy state of stem cells and with a protective intracellular environment [18].
According to the 2021 World Health Organization (WHO) Classification of Tumors of the Central Nervous System (CNS), GBMs are classified as “adult-type diffuse gliomas” (a grade IV tumor) [19]. The presence of extensive microvascular hyperplasia, hypercellularity, proliferation, necrotic foci surrounded by pseudopalisading cells and diffuse infiltrative margins differentiates them from low-grade gliomas [20].
Additionally, immunohistochemistry (IHC) is considered a fast and reliable substitute for molecular genetic tests in diagnostic surgical pathology. A narrow panel of highly sensitive immunohistochemical biomarkers can predict with a high accuracy the GBM type. For this matter, based on the IHC expression of four proteins (CD44, p53, IDH1, and PDGFRA), the study on gliomas published by Jakovlevs et al. indicated the presence of two mutually exclusive molecular signatures similar to proneural and mesenchymal subtypes [21].
Furthermore, integrated algorithms based on IHC can offer high accuracy in predicting glioblastoma transcriptional subtypes. In a study published by Orzan et al., mesenchymal and classical subgroups proved to be well segregated, and the proneural types showed a mixed proneural/classical phenotype, predicted by the algorithm as proneural, but with comparable probability to be a part of a classical subtype [22].
The tissue is protected from harmful molecules by the existence of the blood–brain barrier (BBB) [23]. This is formed by astrocytes and pericytes that surround the endothelial cell tight junctions [24]. Due to the existence of this barrier, the delivery of effective drug concentration is sometimes difficult, as it regulates the extravasation of macromolecules and chemotherapy drugs [24]. Because of the high genetic heterogeneity [25,26,27], it is impossible to target all GBM cells using only one biomarker-targeted therapy. Most of the GSCs are found in the hypoxic and necrotic areas, which are difficult to penetrate by the chemotherapy drugs. Furthermore, their resistance to chemotherapy is facilitated by various mechanisms, such as drug metabolic inactivation, decreased drug influx, overexpression of drug efflux pumps, slow division rate, inhibition of prodrug to bioactive drug conversion, and increased double-strand DNA repair [9,28,29,30]. The mechanism of the drug efflux is energy dependent, and it is facilitated by the increased expression of the ATP-binding cassette superfamily (ABC) of transporters [31], which are overexpressed in GSCs [32].
Aldehyde dehydrogenase 1 (ALDH), an enzyme that detoxifies alkylating agents, reducing their reactivity, and O6-methylguanine-DNA methyltransferase (MGMT), a detoxifier enzyme, are both contributors to GBM chemoresistance and highly expressed by GSCs [33,34,35].
ALDHs are markers of CSCs indicating worse prognosis in GBM [36]. ALDHs have not been proved to be linked to DNA repairing pathways, and the mechanism of how they mediate chemoresistance remains unclear. Clinical data reported increased levels of ALDH1A3 in recurrent GBM tumors [35]. It has been proved that ALDH1A3 is involved in ROS reaction. ROS react with polyunsaturated fatty acids from lipid membranes, inducing lipid peroxidation. ALDH1A3 seems to reduce the extent of toxic aldehydes resulting from lipid peroxidation [37].
Nevertheless, MGMT is directly responsible for the repair of lesion. Epigenetic silencing of the MGMT gene inhibits the synthesis of MGMT, increasing the sensitivity to cytotoxic effects induced by alkylating compounds. MGMT methylation is a predictor of longer survival for diagnosis but not for recurrence, suggesting that other mechanisms are responsible for MGMT upregulation in recurrent tumors [38]. Metastatic mismatch repair gene (MMR) alterations have been described in 10% to 20% of recurrent tumors, but changes in its promoter methylation status have been detected in a few patients [3]. It has been suggested that enhancer hijacking in recurrent GBMs could promote the MGMT expression and, therefore, alkylating compounds’ resistance, but this clinical significance remains to be evaluated [38,39].
Nonetheless, it has been demonstrated that radiotherapy is also insufficiently effective in destroying the GSCs [40,41]. Because of their high resistance to drugs, radiation, and surgery, the GSCs represent an important therapeutic target, intensively studied worldwide in the last years [41].
2. GSCs’ Biomarkers
In order to achieve an optimum treatment efficacy, it is important to identify the GSCs from the rest of the tumoral cells. Glioblastoma stem cells present several biomarkers used for identification, such as CD133, nestin, musashi-1, CXCR4, CD15, CD34, CD44, SOX2, L1CAM, and A2B5, however, neither being exclusively characteristic for GSCs [42,43,44,45,46,47,48,49,50,51,52,53,54] (Table 1). Furthermore, this task becomes even more difficult because GBMs have the ability to remodel their microenvironment by modulating the immune system, vasculature, and stroma [44].

Table 1.
Biomarkers of GSCs.
3. GSCs and Tumor Microenvironment
Even though several hypotheses have been proposed, none can fully explain the origin of the GBM. These hypotheses are based on the dedifferentiation of neural cells, transformation of undifferentiated precursor cells, and proliferation of neural stem cells [55,56,57]. Campos et al. stated that dedifferentiation may occur when an accumulation of genetic mutations in oncogenes appears in normal brain cells [58]. Additionally, genetic mutations in neural stem cells may cause the formation of cancer cells.
It seems that GSCs increase tumorigenesis by recapitulating the normal neural lineage hierarchies of quiescence and self-renewal. By maintaining stemness in specialized niches and by directing differentiation on the appropriate lineages, the microenvironment is controlling the normal neural stem cells’ fate. While GSCs are found in hypoxic and perivascular regions of the tumor bulk, not much is known about the GSCs’ differentiation potential within tumors and neither whether a prodifferentiative niche exists [59].
GBM may develop in the white matter and spread inside the brain via myelinated fibers, but the cellular and molecular processes that support the white matter invasion remain unknown. However, there are studies that revealed that the white matter may suppress malignancy by directing the differentiation of GSCs towards preoligodendrocyte fate [60].
Neural stem cells have two places of origin (also called neurogenic niches): the subventricular zone, located in the forebrain lateral ventricle, and the subgranular zone, located in the hippocampus in the dentate gyrus. Stem cells in quiescent or active mitotic state can be found in both regions [61].
Three major microenvironments have been described in glioblastoma: the hypoxic niche (around the necrotic core), the perivascular niche, and the invasive edge [62].
3.1. The Perivascular Niche
Vasculogenesis is the process of de novo formation of blood vessels that occurs mostly during fetal development. Studies showed that circulating endothelial cells, tumor-associated macrophages (TAMs), Tie-2 monocytes, myeloid-derived suppressor cells (MDSCs), neutrophiles, and GSCs are involved in the formation of new vessels [63,64,65,66]. GBMs are characterized by increased angiogenesis (process of stimulating the formation of new vessels from pre-existing ones) [2,4,5,9,13].
The perivascular niche is formed by nonmalignant cells (astrocytes, fibroblasts, pericytes, immune cells, neural progenitor cells) and malignant cells (GSCs and tumor cells located around disorganized blood vessels) [62]. These cells interact between them, supporting GSCs’ survival and growth. Under hypoxic conditions, the tumor produces angiogenic factors involved in the formation of the tumoral blood vessels.
More than 2 decades ago, the glioma proliferation was linked to growth factors and their receptors [67,68]. These factors include epidermal growth factor (EGF), vascular endothelial growth factor (VEGF), fibroblast growth factor (FGF), platelet-derived growth factor (PDGF), mammalian target of rapamycin (mTOR), protein kinase C (PKC), histone deacetylase, farnesyltransferase, heat shock protein 90 (Hsp90), and histone deacetylase [69]. They are important mediators of angiogenesis, affecting the oxygen supply to tumors [70,71].
A variety of inhibitors became available (Table 2), and their effectiveness was demonstrated on glioblastoma using in vitro and in vivo studies [72,73,74,75,76,77,78,79,80,81,82].

Table 2.
Targeted therapies in glioblastoma.
Furthermore, GBM presents abnormal and fragile blood vessels. The rupture of the vessels leads to the disruption of the BBB [58]. These vessels are the result of the VEGF involvement in pericyte disintegration. After BBB disruption, tumor-derived chemokines attract immunomodulatory cells that enter the brain. They increase the angiogenic factors’ production, suppressing the immune function and leading to tumor progression [62]. Vascular pericytes attach to endothelial cells and play an important role in maintaining the BBB. The depletion of pericyte may disrupt the BBB and may elevate the vascular permeability [83]. In GBM, most of vascular pericytes derive from GSCs via transdifferentiation. They express tumor-specific genetic alterations, differentiating them from normal pericytes; therefore, the neovasculature and tumor growth may be potently inhibited by selective elimination of these GSC-derived pericytes. GSCs are recruited through the SDF-1/CXCR4 axis and are further induced to transform into pericytes by transforming growth factor β (TGF-β). However, GSCs may actively remodel perivascular niches by their contribution to vascular pericytes [84].
Different immune-suppressive mechanisms are used in GBM to prevent its immune detection and eradication. GBM cells secrete a variety of immunosuppressive proteins. Intracellular adhesion molecule 1 (ICAM-1) is a cell–cell interaction key regulator that is commonly upregulated in GBM. ICAM-1 promotes the migration of myeloid cells into tumors by interacting with lymphocyte function-associated antigen 1 (LFA-1) expressed on these cells. The accumulation of MDSC in GBM further contributes to immune suppression. Additionally, GBM overexpresses galectin-1 (Gal-1), which promotes tumor cell proliferation and migration [85].
Furthermore, regulatory T cells (Tregs) that either can originate in the thymus (naturally occurring Tregs) or can be induced by antigens (iTregs) could contribute to GBM-mediated immune suppression. Tregs suppress the immune responses by cytokine secretion, such as interleukin (IL)-10 and TGF-β, or by cell-to-cell mediated contact. Cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) is present on the surface of activated T effector cells. Together with CTLA-4, programmed death 1 (PD-1) is an immune checkpoint that forms a system that regulates the immune activation and proliferation. Studies of tumor microenvironments revealed that this system is capable of inducing T-cell apoptosis [86,87,88].
More recently, an orphan member of the adhesion G-protein-coupled receptor family, EGF, latrophilin, and seven transmembrane domain-containing protein 1 (ELTD1) was reported to be upregulated in high-grade glioma blood vessels, and its expression has been associated with glioma progression and has also been suggested as a potential therapeutic target in glioblastoma [89,90,91,92].
TAMs play a significant role in the perivascular niche. TAMs are attracted in the niche by GSCs’ secreted periostin [93]. They produce heme-oxygenase 1 (HO-1) and thymidine phosphorylase (TP) involved in neovascularization. They express chemoattractants, such as VEGF, interleukin (IL)-6, colony-stimulating factor (CSF), stromal cell-derived factor 1α (SDF-1α), and IL-1β. The attraction of macrophages and monocytes generates an immunosuppressive phenotype and tumor progression. Additionally, TAMs produce TGF-β involved in matrix metalloproteinase 9 (MMP9) expression. This process causes further GSC proliferation [94].
The vasculature and angiogenic factors play a pivotal role in glioblastoma induction and the maintenance of immunosuppression. Proinflammatory repolarization of macrophages in the perivascular and perinecrotic tumor is probably an option to overcome treatment resistance [95].
Similar to VEGF and CSF, the CD34 role has been discussed in the diagnosis of various cancer pathologies. It has been demonstrated that it is involved in promoting a new blood vessel network and in increasing the nutrients and oxygen supply for further tumor growth [96]. In GBM, CD34 has been found to be expressed in the tumor vascular endothelial cell membrane. In gliomas, the CD34-positive cells are recruited by bone-marrow-derived circulating hematopoietic progenitors. In our recent study, we found no correlation between different grades of glioma or tumor vascularization and CD34 expression and with mild distribution of CD34 in CNS tumors [97]. Four types of CD34-labeled microvessel formations in glioblastomas have been detected based on different vascular niche pathologic structures: microvascular sprouting, vascular cluster, vascular garland, and glomeruloid vascular proliferation [98].
Targeting the perivascular niche could represent an effective approach for GBM therapy by using angiogenic factors’ inhibitors to influence the tumor aberrant vascular proliferation. To inhibit GSC differentiation, this strategy may be combined with other treatments, such as repolarizing macrophage-designed immunotherapeutics [99].
3.2. The Perinecrotic or Hypoxic Niche
Hypoxia is one of the main characteristics of glioblastoma. The insufficient blood supply causes hypoxia, leading to pseudo-palisading necrosis. Normal tissular median oxygen saturation is approximately 5%. Instead, in the necrotic regions, the oxygen concentration is less than 2%. This is determined by higher metabolic activity and increased oxygen consumption in the heterogeneous tumor cells [100].
Located around the necrotic core, the hypoxic niche is proved to be involved in tumor growth, cell maintenance, stemness induction, and immune surveillance [101]. The low oxygen concentration upregulates important proteins, such as hypoxia-inducible factors (HIF1 and HIF2), a dimeric protein complex with an important role in the angiogenesis and dedifferentiation process [55,56,57]. It has been found that many GSCs reside in this niche [62,97,101]. Hypoxia can induce stemness characteristics and determine the increased expression of GSC markers [20,29,33]. GSCs and tumor cells may survive in the hypoxic niche after chemo- and radiotherapy. The cellular death in the necrotic area generates proinflammatory signals, converting inflammatory cells into immunosuppressive cells and inducing angiogenesis [102]. Hypoxia can cause the differentiation of GSCs into endothelial cells, promoting the tumor growth from the necrotic area towards the neovascular region [103].
Hypoxia plays a pivotal role in the SRC tyrosine kinase pathway. The increase in SRC activity upregulates the VEGF in low-oxygen conditions. Moreover, a correlation between angiogenesis and hypoxia has been proved by the increased vascularization, with integrin upregulation. Additionally, it has been demonstrated that cell survival mechanisms are activated as a result of increased SRC signaling in response to an anti-VEGF agent [104]. In addition, the hypoxic conditions increase glycolysis because of the MYC oncoprotein. Its regulation has been linked to the SRC pathway in other tumors [105]. Therefore, the SRC–MYC axis may be implicated in metabolic reprogramming also in GBM, apart from the receptor tyrosine kinases’ (RTKs) involvement. The hypoxia-induced SRC pathway results in fostered invasiveness. It consists in integrin β3 and EGFR-vIII interaction, αvβ3 integrin recruitment on cell membranes, and FAK activation, which creates focal adhesion complexes. Furthermore, the EGFRvIII/integrin β3/FAK/SRC axis continues with the intracellular signaling pathway ERK1/2, MAPK, AKT, and STAT3 activation, determining the MMP upregulation, therefore promoting cell invasion [106].
In a recent study, Torrisi et al. evaluated the relationship between hypoxia and radiation-induced radioresistance with the activation of SRC proto-oncogene nonreceptor tyrosine kinase, prospecting potential strategies to overcome the current limitation in glioblastoma treatment. Some promising ways to sensitize GBM cells could be represented by “hypoxia dose painting” (personalized radiation dose as a function of local microenvironmental or phenotypic variations), combination of multiple ion beams in order to deliver high-linear energy transfer radiation in the hypoxic area, reoxygenation strategies, and targeting the molecular mechanisms involved in hypoxia [107].
3.3. The Invasive Niche
GBM tumor cells can migrate along normal blood vessels, invading normal brain tissue [5]. The deletion of MMP2 and MMP9 boosts the perivascular invasiveness and diminishes angiogenesis [108]. The most important cell of the invasive niche is the astrocyte. The close contact between the astrocytes, endothelial cells, and pericytes facilitates the transport of ions and metabolites from the blood vessels to the brain [97]. This microenvironment is heterogeneous, and it seems that cell populations within it are plastic. At the cellular level, malignant cells invade diffusely inside the surrounding normal brain parenchyma, followed by increased proliferation of endothelial cells, generating leaky blood vessels and resulting in a hypoxic microenvironment. The invading GBM cells displace the pre-existing astrocytes and pericytes, thereby disrupting the BBB and resulting in leaky blood vessels [109]. Paracrine interaction at this level can cause astrocyte proliferation and migration. Among the many receptors expressed by the GSCs, integrin-1 and tropomyosin receptor kinase type A (TRKA) can bind to the connective tissue growth factor (CTGF) released by the astrocytes, and produce zinc finger E-box binding homeobox-1 (ZEB1), leading to tumor cell infiltration [110,111].
It has been discovered that astrocytes express Sonic hedgehog (SHH), important glioma-associated oncogene (GLI) activator, and stemness promoter [112]. Therefore, astrocytes play a key role in GSCs’ preservation and in tumor spreading. Thus, it is expected that large tumors have extensive invasive niches, intense hypoxia, and increased neoangiogenesis.
4. Strategies for Targeting GSCs
The GSCs could be targeted directly by blocking their signaling pathways, by promoting GSC differentiation, and by virotherapy, or indirectly by targeting the perivascular, hypoxic, or immune GBMs niche.
Therapeutic targeting of GSC pathways and their receptors implicated in cell proliferation, maintenance, and tumor resistance is of great interest by providing reliable trials to block them. Among these, tyrosine kinase inhibitors (TKIs) have proved less advantages compared with GSC pathways (Notch, Wnt, and SHH) [99]
Notch, SHH, and Wnt are signaling pathways that regulate the cellular processes, including the differentiation, proliferation, and migration of GSCs (Figure 1).
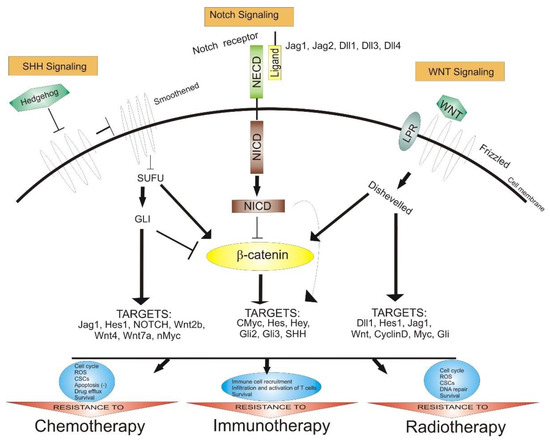
Figure 1.
The role of Notch, SHH, and Wnt signaling pathways in anticancer therapy resistance. Abbreviations: SHH—Sonic hedgehog, SUFU—protein domain suppressor of fused protein, GLI—zinc finger protein, NECD—Notch extracellular domain, NICD—Notch intracellular domain, CSCs—cancer stem cells, ROS—reactive oxygen species, Jag—Notch ligand Jagged, Dll—delta-like ligand, Hes—hairy and enhancer of split, Myc—family of regulator genes and proto-oncogenes, point arrow—activation, block arrow—inhibition.
The Notch pathway (Notch 1–Notch 4 receptors and Jagged-1 and Jagged-2 and delta-like-1, delta-like-3, and delta-like-4 ligands), can modulate the interaction between neighbor cells. This pathway is activated by proteolytic cleavage reactions, and inhibitors of the γ-secretase complex play a pivotal role in blocking the Notch signaling pathway [113].
Additionally, the Wnt/β-catenin pathway is important in modulating the self-renewal and differentiation of GSCs. The nuclear stabilized β-catenin abnormally activates the β-catenin pathway; therefore, it can be correlated to tumorigenesis, growth invasion, and MGMT expression, incriminated for TMZ resistance [114].
Furthermore, the activation of the SHH signaling pathway upregulates the multi-drug-resistance-associated protein-1(ABCC1/MRP1), ABC transporter ABCG2, drug efflux P-glycoprotein (ABCB1), B-cell-specific Moloney murine leukemia virus integration site 1 (BMI1), and MGMT. Additionally, the loss of p53 upregulates the transcription factor Nanog correlated with the SHH signaling pathway, probably contributing to TMZ chemoresistance by regulating the expression of MGMT [115].
RTKs are involved in GSC expansion and differentiation. The EGFR overexpression has been observed in 40–60% of primary human GBM tumors [115]. It is implicated in frame deletions in the extracellular domain and in gain-of-function missense mutations and less in the inhibitor responsiveness [116]. Three types of EGFR inhibitors are currently used in the cancer field: first-generation reversible small-molecule inhibitors that target EGFR and coreceptor HER2 (erlotinib and gefitinib), second-generation inhibitors that irreversibly bind to EGFR (afatinib, dacomitinib, and neratinib), and third-generation irreversible TKIs (AZD9291). Furthermore, in a recent study, SH3 domain containing kinase binding protein 1 (SH3KBP1), an activator and modulator of the EGFR signaling, has been proved to have increased levels of mRNA and protein in GBM. In addition, there is evidence that SH3KBP1 is predominantly expressed in GSCs, which makes it a promising novel GSC marker and a future target [117].
The PDGF receptors are responsible for developing oligodendrocytes and blood vessels during embryonic development. Overexpression and alterations of PDGF ligands are commonly present in GBM tumors, especially in the proneural subtype, contributing to GBM development and GSC functions. Several inhibitors (imatinib, tandutinib, nilotinib, and AG1433) demonstrated their efficacy to decrease the growth of GBM xenografts in vivo, but with no significant effect on recurrent GBM [118].
Furthermore, GBM highly expresses VEGF and its receptors, important regulators of angiogenesis, vasculogenesis, and lymphangiogenesis. There are many VEGFR inhibitors with various treatment effects, such as cediranib, lapatinib, pazopanib, sorafenib, atalanib, tivozanib, and SU1498 [87]. Recently, the human cartilage glycoprotein-39 or chitinase-like protein-1 (YKL-40) has been proved able to upregulate the expression of VEGF. YKL-40 is a molecular marker for a mesenchymal subtype of GBMs found to be responsible for TMZ, resistance that gives it a chance to become an effective target in cancer therapy [119].
Another strategy to directly target the GSCs is by promoting GSC differentiation. It seems that GSCs become more sensitive to various therapies after differentiating in more terminal GBM cells [120]. Therefore, a key in the therapy against GBM could be achieved by promoting the GSC differentiation. Several attempts have been made. For example, the bone morphogenic proteins are involved in the stem cell niche and in GSC differentiation [121]. Another example is the overexpression of miR-128, which influences differentiation and enhances the senescence mediated by axitinib [122]. Moreover, the graphene oxide decreases the expression of stem cell markers and increases the expression of differentiation-related markers [123]. Additionally, the nonsteroidal anti-inflammatory agent sulindac has been proved able to induce GSC differentiation [124].
Among various strategies that target GSCs, oncolytic virotherapy has been proved efficient in preventing GBM recurrence. For this purpose, the oncolytic adenovirus DNX-2401 (BM-hMSCs-DNX2401) has been administered with good results intra-arterially, loaded in allogeneic bone-marrow-derived human mesenchymal stem cells [125].
Instead, the indirect strategy of targeting the GSC niche involves targeting the perivascular niche through angiogenic pathways, the immune niche, and the hypoxic or perinecrotic niche by inducing hypoxia.
The perivascular niche is surrounded by a vasculature with different characteristics compared with normal blood vessels, featuring structure and function abnormalities, hypoxia, and hierarchy impairment. There is a codependent and synergistic relationship between the perivascular niche of GBM and GSCs: the GSC survival, proliferation, and migration are maintained by GBM, which mediates the perivascular region, and GSCs simultaneously act as a cancer-specific vasculature regulator, infiltrating into the tumor [126]. GSCs express high levels of angiogenic factors, providing an optimum environment for GSC survival and proliferation; therefore, the blockage of angiogenesis in the perivascular niche could diminish the original tumor.
Moreover, in GBM therapy, the immune niche could also be targeted. Tumor aggression increased, and immune cells infiltrated in the brain after bevacizumab or cediranib treatment of recurrent GBM in GBM mouse models [127]. The resistance of antiangiogenic therapy could be explained by the direct interaction between GSCs and the immune cells, leading to VEGF-independent angiogenesis and generating tumors without the ability to respond to inhibitors [128]. By targeting the innate immune cells and receptors, tumor sensitivity increased, a strategy that could be a promising therapy for GBM.
Hypoxia is one diagnostic hallmark of GBM. Hypoxia-inducible factor-1α (HIF-1α) and VEGFA are two main regulators of hypoxia response, critically implicated in worse progression-free survival. Thus, bevacizumab, a VEGFA inhibitor, is a promising agent, useful in overcoming both hypoxia and resistance in cancer therapy [87,129].
5. GSCs Biology, Genetic and Epigenetic Changes
GBM features both genetic and epigenetic alterations responsible for cancer cell gene expression regulation. The epigenetic mechanisms cause gene expression alterations in response to the environment. Some of the epigenetic changes that occur in GSCs are:
- The DNA methylation, mostly within gene-promotor area (CpG) islands, which influence gene expression. Hypermethylation of tumor suppressor genes, such as TP53, generally causes tumorigenesis [130], while usually hypomethylation leads to the oncogene activation. Early findings showed that many tumor suppressor genes are targets for DNA hypermethylation in cancer, therefore the idea that aberrant DNA methylation may promote oncogenesis via tumor suppressor gene silencing. However, more recent genome-wide analyses have proved that the classical model requires to be reconsidered [131];
- Changes in the microRNA (miRNA) family, small single-stranded noncoding functional RNA molecules involved in RNA silencing and post-transcriptional regulation. Studies proved the existence of 351 altered expressions of miRNAs in GBM, 256 being overexpressed and 95 underexpressed, demonstrating that the miRNA expression is modified in GSCs compared with the normal brain cells [132];
- Dysregulation of a polycomb group of proteins (PcGs) causes tumor progression and invasion. They can determine gene silencing by remodeling chromatin. One of these proteins, BMI1, a member of PRC1 (polycomb repressive complex 1), blocks the differentiation of GSCs into neurons and inhibits apoptosis [133]. EZH2, a member of PRC2, is suspected to be involved in supporting the GSCs by activating STAT3 [134,135]. This protein group is considered to support GBM progression and invasiveness;
- Additionally, GBM formation explained by post-translational modification of histones [136], influencing the chromatin architecture, with epigenetic changes.
To efficiently target GSCs, the mechanisms behind the genetic and epigenetic alterations, the molecular pathways, and the interactions between the tumor microenvironment and GSCs must be thoroughly understood.
Overall, a permanent epigenetic block supports the self-renewal ability of the GSCs, featuring them with the impairment of differentiation [137]. Still, GSCs can modulate their DNA methylomes and transcriptomes, rapidly adapting to various microenvironments, suggesting that these alterations may be partially reversible, differently from genetic variations. The epigenetic mark’ reversibility has been demonstrated in glioma cells by using a certain combination of transcription factors and by reversing in an early embryonic state, followed by a general resetting of DNA methylation associated with cancer. However, this resetting could not cancel the malignant behavior of the GSCs, proving that epigenetic-related activities need to be thoroughly deciphered in order to explain their malignancy [138,139].
The characteristics of GSCs follow regional variations according to the tumor niche. The regions with BBB disruption are characterized by increased expression of proneural genes (EZH2, SUZ12, H3K27me3), while the hypoxic necrotic regions have a high expression of mesenchymal genes and BMI1 targets, with a strong H2A119ub association, and the presence of CD44 and YKL40 markers. Nonetheless, selective inhibition of BMI1 or EZH2 was proved to be effective against the survival of both mesenchymal and proneural GSCs [139].
GSCs upregulate multiple receptors and signaling pathways that could represent the source of CNS oncogenic alterations. They have been found to be responsible for tumor proliferation, resistance to radiotherapy and chemotherapy, maintenance, and aberrant cell survival [40,57,138]. The signaling pathways and the corresponding receptors have a pivotal role for GSC involvement in oncology. Notch, SHH, and Wnt are the most representative signaling pathways, with large implications in the cancer domain.
Artificially induced replication stress in glioma cells has been associated with radio resistance. Replication stress is represented by an inefficient DNA replication that determines replication forks to evolve slowly or even to stop. Replication stress activates some molecular processes that stabilize the replication forks, preventing DNA damage. Replication stress is considered to be one mechanism for radiotherapy resistance glioma stem cells, proved by the elevated levels of markers, such as replication protein A, DNA damage markers, and single-stranded DNA binding protein [140]. Additionally, tyrosine kinase MET induces radio resistance via activation of AKT kinase and DNA repair downstream effectors and by phosphorylation of the p21 protein with an antiapoptotic effect [141]. Targeting replication stress response with combined ATR (ataxia telangiectasia and Rad3 related) and PARP (poly (ADP-ribose) polymerase) inhibition provides high and specific cytotoxicity in glioma cells. Combined ATR and PARP inhibition leads to a specific vulnerability, resulting in DNA low replication and abrogation of DNA repair [142].
5.1. Notch Signaling Pathway
The Notch signaling pathway is a highly preserved cell signaling system present in most animals. It is involved in cell fate, migration, proliferation, and cellular quiescence maintenance and is also a regulator of neural stem cell differentiation [139]. The multipotency perpetuation is stimulated by the activation of the HES (hairy/enhancer of split) and HEY (hairy/E(spl)-related with YRPW motif) genes and by the formation of an RBPJ (recombining binding protein suppressor of hairless) and MAML (mastermind-like) protein complex in the nucleus. This process is followed by the translocation of NICD (Notch intracellular domain) to the nucleolus caused by the cleavage of the Notch receptor by presenilin/α-secretase, after binding Jagged and delta Notch ligands [143,144]. The high expression of the Notch signaling pathway activators ID4 (inhibitor of differentiation 4) and FABP7 (fatty-acid-binding protein 7) by the CD133-positive GSCs influences the migration of radial ganglion cell (RGC) and stimulates the infiltrating capacity of GBM [145]. It is yet to be discovered how Notch is activating the GSCs’ oncogenic signaling mechanism. It is already known that Notch activation is amplified by tenascin-C, an extracellular matrix protein [146], and it is associated with GSCs’ stemness maintenance. This supports the idea that blocking the Notch signaling pathway may be an effective strategy of limiting tumor progression by GSC inactivation.
5.2. SHH/GLI Signaling Pathway
SHH has an important role in regulating embryonic morphogenesis. In adults, it is involved in tissue repair processes, with great implications in GSCs’ self-renewal mechanism and tumorigenicity [99,147]. Additionally, the SHH/GLI pathway is involved in upregulating proteins that are highly activated in most of the GSCs, such as drug efflux P-glycoprotein (ABCB1), breast cancer resistance protein (BCRP/ABCG2), multi-drug-resistance-associated protein 1 (MRP1/ABCC1), O6-methylguanine-methyltransferase (MGMT), and BMI1 [148,149]. Nanog, a protein-coding gene, considered to be the key to pluripotency, is one principal regulator of the expression of certain stemness factors. The Nanog expression is activated by binding SHH/GLI1/GLI2 to the Nanog promoter. Furthermore, p53 is involved in downregulating the Nanog expression due to the decrease in GLI1 expression and activity. Therefore, the loss of p53 may contribute to the maintenance of stemness by activating SHH signaling and by upregulating Nanog [150]. Studies have shown that targeting this pathway could increase the efficacy of chemotherapy [151].
In a study performed by Torrisi et al., SHH signaling and CX43-based intercellular communication has been modulated using in vitro models of proliferation and migration. They suggested a potential axis between the SHH signaling pathway and CX43 at least on two aspects: “permissive”, in which SHH-GLI signaling favors the intercellular communication and patterning, leading to microenvironmental changes and tumorigenic onset, and “supporting” the GBM stemness signature so that the aberrant SHH-GLI pathway promotes GSC population [152].
5.3. Wnt Signaling Pathway
The Wnt signaling pathway plays an important role during differentiation and self-renewal of the neural stem cells [153] as well as GSCs [154]. Its abnormal activation can lead to the formation of brain tumors. The Wnt pathway is involved in GSC maintenance by means of genetic and epigenetic mechanisms [154]. The overexpression of PLAGL2 (pleomorphic adenoma gene-like 2) increases the expression of various Wnt receptors, such as FZD2 (frizzled), FZD 9, and Wnt6 [155]. FoxM1, a proliferation-associated transcription factor involved in cell proliferation, self-renewal, and tumorigenesis, generates stemness and preservation of the GSCs after binding to Sox2 (SRY-box transcription factor 2) [156]. Evi (evenness interrupted), a transmembrane protein overexpressed in GBM, is regulated by epigenetic mechanism, modifying both canonical and noncanonical Wnt pathways [157]. Wnt5A causes endothelial differentiation of GSCs, leading to neovascularization, a favorable condition for cell growth and invasion [158].
Both canonical (characterized by the intracellular accumulation of β-catenin) and noncanonical (defined by its β-catenin-independent actions) Wnt signaling pathways play a significant role in gliomagenesis and the maintenance of GSCs. Furthermore, the TMZ resistance can be explained by the Wnt-induced expression of MGMT [159].
Because of its involvement in multiple essential functions in the entire body, the Wnt signaling pathway cannot be targeted without various side-effect consequences [153,160,161].
6. Conclusions
Although GBM is continuously being in the focus of researchers worldwide, the gathered information has not led to a major breakthrough in therapy. The standard treatment strategy continues to be the safe maximal surgical resection, followed by radiotherapy and chemotherapy. Unfortunately, these protocols are not able to safely cure cancer patients, but only to prolong their survival rates. Regularly, the treatment is followed by often more aggressive recurrences or metastasis, most probably because of the tumor microenvironment and molecular heterogeneity.
In this review, we focused on GSCs, detailing their vital signal pathways, resistance mechanism, and crosstalk between them and their niche. We also provided different GSC-targeting strategies, a great opportunity for further approaches. Accordingly, more studies need to be performed to have a better framework regarding the properties, origin, and progression of GSCs, as well as their maintenance, resistance, and recurrence. In the future, novel approaches that target GSCs could become useful tools for novel clinical applications that somehow are urgently needed.
Author Contributions
Conceptualization, S.M.B.R. and G.-A.S.; investigation, S.M.B.R. and C.B.; resources, S.M.B.R., G.-A.S. and C.B.; data curation, G.-A.S. and V.C.; writing—original draft preparation, A.-S.S.; writing—review and editing, A.-S.S. and V.C.; visualization, A.-S.S. and A.D.; supervision, L.G.T. and A.D.; project administration, A.D.; funding acquisition, A.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Grant PN-III-P4-ID-PCE2020-1649, by the UEFISCDI Authority, Romania.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Brandes, A.A.; Bartolotti, M.; Tosoni, A.; Franceschi, E. Nitrosoureas in the Management of Malignant Gliomas. Curr. Neurol. Neurosci. Rep. 2016, 16, 13. [Google Scholar] [CrossRef] [PubMed]
- Omuro, A.; DeAngelis, L.M. Glioblastoma and other malignant gliomas: A clinical review. JAMA 2013, 310, 1842–1850. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cazzato, E.; Ladewig, E.; Frattini, V.; Rosenbloom, D.I.; Zairis, S.; Abate, F.; Liu, Z.; Elliott, O.; Shin, Y.-J.; et al. Clonal evolution of glioblastoma under therapy. Nat. Genet. 2016, 48, 768–776. [Google Scholar] [CrossRef] [PubMed]
- Ciurea, M.E.; Georgescu, A.M.; Purcaru, S.O.; Artene, S.A.; Emami, G.H.; Boldeanu, M.V.; Tache, D.E.; Dricu, A. Cancer stem cells: Biological functions and therapeutically targeting. Int. J. Mol. Sci. 2014, 15, 8169–8185. [Google Scholar] [CrossRef] [PubMed]
- Deleanu, R.; Ceafalan, L.C.; Dricu, A. Transcriptomic Crosstalk between Gliomas and Telencephalic Neural Stem and Progenitor Cells for Defining Heterogeneity and Targeted Signaling Pathways. Int. J. Mol. Sci. 2021, 22, 13211. [Google Scholar] [CrossRef]
- Eyüpoglu, I.Y.; Buchfelder, M.; Savaskan, N.E. Surgical resection of malignant gliomas-role in optimizing patient outcome. Nat. Rev. Neurol. 2013, 9, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Yong, R.L.; Lonser, R.R. Surgery for glioblastoma multiforme: Striking a balance. World Neurosurg. 2011, 76, 528–530. [Google Scholar] [CrossRef]
- Chatterji, P.; Douchin, J.; Giroux, V. Demystifying the Differences Between Tu-mor-Initiating Cells and Cancer Stem Cells in Colon Cancer. Curr. Colorectal Cancer Rep. 2018, 14, 242–250. [Google Scholar] [CrossRef]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Cusimano, M.D.; Dirks, P.B. Identification of human brain tumour initiating cells. Nature 2004, 432, 396–401. [Google Scholar] [CrossRef]
- Chen, J.; Li, Y.; Yu, T.S.; McKay, R.M.; Burns, D.K.; Kernie, S.G.; Parada, L.F. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 2012, 488, 522–526. [Google Scholar] [CrossRef]
- Akbari-Birgani, S.; Paranjothy, T.; Zuse, A.; Janikowski, T.; Cieslar-Pobuda, A.; Likus, W.; Urasinska, E.; Schweizer, F.; Ghavami, S.; Klonisch, T.; et al. Cancer stem cells, cancer-initiating cells and methods for their detection. Drug Discov. Today 2016, 21, 836–842. [Google Scholar] [CrossRef]
- Hombach-Klonisch, S.; Mehrpour, M.; Shojaei, S.; Harlos, C.; Pitz, M.; Hamai, A.; Siemianowicz, K.; Likus, W.; Wiechec, E.; Toyota, B.D.; et al. Glioblastoma and chemoresistance to alkylating agents: Involvement of apoptosis, autophagy, and unfolded protein response. Pharmacol. Ther. 2018, 184, 13–41. [Google Scholar] [CrossRef] [PubMed]
- Lathia, J.D.; Mack, S.C.; Mulkearns-Hubert, E.E.; Valentim, C.L.; Rich, J.N. Cancer stem cells in glioblastoma. Genes Dev. 2015, 29, 1203–1217. [Google Scholar] [CrossRef] [PubMed]
- Dricu, A. Recent challenges with stem cell banking. Expert Opin. Biol. Ther. 2018, 18, 355–358. [Google Scholar] [CrossRef] [PubMed]
- Emami Nejad, A.; Najafgholian, S.; Rostami, A.; Shojaeifar, S.; Esparvarinha, M.; Nedaeinia, R.; Javanmard, S.H.; Taherian, M.; Ahmadlou, M.; Salehi, R.; et al. The role of hypoxia in the tumor microenvironment and development of cancer stem cell: A novel approach to devel-oping treatment. Cancer Cell Int. 2021, 21, 62. [Google Scholar] [CrossRef] [PubMed]
- Li, H.S.; Zhou, Y.N.; Li, L.; Li, S.F.; Long, D.; Chen, X.L.; Zhang, J.-B.; Feng, L.; Li, Y.-P. HIF-1alpha protects against oxidative stress by directly targeting mitochondria. Redox Biol. 2019, 25, 101109. [Google Scholar] [CrossRef] [PubMed]
- Li, L.Y.; Guan, Y.D.; Chen, X.S.; Yang, J.M.; Cheng, Y. DNA repair pathways in cancer therapy and resistance. Front. Pharmacol. 2021, 11, 629266. [Google Scholar] [CrossRef]
- Chen, K.; Zhang, C.; Ling, S.; Wei, R.; Wang, J.; Xu, X. The metabolic flexibility of quiescent CSC: Implications for chemotherapy resistance. Cell Death Dis. 2021, 12, 835. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO classification of tumors of the central nervous system: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Rong, Y.; Durden, D.L.; Van Meir, E.G.; Brat, D.J. ‘Pseudopalisading’ necrosis in glioblastoma: A familiar morphologic feature that links vascular pathology, hypoxia, and angiogenesis. J. Neuropathol. Exp. Neurol. 2006, 65, 529–539. [Google Scholar] [CrossRef]
- Jakovlevs, A.; Vanags, A.; Gardovskis, J.; Strumfa, I. Molecular classification of diffuse gliomas. Pol. J. Pathol. 2019, 70, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Orzan, F.; Pagani, F.; Cominelli, M.; Triggiani, L.; Calza, S.; De Bacco, F.; Medicina, D.; Balzarini, P.; Panciani, P.P.; Liserre, R.; et al. A simplified integrated molecular and immunohistochemistry-based algorithm allows high accuracy prediction of glioblastoma transcriptional subtypes. Lab. Investig. 2020, 100, 1330–1344. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.; Manchanda, P.; Vogelbaum, M.A.; Ohlfest, J.R.; Elmquist, W.F. Function of the blood-brain barrier and restriction of drug delivery to invasive glioma cells: Findings in an orthotopic rat xenograft model of glioma. Drug Metab. Dispos. 2013, 41, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Bauer, H.C.; Krizbai, I.A.; Bauer, H.; Traweger, A. “You Shall Not Pass”-tight junctions of the blood brain barrier. Front. Neurosci. 2014, 8, 392. [Google Scholar] [CrossRef]
- Sevastre, A.; Costachi, A.; Tataranu, L.G.; Brandusa, C.; Artene, S.A.; Stovicek, O.; Alexandru, O.; Danoiu, S.; Sfredel, V.; Dricu, A. Glioblastoma pharmacotherapy: A multifaceted perspective of conventional and emerging treatments. Exp. Ther. Med. 2021, 22, 1408. [Google Scholar] [CrossRef]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef]
- Inda, M.M.; Bonavia, R.; Mukasa, A.; Narita, Y.; Sah, D.W.; Vandenberg, S.; Brennan, C.; Johns, T.G.; Bachoo, R.; Hadwiger, P.; et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010, 24, 1731–1745. [Google Scholar] [CrossRef]
- Ahmad, G.; Amiji, M.M. Cancer stem cell-targeted therapeutics and delivery strategies. Expert Opin. Drug Deliv. 2017, 14, 997–1008. [Google Scholar] [CrossRef]
- Heddleston, J.M.; Li, Z.; Lathia, J.D.; Bao, S.; Hjelmeland, A.B.; Rich, J.N. Hypoxia inducible factors in cancer stem cells. Br. J. Cancer 2010, 102, 789–795. [Google Scholar] [CrossRef]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef]
- Hiddingh, L.; Tannous, B.A.; Teng, J.; Tops, B.; Jeuken, J.; Hulleman, E.; Boots-Sprenger, S.H.; Vandertop, W.P.; Noske, D.P.; Kaspers, G.J.; et al. EFEMP1 induces γ-secretase/Notch-mediated temozolomide resistance in glioblastoma. Oncotarget 2014, 5, 363–374. [Google Scholar] [CrossRef]
- Wee, B.; Pietras, A.; Ozawa, T.; Bazzoli, E.; Podlaha, O.; Antczak, C.; Westermark, B.; Nelander, S.; Uhrbom, L.; Forsberg-Nilsson, K.; et al. ABCG2 regulates self-renewal and stem cell marker expression but not tumorigenicity or radiation resistance of glioma cells. Sci. Rep. 2016, 6, 25956. [Google Scholar] [CrossRef] [PubMed]
- Soehngen, E.; Schaefer, A.; Koeritzer, J.; Huelsmeyer, V.; Zimmer, C.; Ringel, F.; Gempt, J.; Schlegel, J. Hypoxia upregulates aldehyde dehydrogenase isoform 1 (ALDH1) expression and induces functional stem cell characteristics in human glioblastoma cells. Brain Tumor Pathol. 2014, 31, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Suwala, A.K.; Koch, K.; Rios, D.H.; Aretz, P.; Uhlmann, C.; Ogorek, I.; Felsberg, J.; Reifenberger, G.; Köhrer, K.; Deenen, R.; et al. Inhibition of Wnt/beta-catenin signaling downregulates expression of aldehyde dehydrogenase isoform 3A1 (ALDH3A1) to reduce resistance against temozolomide in glioblastoma in vitro. Oncotarget 2018, 9, 22703–22716. [Google Scholar] [CrossRef] [PubMed]
- Gan, C.; Pierscianek, D.; El Hindy, N.; Ahmadipour, Y.; Keyvani, K.; Sure, U.; Zhu, Y. The predominant expression of cancer stem cell marker ALDH1A3 in tumor infiltrative area is associated with shorter overall survival of human glioblastoma. BMC Cancer 2020, 20, 672. [Google Scholar] [CrossRef]
- Park, J.; Shim, J.K.; Kang, J.H.; Choi, J.; Chang, J.H.; Kim, S.Y.; Kang, S.G. Regulation of bioenergetics through dual inhibition of aldehyde dehydrogenase and mitochondrial complex I suppresses glioblastoma tumorspheres. Neuro Oncol. 2018, 20, 954–965. [Google Scholar] [CrossRef]
- Wu, W.; Wu, Y.; Mayer, K.; von Rosenstiel, C.; Schecker, J.; Baur, S.; Würstle, S.; Liesche-Starnecker, F.; Gempt, J.; Schlegel, J. Lipid peroxidation plays an important role in chemotherapeutic effects of temozolomide and the development of therapy resistance in human glioblastoma. Transl. Oncol. 2020, 13, 100748. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, M.; Gan, H.; Lee, J.-H.; Fang, D.; Kitange, G.J.; He, L.; Hu, Z.; Parney, I.F.; Meyer, F.B.; et al. A novel enhancer regulates MGMT expression and promotes temozolomide resistance in glioblastoma. Nat. Commun. 2018, 9, 2949. [Google Scholar] [CrossRef]
- Oldrini, B.; Vaquero-Siguero, N.; Mu, Q.; Kroon, P.; Zhang, Y.; Galán-Ganga, M.; Bao, Z.; Wang, Z.; Liu, H.; Sa, J.K.; et al. MGMT genomic rearrangements contribute to chemotherapy resistance in gliomas. Nat. Commun. 2020, 11, 3883. [Google Scholar] [CrossRef]
- Garcés, Á.; Bronk, L.; Bhat, K.; Grosshans, D. ATRX-Loss Increases Sensitivity to Proton Radiotherapy Compared to X-Ray Radiotherapy in Glioma Stem Cells via Secondary Necrosis. Int. J. Radiat. Oncol. Biol. Phys. 2021, 111, e589–e590. [Google Scholar] [CrossRef]
- Crunkhorn, S. Sensitizing glioblastoma to radiotherapy. Nat. Rev. Drug Discov. 2021, 20, 588. [Google Scholar] [CrossRef] [PubMed]
- Polat, B.; Wohlleben, G.; Kosmala, R.; Lisowski, D.; Mantel, F.; Lewitzki, V.; Löhr, M.; Blum, R.; Herud, P.; Flentje, M.; et al. Differences in stem cell marker and osteopontin expression in primary and recurrent glioblastoma. Cancer Cell Int. 2022, 22, 87. [Google Scholar] [CrossRef] [PubMed]
- Erhart, F.; Blauensteiner, B.; Zirkovits, G.; Printz, D.; Soukup, K.; Klingenbrunner, S.; Fischhuber, K.; Reitermaier, R.; Halfmann, A.; Lötsch, D.; et al. Gliomasphere marker combinatorics: Multidimensional flow cytometry detects CD44+/CD133+/ITGA6+/CD36+ signature. J. Cell Mol. Med. 2019, 23, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Hu, B.; Hu, X.; Kim, H.; Squatrito, M.; Scarpace, L.; deCarvalho, A.C.; Lyu, S.; Li, P.; Li, Y.; et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 2017, 32, 42–56.e6. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zhang, M.; Cheng, K.; Li, P.; Han, S.; Li, R.; Su, M.; Zeng, W.; Liu, J.; Guo, J.; et al. Resistance of glioma cells to nutrient-deprived microenvironment can be enhanced by CD133-mediated autophagy. Oncotarget 2016, 7, 76238–76249. [Google Scholar] [CrossRef]
- Sidney, L.E.; Branch, M.J.; Dunphy, S.E.; Dua, H.S.; Hopkinson, A. Concise review: Evidence for CD34 as a common marker for diverse progenitors. Stem Cells 2014, 32, 1380–1389. [Google Scholar] [CrossRef]
- Gu, Q.; Chen, X.; Zhou, L.; Liu, X. Exosome EpCAM promotes the metastasis of glioma by targeting the CD44 signaling molecule on the surface of glioma cells. Adv. Clin. Exp. Med. 2020, 29, 1277–1282. [Google Scholar] [CrossRef]
- Pietras, A.; Katz, A.M.; Ekström, E.J.; Wee, B.; Halliday, J.J.; Pitter, K.L.; Werbeck, J.L.; Amankulor, N.M.; Huse, J.T.; Holland, E.C. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell 2014, 14, 357–369. [Google Scholar] [CrossRef]
- Son, M.J.; Woolard, K.; Nam, D.H.; Lee, J.; Fine, H.A. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell 2009, 4, 440–452. [Google Scholar] [CrossRef]
- Galdieri, L.; Jash, A.; Malkova, O.; Mao, D.D.; DeSouza, P.; Chu, Y.E.; Salter, A.; Campian, J.L.; Naegle, K.M.; Brennan, C.W.; et al. Defining phenotypic and functional heterogeneity of glioblastoma stem cells by mass cytometry. JCI Insight 2021, 6, e128456. [Google Scholar] [CrossRef]
- Yarmishyn, A.A.; Yang, Y.P.; Lu, K.H.; Chen, Y.C.; Chien, Y.; Chou, S.J.; Tsai, P.H.; Ma, H.I.; Chien, C.S.; Chen, M.T.; et al. Musashi-1 promotes cancer stem cell properties of glioblastoma cells via upregulation of YTHDF1. Cancer Cell Int. 2020, 20, 597. [Google Scholar] [CrossRef]
- Prestegarden, L.; Svendsen, A.; Wang, J.; Sleire, L.; Skaftnesmo, K.O.; Bjerkvig, R.; Yan, T.; Askland, L.; Persson, A.; Sakariassen, P.Ø.; et al. Glioma cell populations grouped by different cell type markers drive brain tumor growth. Cancer Res. 2010, 70, 4274–4279. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Yang, H.; Chen, T.; Zhu, X.; Ye, J.; Lv, K. Identification of HMG-box family establishes the significance of SOX6 in the malignant progression of glioblastoma. Aging 2020, 12, 8084–8106. [Google Scholar] [CrossRef] [PubMed]
- Wachowiak, R.; Krause, M.; Mayer, S.; Peukert, N.; Suttkus, A.; Müller, W.C.; Lacher, M.; Meixensberger, J.; Nestler, U. Increased L1CAM (CD171) levels are associated with glioblastoma and metastatic brain tumors. Medicine 2018, 97, e12396. [Google Scholar] [CrossRef] [PubMed]
- Friedmann-Morvinski, D.; Verma, I.M. Dedifferentiation and reprogramming: Origins of cancer stem cells. EMBO Rep. 2014, 15, 244–253. [Google Scholar] [CrossRef]
- Wang, P.; Gong, S.; Liao, B.; Pan, J.; Wang, J.; Zou, D.; Zhao, L.; Xiong, S.; Deng, Y.; Yan, Q.; et al. HIF1?/HIF2? induces glioma cell dedifferentiation into cancer stem cells through Sox2 under hypoxic conditions. J. Cancer 2022, 13, 1–14. [Google Scholar] [CrossRef]
- Lee, G.; Auffinger, B.; Guo, D.; Hasan, T.; Deheeger, M.; Tobias, A.L.; Kim, J.Y.; Atashi, F.; Zhang, L.; Lesniak, M.S.; et al. Dedifferentiation of glioma cells to glioma stem-like cells by therapeutic stress-induced HIF signaling in the recurrent GBM model. Mol. Cancer Ther. 2016, 15, 3064–3076. [Google Scholar] [CrossRef]
- Campos, B.; Olsen, L.R.; Urup, T.; Poulsen, H.S. A comprehensive profile of recurrent glioblastoma. Oncogene 2016, 35, 5819–5825. [Google Scholar] [CrossRef]
- Neftel, C.; Laffy, J.; Filbin, M.G.; Hara, T.; Shore, M.E.; Rahme, G.J.; Richman, A.R.; Silverbush, D.; Shaw, M.L.; Hebert, C.M. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 2019, 178, 835–849.e21. [Google Scholar] [CrossRef]
- Brooks, L.J.; Clements, M.P.; Burden, J.J.; Kocher, D.; Richards, L.; Devesa, S.C.; Zakka, L.; Woodberry, M.; Ellis, M.; Jaunmuktane, Z.; et al. The white matter is a pro-differentiative niche for glioblastoma. Nat. Commun. 2021, 12, 2184. [Google Scholar] [CrossRef]
- Bayin, N.S.; Modrek, A.S.; Placantonakis, D.G. Glioblastoma stem cells: Molecular characteristics and therapeutic implications. World J. Stem Cells 2014, 6, 230–238. [Google Scholar] [CrossRef] [PubMed]
- Hambardzumyan, D.; Bergers, G. Glioblastoma: Defining Tumor Niches. Trends Cancer 2015, 1, 252–265. [Google Scholar] [CrossRef] [PubMed]
- Codrici, E.; Popescu, I.-D.; Tanase, C.; Enciu, A.-M. Friends with Benefits: Chemokines, Glioblastoma-Associated Microglia/Macrophages, and Tumor Microenvironment. Int. J. Mol. Sci. 2022, 23, 2509. [Google Scholar] [CrossRef]
- Turrini, R.; Pabois, A.; Xenarios, I.; Coukos, G.; Delaloye, J.F.; Doucey, M.A. TIE-2 expressing monocytes in human cancers. Oncoimmunology 2017, 6, e1303585. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Ai, X.; Zhao, L.; Fei, F.; Wang, P.; Zhou, S. Phenotypic plasticity of myeloid cells in glioblastoma development, progression, and therapeutics. Oncogene 2021, 40, 6059–6070. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Piao, Y.; Holmes, L.; Fuller, G.N.; Henry, V.; Tiao, N.; de Groot, J.F. Neutrophils promote the malignant glioma phenotype through S100A4. Clin. Cancer Res. 2014, 20, 187–198. [Google Scholar] [CrossRef]
- Hatanpaa, K.J.; Burma, S.; Zhao, D.; Habib, A.A. Epidermal growth factor receptor in glioma: Signal transduction, neuropathology, imaging, and radioresistance. Neoplasia 2010, 12, 675–684. [Google Scholar] [CrossRef]
- Carapancea, M.; Alexandru, O.; Fetea, A.S.; Dragutescu, L.; Castro, J.; Georgescu, A.; Popa-Wagner, A.; Bäcklund, M.L.; Lewensohn, R.; Dricu, A. Growth factor receptors signaling in glioblastoma cells: Therapeutic implications. J. Neurooncol. 2009, 92, 137–147. [Google Scholar] [CrossRef]
- Cruz Da Silva, E.; Mercier, M.C.; Etienne-Selloum, N.; Dontenwill, M.; Choulier, L. A systematic review of glioblastoma-targeted therapies in Phases II, III, IV clinical trials. Cancers 2021, 13, 1795. [Google Scholar] [CrossRef]
- Popescu, A.M.; Alexandru, O.; Brindusa, C.; Purcaru, S.O.; Tache, D.E.; Tataranu, L.G.; Taisescu, C.; Dricu, A. Targeting the VEGF and PDGF signaling pathway in glioblastoma treatment. Int. J. Clin. Exp. Pathol. 2015, 8, 7825–7837. [Google Scholar]
- Ahir, B.K.; Engelhard, H.H.; Lakka, S.S. Tumor Development and Angiogenesis in Adult Brain Tumor: Glioblastoma. Mol. Neurobiol. 2020, 57, 2461–2478. [Google Scholar] [CrossRef] [PubMed]
- Nalkiran, H.S.; McDonald, K.L. Is neuroglial antigen 2 a potential contributor to cilengitide response in glioblastoma? J. Cancer Res. Ther. 2017, 13, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Pan, P.C.; Magge, R.S. Mechanisms of EGFR resistance in glioblastoma. Int. J. Mol. Sci. 2020, 21, 8471. [Google Scholar] [CrossRef] [PubMed]
- Di Cintio, F.; Dal Bo, M.; Baboci, L.; De Mattia, E.; Polano, M.; Toffoli, G. The molecular and microenvironmental landscape of glioblastomas: Implications for the novel treatment choices. Front. Neurosci. 2020, 14, 603647. [Google Scholar] [CrossRef]
- Bolcaen, J.; Nair, S.; Driver, C.H.S.; Boshomane, T.M.G.; Ebenhan, T.; Vandevoorde, C. Novel receptor tyrosine kinase Pathway inhibitors for targeted radionuclide therapy of glioblastoma. Pharmaceuticals 2021, 14, 626. [Google Scholar] [CrossRef]
- Colardo, M.; Segatto, M.; Di Bartolomeo, S. Targeting RTK-PI3K-mTOR axis in gliomas: An update. Int. J. Mol. Sci. 2021, 22, 4899. [Google Scholar] [CrossRef]
- Geribaldi-Doldán, N.; Hervás-Corpión, I.; Gómez-Oliva, R.; Domínguez-García, S.; Ruiz, F.A.; Iglesias-Lozano, I.; Carrascal, L.; Pardillo-Díaz, R.; Gil-Salú, J.L.; Nunez-Abades, P.; et al. Targeting protein kinase C in glioblastoma treatment. Biomedicines 2021, 9, 381. [Google Scholar] [CrossRef]
- Mekala, J.R.; Ramalingam, P.S.; Mathavan, S.; Yamajala, R.B.R.D.; Moparthi, N.R.; Kurappalli, R.K.; Manyam, R.R. Synthesis, in vitro and structural aspects of cap substituted Suberoylanilide hydroxamic acid analogs as potential inducers of apoptosis in Glioblastoma cancer cells via HDAC/microRNA regulation. Chem. Biol. Interact. 2022, 357, 109876. [Google Scholar] [CrossRef]
- Nghiemphu, P.L.; Ebiana, V.A.; Wen, P.; Gilbert, M.; Abrey, L.E.; Lieberman, F.; DeAngelis, L.M.; Robins, H.I.; Yung, W.K.A.; Chang, S.; et al. Phase I study of sorafenib and tipifarnib for recurrent glioblastoma: NABTC 05-02. J. Neuro-Oncol. 2018, 136, 79–86. [Google Scholar] [CrossRef]
- Hanna, R.; Abdallah, J.; Abou-Antoun, T. A novel mechanism of 17-AAG therapeutic efficacy on HSP90 inhibition in MYCN-amplified neuroblastoma cells. Front. Oncol. 2021, 10, 624560. [Google Scholar] [CrossRef]
- Chen, R.; Zhang, M.; Zhou, Y.; Guo, W.; Yi, M.; Zhang, Z.; Ding, Y.; Wang, Y. The application of histone deacetylases inhibitors in glioblastoma. J. Exp. Clin. Cancer Res. 2020, 39, 138. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Han, S.; Jin, Q.; Zhou, N.; Lu, J.; Shangguan, F.; Yu, S.; Liu, Y.; Wang, L.; Lu, J.; et al. Ciclopirox and bortezomib synergistically inhibits glioblastoma multiforme growth via simultaneously enhancing JNK/p38 MAPK and NF-κB signaling. Cell Death Dis. 2021, 12, 251. [Google Scholar] [CrossRef] [PubMed]
- Villaseñor, R.; Kuennecke, B.; Ozmen, L.; Ammann, M.; Kugler, C.; Grüninger, F.; Loetscher, H.; Freskgård, P.O.; Collin, L. Region-specific permeability of the blood-brain barrier upon pericyte loss. J. Cereb. Blood Flow Metab. 2017, 37, 3683–3694. [Google Scholar] [CrossRef] [PubMed]
- Girolamo, F.; de Trizio, I.; Errede, M.; Longo, G.; D’Amati, A.; Virgintino, D. Neural crest cell-derived pericytes act as pro-angiogenic cells in human neocortex development and gliomas. Fluids Barriers CNS 2021, 18, 14. [Google Scholar] [CrossRef] [PubMed]
- Pearson, J.R.D.; Cuzzubbo, S.; McArthur, S.; Durrant, L.G.; Adhikaree, J.; Tinsley, C.J.; Pockley, A.G.; McArdle, S.E.B. Immune escape in glioblastoma multiforme and the adaptation of immunotherapies for treatment. Front. Immunol. 2020, 11, 582106. [Google Scholar] [CrossRef]
- Nduom, E.K.; Weller, M.; Heimberger, A.B. Immunosuppressive mechanisms in glioblastoma. Neuro Oncol. 2015, 17 (Suppl. S7), vii9–vii14. [Google Scholar] [CrossRef]
- Grabowski, M.M.; Sankey, E.W.; Ryan, K.J.; Chongsathidkiet, P.; Lorrey, S.J.; Wilkinson, D.; Fecci, P.E. Immune suppression in gliomas. J. Neurooncol. 2021, 151, 3–12. [Google Scholar] [CrossRef]
- Towner, R.A.; Jensen, R.L.; Vaillant, B.; Colman, H.; Saunders, D.; Giles, C.B.; Wren, J.D. Experimental validation of 5 in-silico predicted glioma biomarkers. Neuro Oncol. 2013, 15, 1625–1634. [Google Scholar] [CrossRef][Green Version]
- Serban, F.; Artene, S.A.; Georgescu, A.M.; Purcaru, S.O.; Tache, D.E.; Alexandru, O.; Dricu, A. Epidermal growth factor, latrophilin, and seven transmembrane domain-containing protein 1 marker, a novel angiogenesis marker. Onco Targets Ther. 2015, 8, 3767–3774. [Google Scholar] [CrossRef]
- Sevastre, A.S.; Buzatu, I.M.; Baloi, C.; Oprita, A.; Dragoi, A.; Tataranu, L.G.; Alexandru, O.; Tudorache, S.; Dricu, A. ELTD1-An Emerging Silent Actor in Cancer Drama Play. Int. J. Mol. Sci. 2021, 22, 5151. [Google Scholar] [CrossRef]
- Serban, F.; Daianu, O.; Tataranu, L.G.; Artene, S.A.; Emami, G.; Georgescu, A.M.; Alexandru, O.; Purcaru, S.O.; Tache, D.E.; Danciulescu, M.M.; et al. Silencing of epidermal growth factor, latrophilin and seven transmembrane domain-containing protein 1 (ELTD1) via siRNA-induced cell death in glioblastoma. J. Immunoass. Immunochem. 2017, 38, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Ke, S.Q.; Huang, Z.; Flavahan, W.; Fang, X.; Paul, J.; Wu, L.; Sloan, A.E.; McLendon, R.E.; Li, X.; et al. Periostin secreted by glioblastoma stem cells recruits M2 tumour-associated macrophages and promotes malignant growth. Nat. Cell Biol. 2015, 17, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Hambardzumyan, D.; Gutmann, D.H.; Kettenmann, H. The role of microglia and macrophages in glioma maintenance and progression. Nat. Neurosci. 2016, 19, 20–27. [Google Scholar] [CrossRef]
- Wirsching, H.-G.; Roth, P.; Weller, M. A vasculature centric approach to developing novel treatment options for glioblastoma. Expert Opin. Ther. Targets 2021, 25, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Mathiisen, T.M.; Lehre, K.P.; Danbolt, N.C.; Ottersen, O.P. The perivascular astroglial sheath provides a complete covering of the brain microvessels: An electron microscopic 3D reconstruction. Glia 2010, 58, 1094–1103. [Google Scholar] [CrossRef]
- Chen, J.; Mao, S.; Li, H.; Zheng, M.; Yi, L.; Lin, J.M.; Lin, Z.X. The pathological structure of the perivascular niche in different microvascular patterns of glioblastoma. PLoS ONE 2017, 12, e0182183. [Google Scholar] [CrossRef]
- Baloi, C.; Oprita, A. Estimation of immunohistochemical expression of CD34 in nervous system tumours. Med. Oncol. 2021, 2, 80–88. [Google Scholar] [CrossRef]
- Tang, X.; Zuo, C.; Fang, P.; Liu, G.; Qiu, Y.; Huang, Y.; Tang, R. Targeting glioblastoma stem cells: A review on biomarkers, signal pathways and targeted therapy. Front. Oncol. 2021, 11, 701291. [Google Scholar] [CrossRef]
- Leone, R.D.; Powell, J.D. Metabolism of immune cells in cancer. Nat. Rev. Cancer 2020, 20, 516–531. [Google Scholar] [CrossRef]
- Ishii, A.; Kimura, T.; Sadahiro, H.; Kawano, H.; Takubo, K.; Suzuki, M.; Ikeda, E. Histological characterization of the tumorigenic “Peri-Necrotic Niche” harboring quiescent stem-like tumor cells in glioblastoma. PLoS ONE 2016, 11, e0147366. [Google Scholar] [CrossRef]
- Casazza, A.; Laoui, D.; Wenes, M.; Rizzolio, S.; Bassani, N.; Mambretti, M.; Deschoemaeker, S.; Van Ginderachter, J.A.; Tamagnone, L.; Mazzone, M. Impeding macrophage entry into hypoxic tumor areas by Sema3A/Nrp1 signaling blockade inhibits angiogenesis and restores antitumor immunity. Cancer Cell 2013, 24, 695–709. [Google Scholar] [CrossRef] [PubMed]
- Soda, Y.; Marumoto, T.; Friedmann-Morvinski, D.; Soda, M.; Liu, F.; Michiue, H.; Pastorino, S.; Yang, M.; Hoffman, R.M.; Kesari, S.; et al. Transdifferentiation of glioblastoma cells into vascular endothelial cells. Proc. Natl. Acad. Sci. USA 2011, 108, 4274–4280. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.; Sewell-Loftin, M.K. Mechanoregulation of vascular endothelial growth factor receptor 2 in angiogenesis. Front. Cardiovasc. Med. 2022, 8, 804934. [Google Scholar] [CrossRef]
- Abdullah, C.; Korkaya, H.; Iizuka, S.; Courtneidge, S.A. SRC increases MYC mRNA expression in estrogen receptor-positive breast cancer via mRNA stabilization and inhibition of p53 function. Mol. Cell Biol. 2018, 38, e00463-17. [Google Scholar] [CrossRef]
- Liu, Z.; Han, L.; Dong, Y.; Tan, Y.; Li, Y.; Zhao, M.; Xie, H.; Ju, H.; Wang, H.; Zhao, Y.; et al. EGFRvIII/integrin β3 interaction in hypoxic and vitronectin enriching microenvironment promote GBM progression and metastasis. Oncotarget 2016, 7, 4680–4694. [Google Scholar] [CrossRef]
- Torrisi, F.; Vicario, N.; Spitale, F.M.; Cammarata, F.P.; Minafra, L.; Salvatorelli, L.; Russo, G.; Cuttone, G.; Valable, S.; Gulino, R.; et al. The role of hypoxia and SRC tyrosine kinase in glioblastoma invasiveness and radioresistance. Cancers 2020, 12, 2860. [Google Scholar] [CrossRef] [PubMed]
- Du, R.; Petritsch, C.; Lu, K.; Liu, P.; Haller, A.; Ganss, R.; Song, H.; Vandenberg, S.; Bergers, G. Matrix metalloproteinase-2 regulates vascular patterning and growth affecting tumor cell survival and invasion in GBM. Neuro Oncol. 2008, 10, 254–264. [Google Scholar] [CrossRef]
- Sattiraju, A.; Mintz, A. Pericytes in glioblastomas: Multifaceted role within tumor microenvironments and potential for therapeutic interventions. Adv. Exp. Med. Biol. 2019, 1147, 65–91. [Google Scholar] [CrossRef]
- Edwards, L.A.; Woolard, K.; Son, M.J.; Li, A.; Lee, J.; Ene, C.; Mantey, S.A.; Maric, D.; Song, H.; Belova, G.; et al. Effect of brain- and tumor-derived connective tissue growth factor on glioma invasion. J. Natl. Cancer Inst. 2011, 103, 1162–1178. [Google Scholar] [CrossRef]
- Alexandru, O.; Horescu, C.; Sevastre, A.S.; Cioc, C.E.; Baloi, C.; Oprita, A.; Dricu, A. Receptor tyrosine kinase targeting in glioblastoma: Performance, limitations and future approaches. Contemp. Oncol. 2020, 24, 55–66. [Google Scholar] [CrossRef]
- Petrova, R.; Joyner, A.L. Roles for Hedgehog signaling in adult organ homeostasis and repair. Development 2014, 141, 3445–3457. [Google Scholar] [CrossRef] [PubMed]
- Negri, V.A.; Logtenberg, M.E.W.; Renz, L.M.; Oules, B.; Walko, G.; Watt, F.M. Delta-like 1-mediated cis-inhibition of Jagged1/2 signalling inhibits differentiation of human epidermal cells in culture. Sci. Rep. 2019, 9, 10825. [Google Scholar] [CrossRef] [PubMed]
- Rajakulendran, N.; Rowland, K.J.; Selvadurai, H.J.; Ahmadi, M.; Park, N.I.; Naumenko, S.; Dolma, S.; Ward, R.J.; So, M.; Lee, L.; et al. Wnt and Notch signaling govern self-renewal and differentiation in a subset of human glioblastoma stem cells. Genes Dev. 2019, 33, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Zheng, Y.; Ma, L.; Tian, L.; Sun, Q. Clinically-relevant ABC transporter for anti-cancer drug resistance. Front. Pharmacol. 2021, 12, 648407. [Google Scholar] [CrossRef] [PubMed]
- Ohgaki, H. Genetic pathways to glioblastoma: A population-based study. Cancer Res. 2004, 64, 6892–6899. [Google Scholar] [CrossRef] [PubMed]
- Hoogstrate, Y.; Vallentgoed, W.; Kros, J.M.; de Heer, I.; de Wit, M.; Eoli, M.; Sepulveda, J.M.; Walenkamp, A.M.E.; Frenel, J.S.; Franceschi, E.; et al. EGFR mutations are associated with response to depatux-m in combination with temozolomide and result in a receptor that is hypersensitive to ligand. Neuro-Oncol. Adv. 2020, 2, 51. [Google Scholar] [CrossRef]
- Song, H.; Wang, Y.; Shi, C.; Lu, J.; Yuan, T.; Wang, X. SH3KBP1 promotes glioblastoma tumorigenesis by activating EGFR signaling. Front. Oncol. 2021, 10, 583984. [Google Scholar] [CrossRef]
- Tilak, M.; Holborn, J.; New, L.A.; Lalonde, J.; Jones, N. Receptor tyrosine kinase signaling and targeting in glioblastoma multiform. Int. J. Mol. Sci. 2021, 22, 1831. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, X.; Han, H.; Lv, J.-N.; Kang, E.-M.; Zhang, Y.-L.; Liu, W.-P.; He, X.-S.; Wang, J.; Wang, G.-H.; et al. The different role of YKL-40 in glioblastoma is a function of MGMT promoter methylation status. Cell Death Dis. 2020, 11, 668. [Google Scholar] [CrossRef]
- Stevanovic, M.; Kovacevic-Grujicic, N.; Mojsin, M.; Milivojevic, M.; Drakulic, D. SOX transcription factors and glioma stem cells: Choosing between stemness and differentiation. World J. Stem Cells 2021, 13, 1417–1445. [Google Scholar] [CrossRef]
- Sachdeva, R.; Wu, M.; Johnson, K.; Kim, H.; Celebre, A.; Shahzad, U.; Graham, M.S.; Kessler, J.A.; Chuang, J.H.; Karamchandani, J.; et al. BMP signaling mediates glioma stem cell quiescence and confers treatment resistance in glioblastoma. Sci. Rep. 2019, 9, 14569. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, A.M.; Morais, C.M.; Pena, F.; Marante, T.; Cunha, P.P.; Jurado, A.S.; Pedroso de Lima, M.C. Differentiation of glioblastoma stem cells promoted by miR-128 or miR-302a overexpression enhances senescence-associated cytotoxicity of axitinib. Hum. Mol. Genet. 2021, 30, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhou, W.; Li, X.; Ren, J.; Ji, G.; Du, J.; Tian, W.; Liu, Q.; Hao, A. Graphene oxide suppresses the growth and malignancy of glioblastoma stem cell-like spheroids via epigenetic mechanisms. J. Transl. Med. 2020, 18, 200. [Google Scholar] [CrossRef]
- Allani, S.K.; Weissbach, H.; Lopez Toledano, M.A. Sulindac induces differentiation of glioblastoma stem cells making them more sensitive to oxidative stress. Neoplasma 2018, 65, 376–388. [Google Scholar] [CrossRef]
- Chen, S.R.; Chen, M.M.; Ene, C.; Lang, F.F.; Kan, P. Perfusion-guided endovascular super-selective intra-arterial infusion for treatment of malignant brain tumors. J. Neurointerv. Surg. 2021. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, N.; Hayashi, T.; Fujiki, K.; Shirahige, K.; Akiyama, T.; Akutsu, T.; Nakato, R. Codependency and mutual exclusivity for gene community detection from sparse single-cell transcriptome data. bioRxiv 2021. [Google Scholar] [CrossRef]
- Wouters, R.; Bevers, S.; Riva, M.; De Smet, F.; Coosemans, A. Immunocompetent Mouse Models in the Search for Effective Immunotherapy in Glioblastoma. Cancers 2021, 13, 19. [Google Scholar] [CrossRef]
- Jain, S.; Chalif, E.J.; Aghi, M.K. Interactions between anti-angiogenic therapy and immunotherapy in glioblastoma. Front. Oncol. 2022, 11, 812916. [Google Scholar] [CrossRef]
- Zhu, H.; Zhang, S. Hypoxia inducible factor-1α/vascular endothelial growth factor signaling activation correlates with response to radiotherapy and its inhibition reduces hypoxia-induced angiogenesis in lung cancer. J. Cell Biochem. 2018, 119, 7707–7718. [Google Scholar] [CrossRef]
- Noushmehr, H.; Weisenberger, D.J.; Diefes, K.; Phillips, H.S.; Pujara, K.; Berman, B.P.; Pan, F.; Pelloski, C.E.; Sulman, E.P.; Bhat, K.P. Cancer Genome Atlas Research Network. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010, 17, 510–522. [Google Scholar] [CrossRef]
- Nishiyama, A.; Nakanishi, M. Navigating the DNA methylation landscape of cancer. Trends Genet. 2021, 37, 1012–1027. [Google Scholar] [CrossRef]
- Henriksen, M.; Johnsen, K.B.; Andersen, H.H.; Pilgaard, L.; Duroux, M. MicroRNA expression signatures determine prognosis and survival in glioblastoma multiforme--a systematic overview. Mol. Neurobiol. 2014, 50, 896–913. [Google Scholar] [CrossRef] [PubMed]
- Abdouh, M.; Hanna, R.; El Hajjar, J.; Flamier, A.; Bernier, G. The Polycomb Repressive Complex 1 Protein BMI1 Is Required for Constitutive Heterochromatin Formation and Silencing in Mammalian Somatic Cells. J. Biol. Chem. 2016, 291, 182–197. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Kim, M.; Woo, D.H.; Shin, Y.; Shin, J.; Chang, N.; Oh, Y.T.; Kim, H.; Rheey, J.; Nakano, I.; et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell 2013, 23, 839–852. [Google Scholar] [CrossRef] [PubMed]
- Laugesen, A.; Højfeldt, J.W.; Helin, K. Role of the Polycomb Repressive Complex 2 (PRC2) in Transcriptional Regulation and Cancer. Cold Spring Harb. Perspect. Med. 2016, 6, a026575. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, M.; Wang, D.; Hou, P.; Chen, X.; Chu, S.; Chai, D.; Zheng, J.; Bai, J. Post-translational modifications of EZH2 in cancer. Cell Biosci. 2020, 10, 143. [Google Scholar] [CrossRef]
- Valor, L.M.; Hervás-Corpión, I. The Epigenetics of Glioma Stem Cells: A Brief Overview. Front. Oncol. 2020, 10, 602378. [Google Scholar] [CrossRef]
- Jin, X.; Kim, L.J.Y.; Wu, Q.; Wallace, L.C.; Prager, B.C.; Sanvoranart, T.; Gimple, R.C.; Wang, X.; Mack, S.C.; Miller, T.; et al. Targeting glioma stem cells through combined BMI1 and EZH2 inhibition. Nat. Med. 2017, 23, 1352–1361. [Google Scholar] [CrossRef]
- Saito, N.; Aoki, K.; Hirai, N.; Fujita, S.; Iwama, J.; Ikota, M.; Nakayama, H.; Hayashi, M.; Ito, K.; Sakurai, T.; et al. Notch Pathway Activation Predicts Resistance to Bevacizumab Therapy in Glioblastoma. Cancer Res. 2017, 77 (Suppl. S13), 774. [Google Scholar] [CrossRef]
- Carruthers, R.D.; Ahmed, S.U.; Ramachandran, S.; Strathdee, K.; Kurian, K.M.; Hedley, A.; Gomez-Roman, N.; Kalna, G.; Neilson, M.; Gilmour, L.; et al. Replication stress drives constitutive activation of the DNA damage response and radioresistance in glioblastoma stem-like cells. Cancer Res. 2018, 78, 5060–5071. [Google Scholar] [CrossRef]
- Weiss, F.; Lauffenburger, D.; Friedl, P. Towards targeting of shared mechanisms of cancer metastasis and therapy resistance. Nat. Rev. Cancer 2022, 22, 157–173. [Google Scholar] [CrossRef] [PubMed]
- Clough, E.; Ballinger, T.; Semple, C.; Strathdee, K.; Carruthers, R. STEM-22. Targeting replication stress response for glioma stem cell specific cytotoxicity. Neuro-Oncology 2021, 23, vi25–vi26. [Google Scholar] [CrossRef]
- Bayin, N.S.; Frenster, J.D.; Sen, R.; Si, S.; Modrek, A.S.; Galifianakis, N.; Dolgalev, I.; Ortenzi, V.; Illa-Bochaca, I.; Khahera, A.; et al. Notch signaling regulates metabolic heterogeneity in glioblastoma stem cells. Oncotarget 2017, 8, 64932–64953. [Google Scholar] [CrossRef] [PubMed]
- Dantas-Barbosa, C.; Bergthold, G.; Daudigeos-Dubus, E.; Blockus, H.; Boylan, J.F.; Ferreira, C.; Puget, S.; Abely, M.; Vassal, G.; Grill, J.; et al. Inhibition of the NOTCH pathway using γ-secretase inhibitor RO4929097 has limited antitumor activity in established glial tumors. Anticancer Drugs 2015, 26, 272–283. [Google Scholar] [CrossRef]
- Kaloshi, G.; Mokhtari, K.; Carpentier, C.; Taillibert, S.; Lejeune, J.; Marie, Y.; Delattre, J.Y.; Godbout, R.; Sanson, M. FABP7 expression in glioblastomas: Relation to prognosis, invasion and EGFR status. J. Neurooncol. 2007, 84, 245–248. [Google Scholar] [CrossRef]
- Sarkar, S.; Mirzaei, R.; Zemp, F.J.; Wei, W.; Senger, D.L.; Robbins, S.M.; Yong, V.W. Activation of NOTCH Signaling by Tenascin-C Promotes Growth of Human Brain Tumor-Initiating Cells. Cancer Res. 2017, 77, 3231–3243. [Google Scholar] [CrossRef]
- Clement, V.; Sanchez, P.; de Tribolet, N.; Radovanovic, I.; Ruiz i Altaba, A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr. Biol. 2007, 17, 165–172. [Google Scholar] [CrossRef]
- Daood, M.; Tsai, C.; Ahdab-Barmada, M.; Watchko, J.F. ABC transporter (P-gp/ABCB1, MRP1/ABCC1, BCRP/ABCG2) expression in the developing human CNS. Neuropediatrics 2008, 39, 211–218. [Google Scholar] [CrossRef]
- Al-Eitan, L.N.; Rababa’h, D.M.; Alghamdi, M.A.; Khasawneh, R.H. Role of Four ABC Transporter Genes in Pharmacogenetic Susceptibility to Breast Cancer in Jordanian Patients. J. Oncol. 2019, 2019, 6425708. [Google Scholar] [CrossRef]
- Zbinden, M.; Duquet, A.; Lorente-Trigos, A.; Ngwabyt, S.N.; Borges, I.; Ruiz i Altaba, A. NANOG regulates glioma stem cells and is essential in vivo acting in a cross-functional network with GLI1 and p53. EMBO J. 2010, 29, 2659–2674. [Google Scholar] [CrossRef]
- Ma, Y.; Yu, W.; Shrivastava, A.; Alemi, F.; Lankachandra, K.; Srivastava, R.K.; Shankar, S. Sanguinarine inhibits pancreatic cancer stem cell characteristics by inducing oxidative stress and suppressing sonic hedgehog-Gli-Nanog pathway. Carcinogenesis 2017, 38, 1047–1056. [Google Scholar] [CrossRef] [PubMed]
- Torrisi, F.; Alberghina, C.; Lo Furno, D.; Zappalà, A.; Valable, S.; Li Volti, G.; Tibullo, D.; Vicario, N.; Parenti, R. Connexin 43 and Sonic Hedgehog pathway interplay in glioblastoma cell proliferation and migration. Biology 2021, 10, 767. [Google Scholar] [CrossRef] [PubMed]
- Kalani, M.Y.; Cheshier, S.H.; Cord, B.J.; Bababeygy, S.R.; Vogel, H.; Weissman, I.L.; Palmer, T.D.; Nusse, R. Wnt-mediated self-renewal of neural stem/progenitor cells. Proc. Natl. Acad. Sci. USA 2008, 105, 16970–16975. [Google Scholar] [CrossRef] [PubMed]
- Binda, E.; Visioli, A.; Giani, F.; Trivieri, N.; Palumbo, O.; Restelli, S.; Dezi, F.; Mazza, T.; Fusilli, C.; Legnani, F.; et al. Wnt5a Drives an Invasive Phenotype in Human Glioblastoma Stem-like Cells. Cancer Res. 2017, 77, 996–1007. [Google Scholar] [CrossRef]
- Zheng, H.; Ying, H.; Wiedemeyer, R.; Yan, H.; Quayle, S.N.; Ivanova, E.V.; Paik, J.H.; Zhang, H.; Xiao, Y.; Perry, S.R.; et al. PLAGL2 regulates Wnt signaling to impede differentiation in neural stem cells and gliomas. Cancer Cell 2010, 17, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, K.H.; Kim, D.G.; Cho, H.J.; Kim, Y.; Rheey, J.; Shin, K.; Seo, Y.J.; Choi, Y.S.; Lee, J.I.; et al. FoxM1 Promotes Stemness and Radio-Resistance of Glioblastoma by Regulating the Master Stem Cell Regulator Sox2. PLoS ONE 2015, 10, e0137703. [Google Scholar] [CrossRef]
- Augustin, I.; Goidts, V.; Bongers, A.; Kerr, G.; Vollert, G.; Radlwimmer, B.; Hartmann, C.; Herold-Mende, C.; Reifenberger, G.; von Deimling, A.; et al. The Wnt secretion protein Evi/Gpr177 promotes glioma tumourigenesis. EMBO Mol. Med. 2012, 4, 38–51. [Google Scholar] [CrossRef]
- Hu, B.; Wang, Q.; Wang, Y.A.; Hua, S.; Sauvé, C.G.; Ong, D.; Lan, Z.D.; Chang, Q.; Ho, Y.W.; Monasterio, M.M.; et al. Epigenetic Activation of WNT5A Drives Glioblastoma Stem Cell Differentiation and Invasive Growth. Cell 2016, 167, 1281–1295.e18. [Google Scholar] [CrossRef]
- Wickström, M.; Dyberg, C.; Milosevic, J.; Einvik, C.; Calero, R.; Sveinbjörnsson, B.; Sandén, E.; Darabi, A.; Siesjö, P.; Kool, M.; et al. Wnt/β-catenin pathway regulates MGMT gene expression in cancer and inhibition of Wnt signalling prevents chemoresistance. Nat. Commun. 2015, 6, 8904. [Google Scholar] [CrossRef]
- El-Khayat, S.M.; Arafat, W.O. Therapeutic strategies of recurrent glioblastoma and its molecular pathways ‘Lock up the beast’. Ecancermedicalscience 2021, 15, 1176. [Google Scholar] [CrossRef]
- Kumar, V.; Vashishta, M.; Kong, L.; Wu, X.; Lu, J.J.; Guha, C.; Dwarakanath, B.S. The role of Notch, Hedgehog, and Wnt signaling pathways in the resistance of tumors to anticancer therapies. Front. Cell Dev. Biol. 2021, 9, 650772. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).