Integrative Physiological and Transcriptomic Analysis Reveals the Transition Mechanism of Sugar Phloem Unloading Route in Camellia oleifera Fruit
Abstract
1. Introduction
2. Results
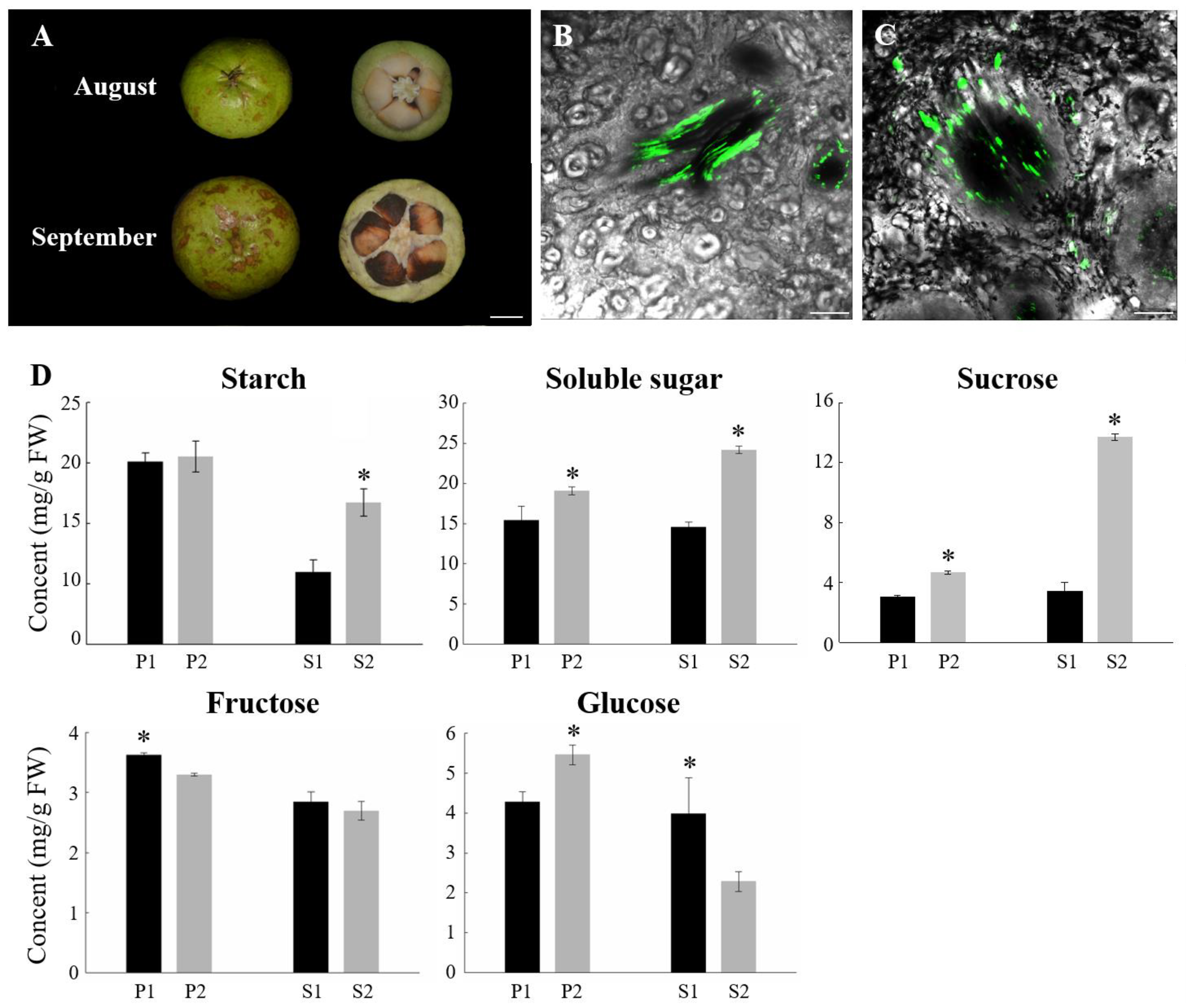
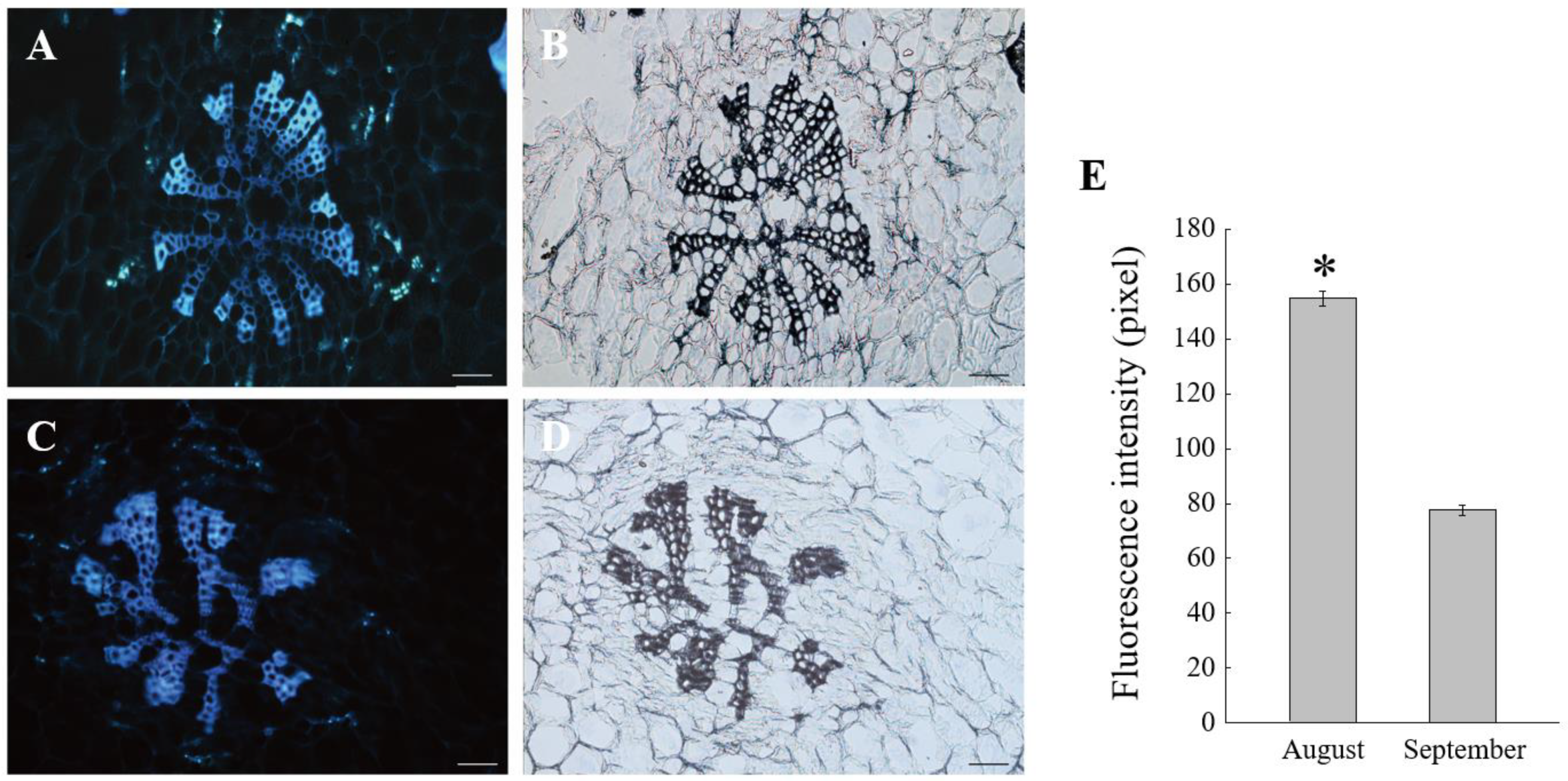
2.1. Phloem Unloading Pathway Altered during the Late Period of C. oleifera Fruit Development
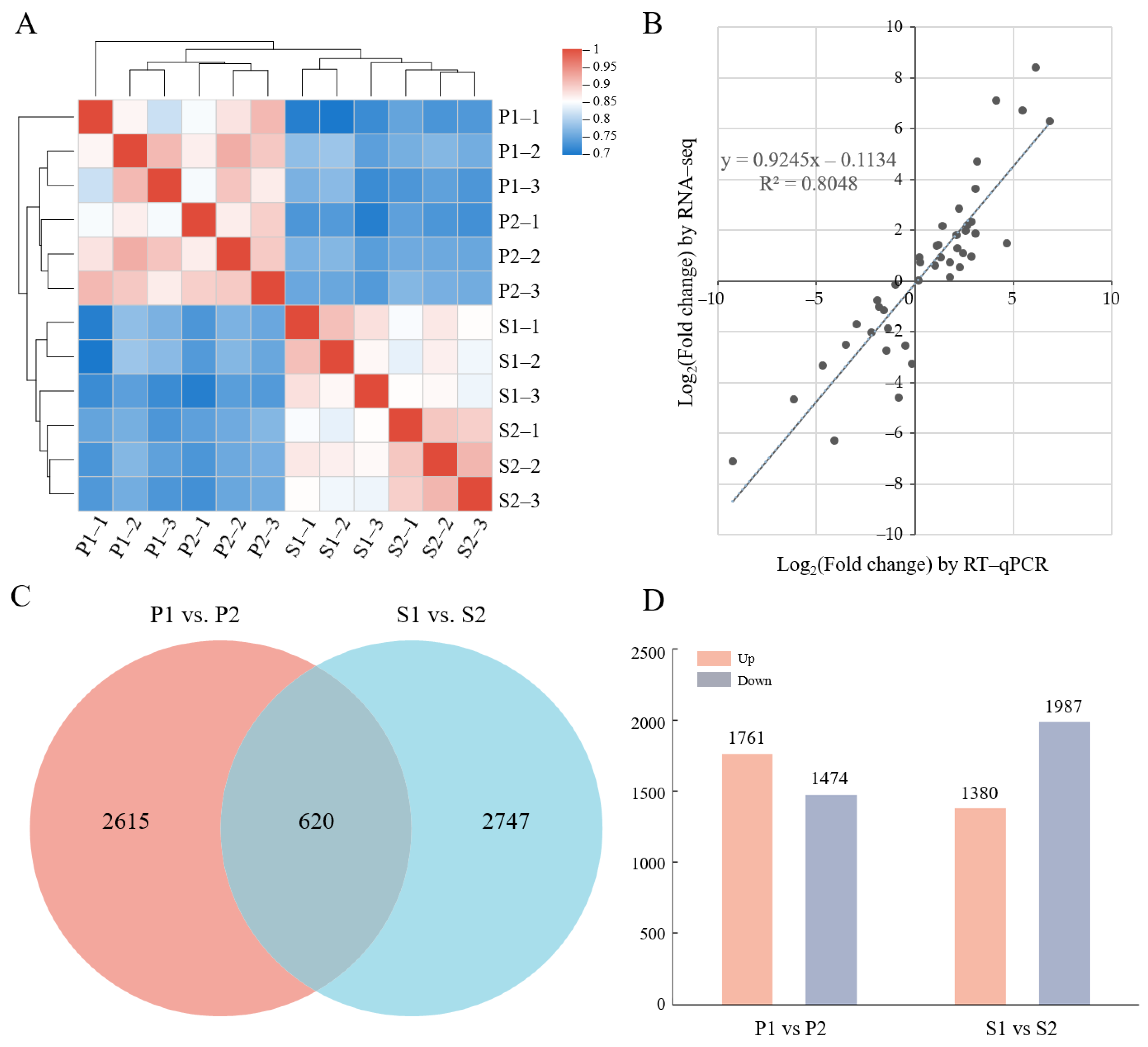
2.2. Transcriptional Profiling of C. oleifera Pericarp and Seeds at Different Stages of Phloem Unloading
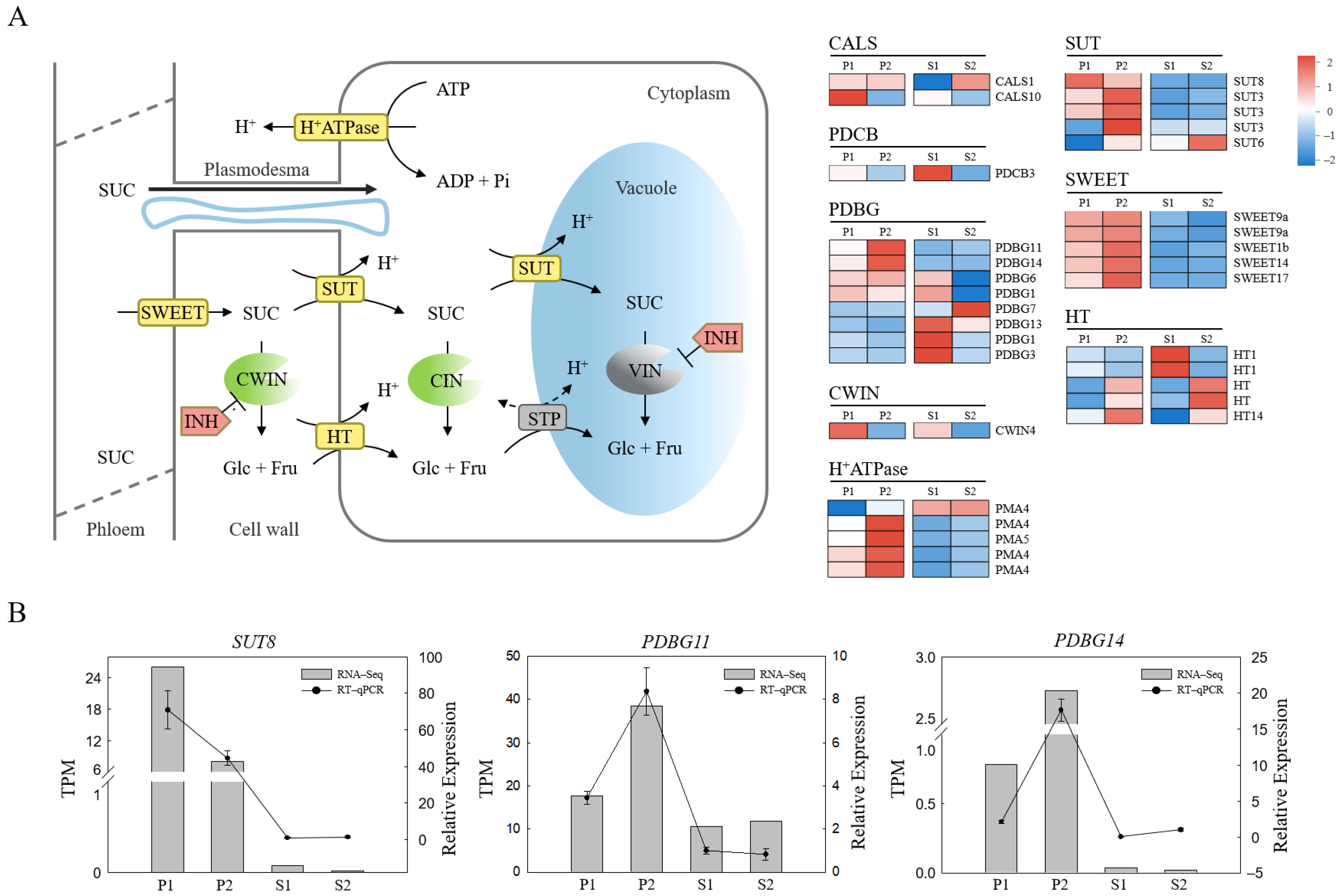
2.3. Screening of Differentially Expressed Structural Genes Involved in Phloem Unloading
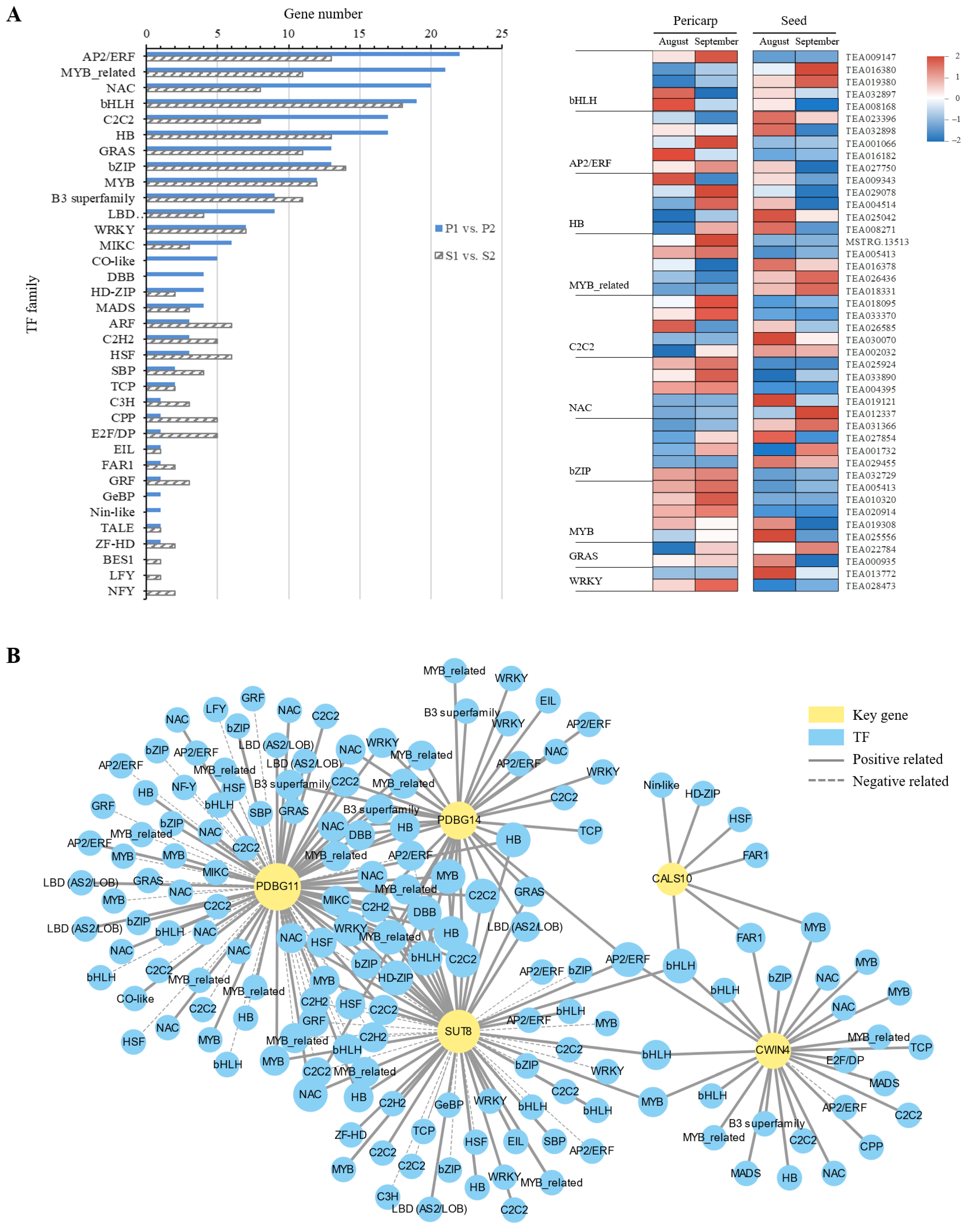
2.4. Differentially Expressed TFs and Their Co-Expression Network with Structural Genes
2.5. Biological Processes Change with the Shift of the Phloem Unloading Route in Oil Tea
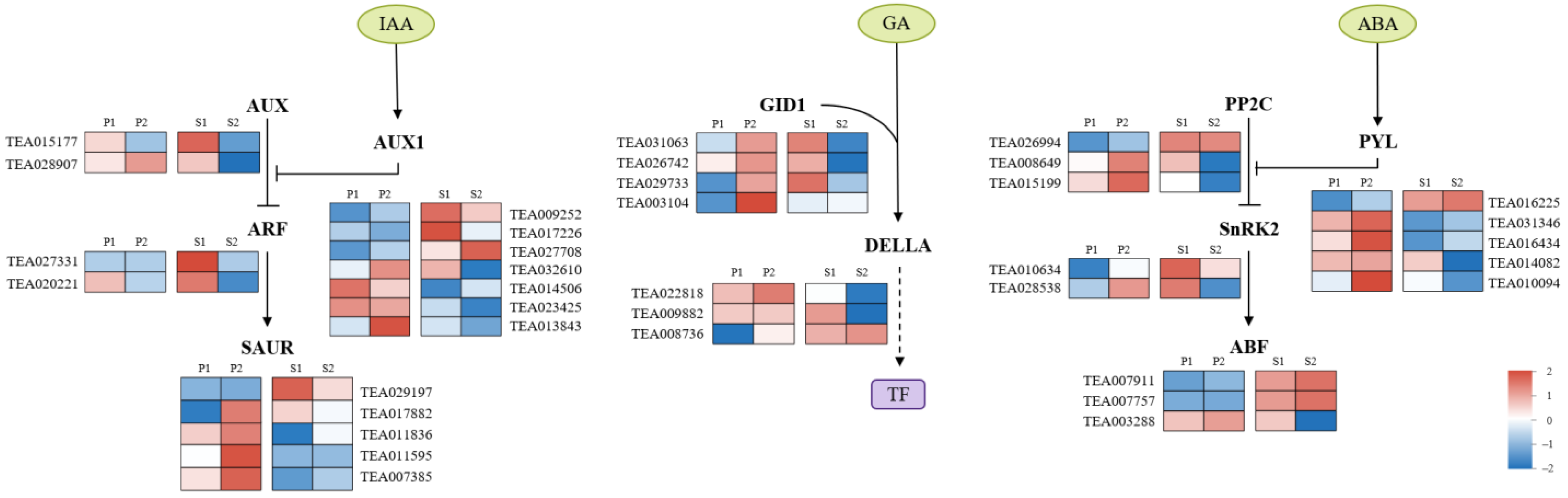
2.6. Plant Hormone Metabolism Responding to the Change of Phloem Unloading Pathways in C. oleifera Fruit
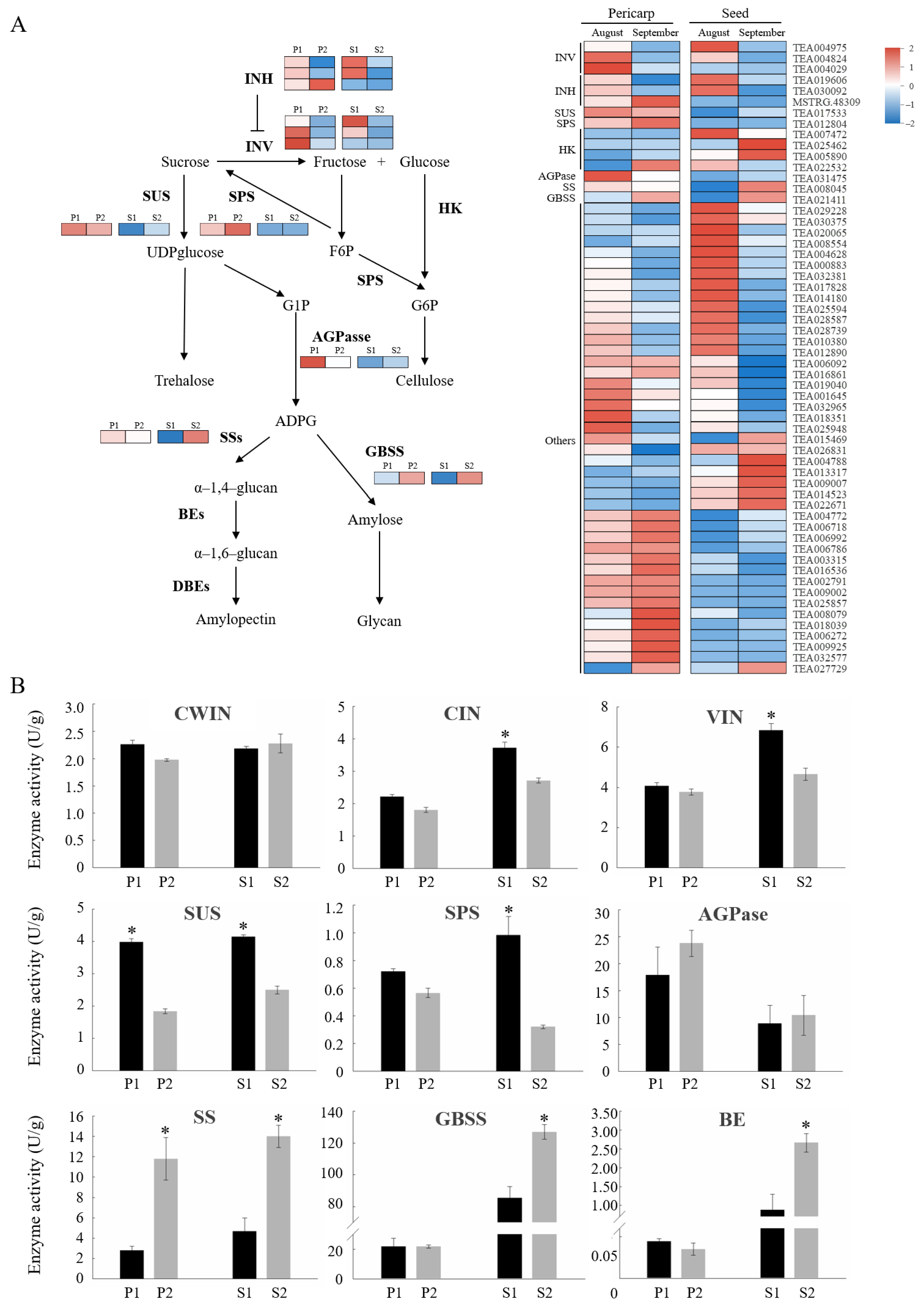
2.7. Starch and Sucrose Metabolism Contributed to Sucrose Phloem Unloading in C. oleifera Fruit
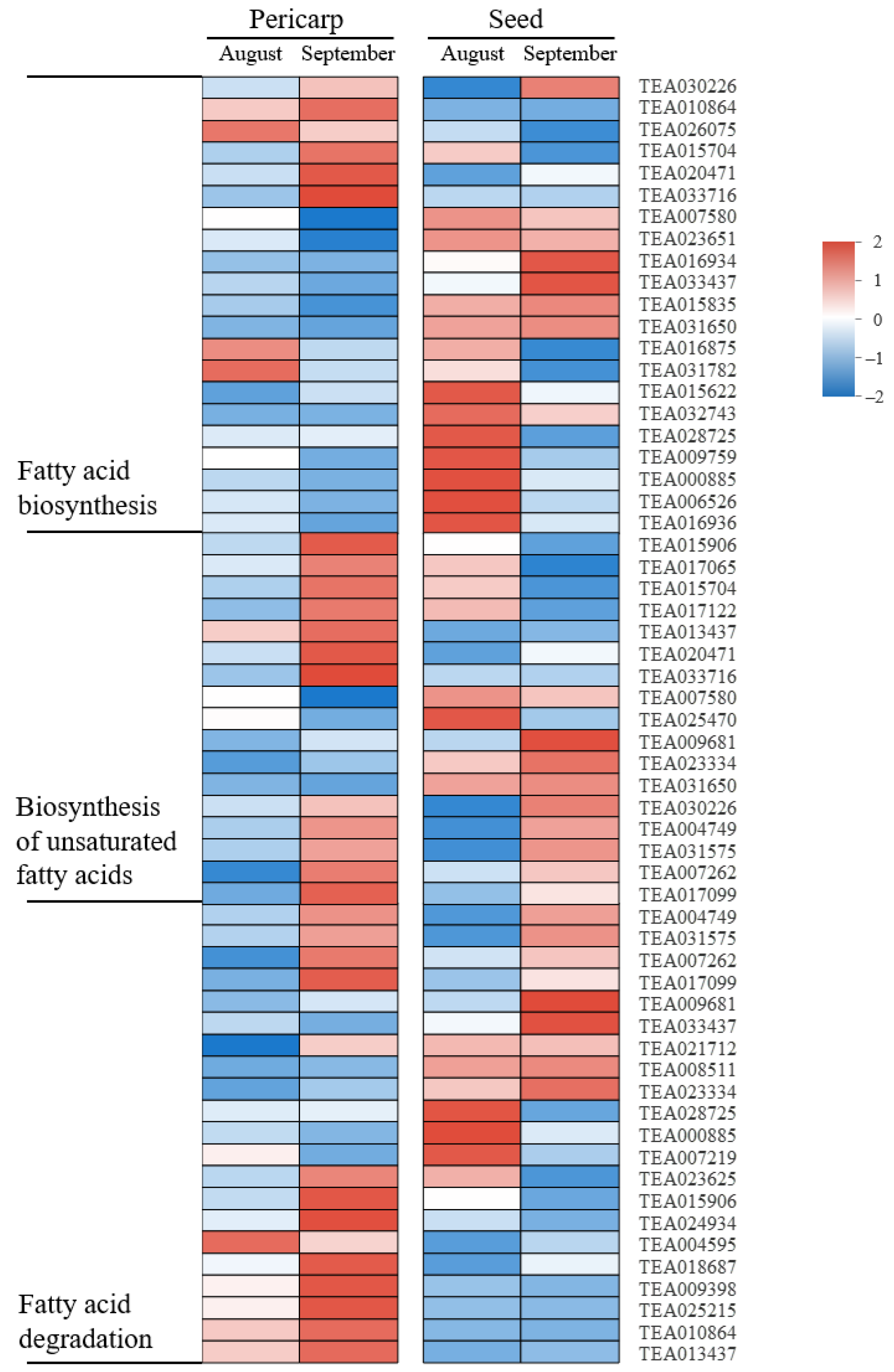
2.8. Lipid Metabolism-Related Pathways at the Two Developmental Stages of C. oleifera Fruit
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Carboxy Fluorescein Diacetate Labeling
4.3. Paraffin Sections and Fluorescent Labeling of Callose
4.4. Paraffin Sections and Fluorescent Labeling of Callose
4.5. Physiological Measurements
4.6. RNA Extraction and Sequencing (RNA-Seq)
4.7. RNA-Seq Data Processing and Analysis
4.8. Quantitative Real-Time PCR (qRT-PCR) Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| ABF | ABA responsive element binding factor |
| ADP | Adenosine diphosphate |
| ADPG | Adenosine diphosphoglu |
| AGPase | ADP-glucose pyrophosphorylase |
| ARF | Auxin response factor |
| ATP | Adenosine triphosphate |
| AUX | Auxin-responsive protein IAA |
| AUX1 | Auxin influx carrier (AUX1 LAX family) |
| BE | Branching enzyme |
| CIN | Cytoplasmic invertases |
| CWIN | Cell wall invertase |
| DBE | Debranching enzyme |
| DELLA | DELLA protein |
| F6P | Fructose−6−phosphate |
| Fru | Fructose |
| G1P | Alpha-D-Glucose 1-phosphate |
| G6P | Glucose−6−phosphate |
| GA | Gibberellic acid |
| GBSS | Granule-bound starch synthase |
| GID1 | Gibberellin receptor GID1 |
| Glc | Glucose |
| H+-ATPase | H+adenosine triphosphatase ATPase |
| HK | Hexokinase |
| HT | Hexose transporter |
| IAA | Auxin |
| INH | Invertase inhibitor |
| INV | Invertases |
| PP2C | Protein phosphatase 2C |
| PYL | Abscisic acid receptor PYR/PYL family |
| SAUR | SAUR family protein |
| SnRK2 | Serine/Threonine-protein kinase SRK2 |
| SPS | Sucrose phosphate synthase |
| SS | Starch synthase |
| STP | Sugar transport protein |
| SUS | Sucrose synthase |
| SUT | Sucrose transporters |
| UDPglucose | Uridine diphosphate glucose |
| VIN | Vacuolar invertases |
References
- De Schepper, V.; De Swaef, T.; Bauweraerts, I.; Steppe, K. Phloem transport: A review of mechanisms and controls. J. Exp. Bot. 2013, 64, 4839–4850. [Google Scholar] [CrossRef] [PubMed]
- Turgeon, R.; Wolf, S. Phloem transport: Cellular pathways and molecular trafficking. Annu. Rev. Plant Biol. 2009, 60, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Feas, X.; Estevinho, L.M.; Salinero, C.; Vela, P.; Sainz, M.J.; Vazquez-Tato, M.P.; Seijas, J.A. Triacylglyceride, antioxidant and antimicrobial features of virgin Camellia oleifera, C. reticulata and C. sasanqua oils. Molecules 2013, 18, 4573–4587. [Google Scholar] [CrossRef]
- Ye, Z.; Yu, J.; Yan, W.; Zhang, J.; Yang, D.; Yao, G.; Liu, Z.; Wu, Y.; Hou, X. Integrative iTRAP-based proteomic and transcriptomic analysis reveals the accumulation patterns of key metabolites associated with oil quality during seed ripening of Camellia oleifera. Hortic. Res. 2021, 8, 157. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.L.; Chen, R.F.; Zhao, X.Q.; Shen, R.F.; Noguchi, A.; Shinmachi, F.; Hasegawa, I. Aluminum could be transported via phloem in Camellia oleifera Abel. Tree Physiol. 2013, 33, 96–105. [Google Scholar] [CrossRef]
- Wang, X.; You, H.; Yuan, Y.; Zhang, H.; Zhang, L. The cellular pathway and enzymatic activity for phloem-unloading transition in developing Camellia oleifera Abel. Fruit. Acta Physiol. Plant. 2018, 40, 1–11. [Google Scholar] [CrossRef]
- Hu, L.; Sun, H.; Li, R.; Zhang, L.; Wang, S.; Sui, X.; Zhang, Z. Phloem unloading follows an extensive apoplasmic pathway in cucumber (Cucumis sativus L.) fruit from anthesis to marketable maturing stage. Plant Cell Environ. 2011, 34, 1835–1848. [Google Scholar] [CrossRef]
- Yang, J.L.; Luo, D.P.; Yang, B.; Frommer, W.B.; Eom, J.S. Sweet11 and 15 as key players in seed filling in rice. New Phytol. 2018, 218, 604–615. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Wang, X.L.; Wang, X.F.; Xia, G.H.; Pan, Q.H.; Fan, R.C.; Wu, F.Q.; Yu, X.C.; Zhang, D.P. A shift of phloem unloading from symplasmic to apoplasmic pathway is involved in developmental onset of ripening in grape berry. Plant Physiol. 2006, 142, 220–232. [Google Scholar] [CrossRef]
- Fishman, S.; Génard, M. A biophysical model of fruit growth: Simulation of seasonal and diurnal dynamics of mass. Plant Cell Environ. 2010, 21, 739–752. [Google Scholar] [CrossRef]
- Grappadelli, R.L.C. Vascular flows and transpiration affect peach (Prunus persica Batsch.) fruit daily growth. J. Exp. Bot. 2007, 58, 3941–3947. [Google Scholar]
- Nie, P.; Wang, X.; Hu, L.; Zhang, H.; Zhang, J.; Zhang, Z.; Zhang, L. The predominance of the apoplasmic phloem-unloading pathway is interrupted by a symplasmic pathway during Chinese jujube fruit development. Plant Cell Physiol. 2010, 51, 1007–1018. [Google Scholar] [CrossRef] [PubMed]
- Zanon, L.; Falchi, R.; Hackel, A.; Kühn, C.; Vizzotto, G. Expression of peach sucrose transporters in heterologous systems points out their different physiological role. Plant Sci. 2015, 238, 262–272. [Google Scholar] [CrossRef]
- Zanon, L.; Falchi, R.; Santi, S.; Vizzotto, G. Sucrose transport and phloem unloading in peach fruit: Potential role of two transporters localized in different cell types. Physiol. Plant. 2015, 154, 179–193. [Google Scholar] [CrossRef]
- Guo, Y.; Song, H.; Zhao, Y.; Qin, X.; Cao, Y.; Zhang, L. Switch from symplasmic to aspoplasmic phloem unloading in Xanthoceras sorbifolia fruit and sucrose influx XsSWEET10 as a key candidate for sugar transport. Plant Sci. Int. J. Exp. Plant Biol. 2021, 313, 111089. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.W.; Kumar, R.; Iswanto, A.B.B.; Kim, J.Y. Callose balancing at plasmodesmata. J. Exp. Bot. 2018, 69, 5325–5339. [Google Scholar] [CrossRef]
- De Storme, N.; Geelen, D. Callose homeostasis at plasmodesmata: Molecular regulators and developmental relevance. Front. Plant Sci. 2014, 5, 138. [Google Scholar] [CrossRef]
- Han, X.; Huang, L.J.; Feng, D.; Jiang, W.; Miu, W.; Li, N. Plasmodesmata-related structural and functional proteins: The long sought-after secrets of a cytoplasmic channel in plant cell walls. Int. J. Mol. Sci. 2019, 20, 2946. [Google Scholar] [CrossRef]
- Xie, B.; Wang, X.; Zhu, M.; Zhang, Z.; Hong, Z. CALS7 encodes a callose synthase responsible for callose deposition in the phloem. Plant J. 2011, 65, 1–14. [Google Scholar] [CrossRef]
- Levy, A.; Guenoune-Gelbart, D.; Epel, B.L. Beta-1,3-glucanases: Plasmodesmal gate keepers for intercellular communication. Plant Signal Behav. 2007, 2, 404–407. [Google Scholar] [CrossRef]
- Amari, K.; Boutant, E.; Hofmann, C.; Schmitt-Keichinger, C.; Fernandez-Calvino, L.; Didier, P.; Lerich, A.; Mutterer, J.; Thomas, C.L.; Heinlein, M.; et al. A family of plasmodesmal proteins with receptor-like properties for plant viral movement proteins. PLoS Pathog. 2010, 6, e1001119. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Hyun, T.K.; Zhang, M.H.; Kumar, R.; Koh, E.J.; Kang, B.H.; Lucas, W.J.; Kim, J.Y. Auxin-callose-mediated plasmodesmal gating is essential for tropic auxin gradient formation and signaling. Dev. Cell 2014, 28, 132–146. [Google Scholar] [CrossRef] [PubMed]
- Levy, A.; Erlanger, M.; Rosenthal, M.; Epel, B.L. A plasmodesmata-associated beta-1,3-glucanase in Arabidopsis. Plant J. 2007, 49, 669–682. [Google Scholar] [CrossRef] [PubMed]
- Simpson, C.; Thomas, C.; Findlay, K.; Bayer, E.; Maule, A.J. An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 2009, 21, 581–594. [Google Scholar] [CrossRef]
- Vaten, A.; Dettmer, J.; Wu, S.; Stierhof, Y.D.; Miyashima, S.; Yadav, S.R.; Roberts, C.J.; Campilho, A.; Bulone, V.; Lichtenberger, R.; et al. Callose biosynthesis regulates symplastic trafficking during root development. Dev. Cell 2011, 21, 1144–1155. [Google Scholar] [CrossRef] [PubMed]
- Abelenda, J.A.; Bergonzi, S.; Oortwijn, M.; Sonnewald, S.; Du, M.; Visser, R.G.F.; Sonnewald, U.; Bachem, C.W.B. Source-sink regulation is mediated by interaction of an FT homolog with a SWEET protein in potato. Curr. Biol. 2019, 29, 1178–1186.e1176. [Google Scholar] [CrossRef] [PubMed]
- Buttner, M. The monosaccharide transporter(-like) gene family in Arabidopsis. FEBS Lett. 2007, 581, 2318–2324. [Google Scholar] [CrossRef]
- Buttner, M. The Arabidopsis sugar transporter (AtSTP) family: An update. Plant Biol. 2010, 12, 35–41. [Google Scholar] [CrossRef]
- Milne, R.J.; Perroux, J.M.; Rae, A.L.; Reinders, A.; Ward, J.M.; Offler, C.E.; Patrick, J.W.; Grof, C.P. Sucrose transporter localization and function in phloem unloading in developing stems. Plant Physiol. 2017, 173, 1330–1341. [Google Scholar] [CrossRef]
- Bi, Y.J.; Sun, Z.C.; Zhang, J.; Liu, E.Q.; Shen, H.M.; Lai, K.L.; Zhang, S.; Guo, X.T.; Sheng, Y.T.; Yu, C.Y.; et al. Manipulating the expression of a cell wall invertase gene increases grain yield in maize. Plant Growth Regul. 2018, 84, 37–43. [Google Scholar] [CrossRef]
- Hirose, T.; Takano, M.; Terao, T. Cell wall invertase in developing rice caryopsis: Molecular cloning of OsCIN1 and analysis of its expression in relation to its role in grain filling. Plant Cell Physiol. 2002, 43, 452–459. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Ni, D.A.; Ruan, Y.L. Posttranslational elevation of cell wall invertase activity by silencing its inhibitor in tomato delays leaf senescence and increases seed weight and fruit hexose level. Plant Cell 2009, 21, 2072–2089. [Google Scholar] [CrossRef] [PubMed]
- McCurdy, D.W.; Dibley, S.; Cahyanegara, R.; Martin, A.; Patrick, J.W. Functional characterization and RNAi-mediated suppression reveals roles for hexose transporters in sugar accumulation by tomato fruit. Mol. Plant 2010, 3, 1049–1063. [Google Scholar] [CrossRef]
- Zanor, M.I.; Osorio, S.; Nunes-Nesi, A.; Carrari, F.; Lohse, M.; Usadel, B.; Kuhn, C.; Bleiss, W.; Giavalisco, P.; Willmitzer, L.; et al. RNA interference of LIN5 in tomato confirms its role in controlling brix content, uncovers the influence of sugars on the levels of fruit hormones, and demonstrates the importance of sucrose cleavage for normal fruit development and fertility. Plant Physiol. 2009, 150, 1204–1218. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yao, Q.; Gao, X.; Jiang, C.; Harberd, N.P.; Fu, X. Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr. Biol. 2016, 26, 640–646. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Gao, Z.; Wang, J.; Huang, Y.; Chen, Y.; Li, J.; Lv, M.; Wang, J.; Luo, M.; Zuo, K. Cotton fiber elongation requires the transcription factor GhMYB212 to regulate sucrose transportation into expanding fibers. New Phytol. 2019, 222, 864–881. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.J.; Sun, M.H.; Lu, J.; Liu, Y.J.; Hu, D.G.; Hao, Y.J. Transcription factor AREB2 is involved in soluble sugar accumulation by activating sugar transporter and amylase genes. Plant Physiol. 2017, 174, 2348–2362. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Lee, S.K.; Yoo, Y.; Wei, J.; Kwon, S.Y.; Lee, S.W.; Jeon, J.S.; An, G. Rice transcription factor OsDOF11 modulates sugar transport by promoting expression of Sucrose Transporter and SWEET genes. Mol. Plant 2018, 11, 833–845. [Google Scholar] [CrossRef]
- He, Y.; Chen, R.; Yang, Y.; Liang, G.; Zhang, H.; Deng, X.; Xi, R. Sugar metabolism and transcriptome analysis reveal key sugar transporters during Camellia oleifera fruit development. Int. J. Mol. Sci. 2022, 23, 822. [Google Scholar] [CrossRef]
- Guenoune-Gelbart, D.; Elbaum, M.; Sagi, G.; Levy, A.; Epel, B.L. Tobacco mosaic virus (TMV) replicase and movement protein function synergistically in facilitating TMV spread by lateral diffusion in the plasmodesmal desmotubule of Nicotiana benthamiana. Mol. Plant Microbe Interact. 2008, 21, 335–345. [Google Scholar] [CrossRef]
- Heinlein, M.; Epel, B.L. Macromolecular transport and signaling through plasmodesmata. Int. Rev. Cytol. 2004, 235, 93–164. [Google Scholar] [PubMed]
- Misra, A.; Srivastava, N.K.; Srivastava, A.K.; Khan, A. Influence of etherel and gibberellic acid on carbon metabolism, growth, and alkaloids accumulation in Catharanthus roseus L. Afr. J. Pharm. Pharmacol. 2009, 3, 515–520. [Google Scholar]
- Ofosu-Anim, J.; Kanayama, Y.; Yamaki, S. Sugar uptake into strawberry fruit is stimulated by abscisic acid and indoleacetic acid. Physiol. Plant. 2010, 97, 169–174. [Google Scholar] [CrossRef]
- Zhang, Z.; Luo, Y.; Wang, X.; Yu, F. Fruit spray of 24-epibrassinolide and fruit shade alter pericarp photosynthesis activity and seed lipid accumulation in Styrax tonkinensis. J. Plant Growth Regul. 2018, 37, 1066–1084. [Google Scholar] [CrossRef]
- Hua, W.; Li, R.J.; Zhan, G.M.; Liu, J.; Li, J.; Wang, X.F.; Liu, G.H.; Wang, H.Z. Maternal control of seed oil content in Brassica napus: The role of silique wall photosynthesis. Plant J. 2012, 69, 432–444. [Google Scholar] [CrossRef]
- Liu, J.; Hua, W.; Yang, H.; Guo, T.; Sun, X.; Wang, X.; Liu, G.; Wang, H. Effects of specific organs on seed oil accumulation in Brassica napus L. Plant Sci. 2014, 227, 60–68. [Google Scholar] [CrossRef]
- Wu, G.L.; Zhang, X.Y.; Zhang, L.Y.; Pan, Q.H.; Shen, Y.Y.; Zhang, D.P. Phloem unloading in developing walnut fruit is symplasmic in the seed pericarp, and apoplasmic in the fleshy pericarp. Plant Cell Physiol. 2004, 45, 1461–1470. [Google Scholar] [CrossRef]
- Zhang, F.; Li, Z.; Zhou, J.; Gu, Y.; Tan, X. Comparative study on fruit development and oil synthesis in two cultivars of Camellia oleifera. BMC Plant Biol. 2021, 21, 348. [Google Scholar] [CrossRef]
- Amsbury, S.; Kirk, P.; Benitez-Alfonso, Y. Emerging models on the regulation of intercellular transport by plasmodesmata-associated callose. J. Exp. Bot. 2018, 69, 105–115. [Google Scholar] [CrossRef]
- Chen, X.Y.; Kim, J.Y. Callose synthesis in higher plants. Plant Signal Behav. 2009, 4, 489–492. [Google Scholar] [CrossRef]
- Zavaliev, R.; Ueki, S.; Epel, B.L.; Citovsky, V. Biology of callose (β-1,3-glucan) turnover at plasmodesmata. Protoplasma 2011, 248, 117–130. [Google Scholar] [CrossRef]
- Ruan, Y.L.; Llewellyn, D.J.; Furbank, R.T. The control of single-celled cotton fiber elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K+ transporters and expansin. Plant Cell 2001, 13, 47–60. [Google Scholar] [PubMed]
- Ruan, Y.L.; Xu, S.M.; White, R.; Furbank, R.T. Genotypic and developmental evidence for the role of plasmodesmatal regulation in cotton fiber elongation mediated by callose turnover. Plant Physiol. 2004, 136, 4104–4113. [Google Scholar] [CrossRef] [PubMed]
- Fan, R.C.; Peng, C.C.; Xu, Y.H.; Wang, X.F.; Li, Y.; Shang, Y.; Du, S.Y.; Zhao, R.; Zhang, X.Y.; Zhang, L.Y.; et al. Apple sucrose transporter SUT1 and sorbitol transporter SOT6 interact with cytochrome b5 to regulate their affinity for substrate sugars. Plant Physiol. 2009, 150, 1880–1901. [Google Scholar] [CrossRef] [PubMed]
- Gould, N.; Thorpe, M.R.; Pritchard, J.; Christeller, J.T.; Williams, L.E.; Roeb, G.; Schurr, U.; Minchin, P. AtSUC2 has a role for sucrose retrieval along the phloem pathway: Evidence from carbon-11 tracer studies. Plant Sci. 2012, 188, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.C.; Ganesan, S.; Ismail, I.O.; Ayre, B.G. Functional characterization of the Arabidopsis AtSUC2 sucrose/H+ symporter by tissue-specific complementation reveals an essential role in phloem loading but not in long-distance transport. Plant Physiol. 2008, 148, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Ru, L.; He, Y.; Zhu, Z.; Patrick, J.W.; Ruan, Y.L. Integrating sugar metabolism with transport: Elevation of endogenous cell wall invertase activity up-regulates SlHT2 and SlSWEET12c expression for early fruit development in tomato. Front. Genet. 2020, 11, 592596. [Google Scholar] [CrossRef]
- Ruan, Y.L.; Jin, Y.; Yang, Y.J.; Li, G.J.; Boyer, J.S. Sugar input, metabolism, and signaling mediated by invertase: Roles in development, yield potential, and response to drought and heat. Mol. Plant 2010, 3, 942–955. [Google Scholar] [CrossRef]
- Rae, A.L.; Perroux, J.M.; Grof, C.P. Sucrose partitioning between vascular bundles and storage parenchyma in the sugarcane stem: A potential role for the ShSUT1 sucrose transporter. Planta 2005, 220, 817–825. [Google Scholar] [CrossRef]
- Zhang, L.Y.; Peng, Y.B.; Pelleschi-Travier, S.; Fan, Y.; Lu, Y.F.; Lu, Y.M.; Gao, X.P.; Shen, Y.Y.; Delrot, S.; Zhang, D.P. Evidence for apoplasmic phloem unloading in developing apple fruit. Plant Physiol. 2004, 135, 574–586. [Google Scholar] [CrossRef]
- Reinhold, H.; Soyk, S.; Šimková, K.; Hostettler, C.; Marafino, J.; Mainiero, S.; Vaughan, C.K.; Monroe, J.D.; Zeeman, S.C. Β-amylase-like proteins function as transcription factors in Arabidopsis, controlling shoot growth and development. Plant Cell 2011, 23, 1391–1403. [Google Scholar] [CrossRef] [PubMed]
- Lara, M.E.B.; Garcia, M.C.G.; Fatima, T.; Ehness, R.; Lee, T.K.; Proels, R.; Tanner, W.; Roitsch, T. Extracellular invertase is an essential component of cytokinin-mediated delay of senescence. Plant Cell 2004, 16, 1276–1287. [Google Scholar] [CrossRef] [PubMed]
- Li, M.J.; Feng, F.J.; Cheng, L.L. Expression patterns of genes involved in sugar metabolism and accumulation during apple fruit development. PLoS ONE 2012, 7, e33055. [Google Scholar] [CrossRef]
- Moscatello, S.; Famiani, F.; Proietti, S.; Farinelli, D.; Battistelli, A. Sucrose synthase dominates carbohydrate metabolism and relative growth rate in growing kiwifruit (Actinidia deliciosa, cv Hayward). Sci. Hortic. 2011, 128, 197–205. [Google Scholar] [CrossRef]
- Sun, J.; Loboda, T.; Sung, S.J.; Black, C.C. Sucrose synthase in wild tomato, Lycopersicon chmielewskii, and tomato fruit sink strength. Plant Physiol. 1992, 98, 1163–1169. [Google Scholar] [CrossRef]
- David, A.M. Hormonal regulation of source-sink relationships: An overview of potential control mechanisms. In Photoassimilate Distribution in Plants and Crops: Source-Sink Relationships; Zamski, E., Schaffer, A.A., Eds.; Marcel Dekker, Inc.: New York, NY, USA, 1996; pp. 441–465. [Google Scholar]
- Zhang, C.; Tanabe, K.; Tamura, F.; Itai, A.; Yoshida, M. Roles of gibberellins in increasing sink demand in Japanese pear fruit during rapid fruit growth. Plant Growth Regul. 2007, 52, 161–172. [Google Scholar] [CrossRef]
- Cao, Y.Q.; Yao, X.H.; Ren, H.D.; Wang, K.L. Changes in contents of endogenous hormones and main mineral elements in oil-tea camellia fruit during maturation. J. Beijing For. Univ. 2015, 37, 76–81. [Google Scholar]
- Cole, M.A.; Patrick, J.W. Auxin control of photoassimilate transport to and within developing grains of wheat. Aust. J. Plant Physiol. 1998, 25, 69–77. [Google Scholar]
- Holdaway-Clarke, T.L.; Walker, N.A.; Hepler, P.K.; Overall, R.L. Physiological elevations in cytoplasmic free calcium by cold or ion injection result in transient closure of higher plant plasmodesmata. Planta 2000, 210, 329–335. [Google Scholar] [CrossRef]
- Baluska, F.; Cvrcková, F.; Kendrick-Jones, J.; Volkmann, D. Sink plasmodesmata as gateways for phloem unloading. Myosin viii and calreticulin as molecular determinants of sink strength? Plant Physiol. 2001, 126, 39–46. [Google Scholar] [CrossRef]
- Botha, C.; Cross, R. Towards reconciliation of structure with function in plasmodesmata—Who is the gatekeeper? Micron 2000, 31, 713–721. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Enzymes and other agents that enhance cell wall extensibility. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 391–417. [Google Scholar] [CrossRef] [PubMed]
- Du, B.S.; Zhang, Q.; Cao, Q.Q.; Xing, Y.; Qin, L.; Fang, K.F. Changes of cell wall components during embryogenesis of Castanea mollissima. J. Plant Res. 2020, 133, 257–270. [Google Scholar] [CrossRef]
- Jan, B.; Roel, M. An improved colorimetric method to quantify sugar content of plant tissue. J. Exp. Bot. 1993, 44, 1627–1629. [Google Scholar]
- Jin, Z.; Zhu, F.; Guo, X.; Zhang, Z.; Agriculture, S.O.; University, N.A. Effect of different nitrogen application methods on activity of sucrose metabolism correlation enzymes and quality in rice at filling stage. J. Northeast. Agric. Univ. 2016, 47, 1–7. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Yu, M.; Liu, J.; Du, B.; Zhang, M.; Wang, A.; Zhang, L. NAC transcription factor PwNAC11 activates ERD1 by interaction with ABF3 and DREB2A to enhance drought tolerance in transgenic Arabidopsis. Int. J. Mol. Sci. 2021, 22, 6952. [Google Scholar] [CrossRef]
- Yang, S.; Liang, K.; Wang, A.; Zhang, M.; Zhang, L. Physiological characterization and transcriptome analysis of Camellia oleifera Abel. During leaf senescence. Forests 2020, 11, 812. [Google Scholar] [CrossRef]
| Sample | Total Reads | Total Mapped | Q30 (%) |
|---|---|---|---|
| P1_1 | 60,538,690 | 46,670,044 (77.09%) | 93.14 |
| P1_2 | 44,632,706 | 33,808,440 (75.75%) | 93.09 |
| P1_3 | 54,987,824 | 43,454,320 (79.03%) | 92.94 |
| P2_1 | 50,331,646 | 38,615,427 (76.72%) | 93.19 |
| P2_2 | 56,935,162 | 48,089,102 (84.46%) | 93.43 |
| P2_3 | 53,874,354 | 42,709,833 (79.28%) | 93.21 |
| S1_1 | 46,444,352 | 35,140,094 (75.66%) | 93.55 |
| S1_2 | 66,249,036 | 50,478,873 (76.2%) | 93.58 |
| S1_3 | 62,077,406 | 46,570,332 (75.02%) | 93.6 |
| S2_1 | 52,061,320 | 39,827,703 (76.5%) | 93.67 |
| S2_2 | 52,807,790 | 39,974,735 (75.7%) | 93.73 |
| S2_3 | 47,665,372 | 35,524,004 (74.53%) | 93.1 |
| Database Type | Gene Number (Percent) | Transcript Number (Percent) |
|---|---|---|
| GO | 23,727 (0.3581) | 43,987 (0.4111) |
| KEGG | 30,413 (0.459) | 48,065 (0.4492) |
| COG | 49,385 (0.7453) | 85,076 (0.795) |
| NR | 51,682 (0.78) | 88,293 (0.8251) |
| Swiss-Prot | 39,734 (0.5997) | 69,788 (0.6522) |
| Pfam | 37,584 (0.5672) | 66,079 (0.6175) |
| Total annotated | 52,014 (0.785) | 88,727 (0.8292) |
| Total | 66,261 (1) | 107,009 (1) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, J.; Du, B.; Chen, Y.; Cao, Y.; Yu, M.; Zhang, L. Integrative Physiological and Transcriptomic Analysis Reveals the Transition Mechanism of Sugar Phloem Unloading Route in Camellia oleifera Fruit. Int. J. Mol. Sci. 2022, 23, 4590. https://doi.org/10.3390/ijms23094590
Zhou J, Du B, Chen Y, Cao Y, Yu M, Zhang L. Integrative Physiological and Transcriptomic Analysis Reveals the Transition Mechanism of Sugar Phloem Unloading Route in Camellia oleifera Fruit. International Journal of Molecular Sciences. 2022; 23(9):4590. https://doi.org/10.3390/ijms23094590
Chicago/Turabian StyleZhou, Jing, Bingshuai Du, Yuqing Chen, Yibo Cao, Mingxin Yu, and Lingyun Zhang. 2022. "Integrative Physiological and Transcriptomic Analysis Reveals the Transition Mechanism of Sugar Phloem Unloading Route in Camellia oleifera Fruit" International Journal of Molecular Sciences 23, no. 9: 4590. https://doi.org/10.3390/ijms23094590
APA StyleZhou, J., Du, B., Chen, Y., Cao, Y., Yu, M., & Zhang, L. (2022). Integrative Physiological and Transcriptomic Analysis Reveals the Transition Mechanism of Sugar Phloem Unloading Route in Camellia oleifera Fruit. International Journal of Molecular Sciences, 23(9), 4590. https://doi.org/10.3390/ijms23094590






