Prognostic Significance of GPR55 mRNA Expression in Colon Cancer
Abstract
:1. Introduction
2. Results
2.1. GPR55 mRNA Expression Levels in Primary Colon Cancer Tumors and Colon Cancer Cell Lines
2.2. GPR55 mRNA Expression Levels in Regional Lymph Nodes of Colon Cancer Patients
2.3. Correlation between mRNA Expression Levels of GPR55 and Each of CEA, CXCL17, CXCL16, GPR35 V2/3 and LGR5 in Regional Lymph Nodes of Colon Cancer Patients
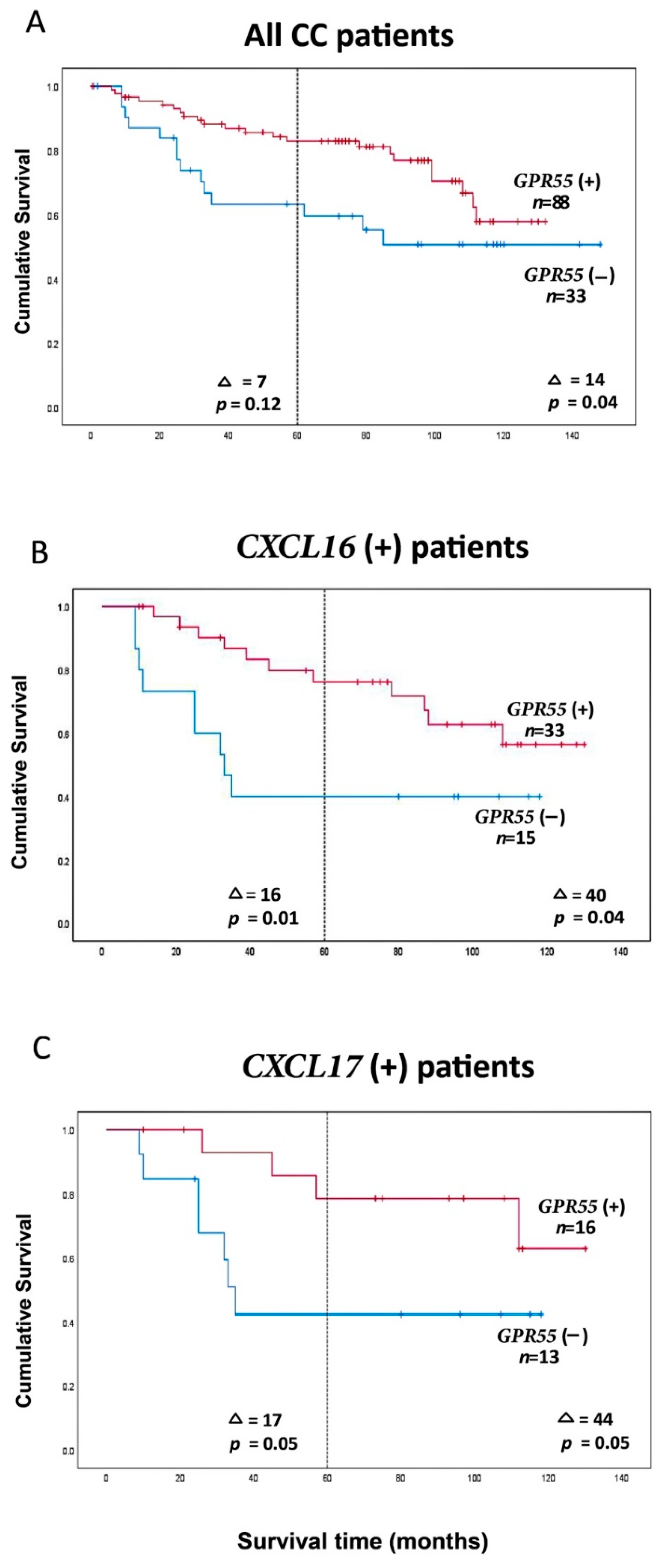
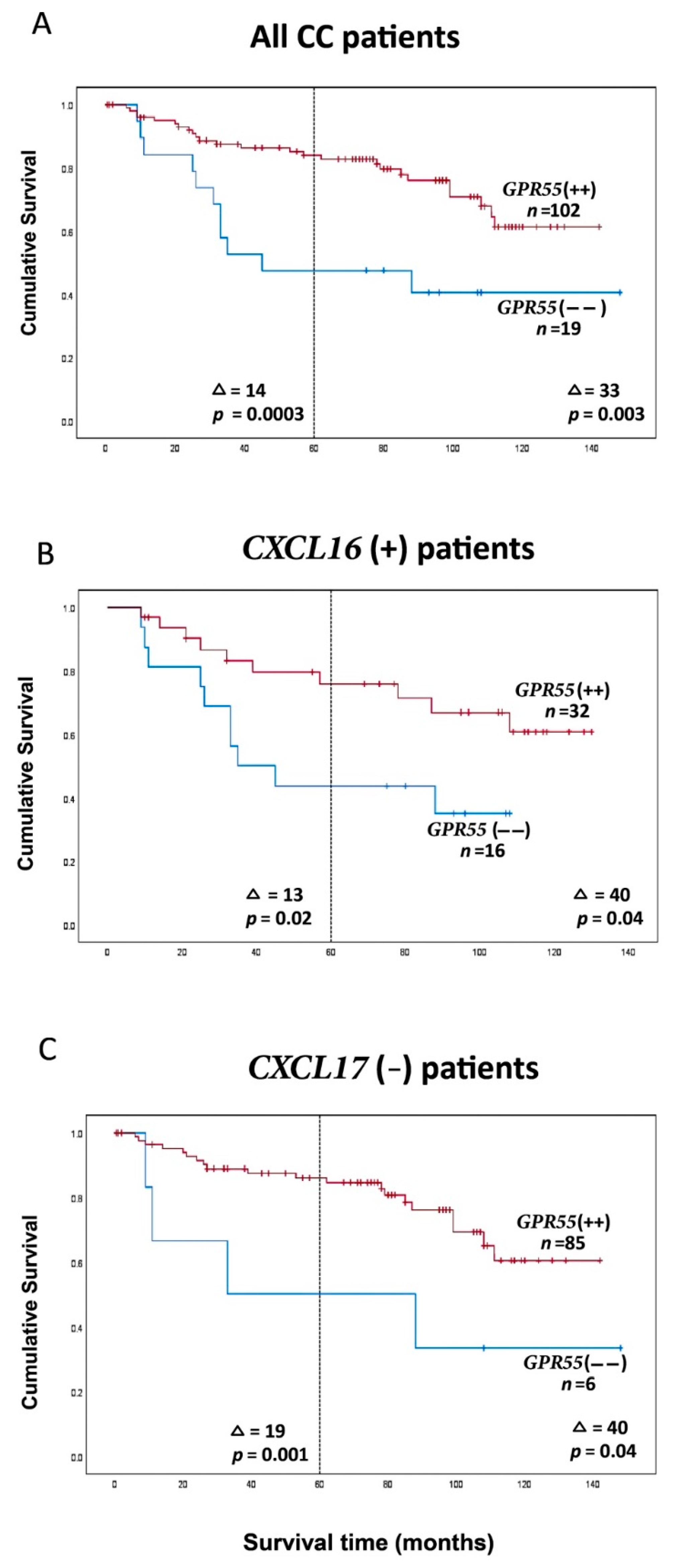
2.4. Clinical Significance of GPR55 mRNA Expression Levels and of GPR55/CEA mRNA Expression Ratios in Lymph Nodes for Colon Cancer Patients: Prediction of Recurrence
2.5. Absence of Association between Risk of Recurrence or Survival Time after Surgery and Levels GPR55 mRNA Expression in Primary Colon Cancer Tumors
3. Discussion
4. Materials and Methods
4.1. Patients and Tissue Specimens for mRNA Analysis
4.2. Cell Lines
4.3. Real-Time qRT-PCR
4.4. Ethical Considerations
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sriram, K.; Insel, P.A. G Protein-coupled receptors as targets for approved drugs: How many targets and how many drugs? Mol. Pharmacol. 2018, 93, 251–258. [Google Scholar] [CrossRef] [Green Version]
- Fredriksson, R.; Lagerström, M.C.; Lundin, L.G.; Schiöth, H.B. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 2003, 63, 1256–1272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Insel, P.A.; Zhang, L.; Murray, F.; Yokouchi, H.; Zambon, A.C. Cyclic AMP is both a pro-apoptotic and anti-apoptotic second messenger. Acta Physiol. 2012, 204, 277–287. [Google Scholar] [CrossRef]
- Chiurchiu, V.; Lanuti, M.; De Bardi, M.; Battistini, L.; Maccarrone, M. The differential characterization of GPR55 receptor in human peripheral blood reveals a distinctive expression in monocytes and NK cells and a proinflammatory role in these innate cells. Int. Immunol. 2015, 27, 153–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sumida, H.; Lu, E.; Chen, H.; Yang, Q.; Mackie, K.; Cyster, J.G. GPR55 regulates intraepithelial lymphocyte migration dynamics and susceptibility to intestinal damage. Sci. Immunol. 2017, 2, eaao1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falasca, M.; Ferro, R. Role of the lysophosphatidylinositol/GPR55 axis in cancer. Adv. Biol. Regul. 2016, 60, 88–93. [Google Scholar] [CrossRef]
- Hasenoehrl, C.; Feuersinger, D.; Sturm, E.M.; Barnthaler, T.; Heitzer, E.; Graf, R.; Grill, M.; Pichler, M.; Beck, S.; Butcher, L.; et al. G protein-coupled receptor GPR55 promotes colorectal cancer and has opposing effects to cannabinoid receptor 1. Int. J. Cancer 2018, 142, 121–132. [Google Scholar] [CrossRef]
- Kitamura, C.; Sonoda, H.; Anzai, H.; Nagai, Y.; Abe, S.; Yokoyama, Y.; Ishii, H.; Kishikawa, J.; Emoto, S.; Murono, K.; et al. Expression of Lysophosphatidylinositol Signaling-relevant Molecules in Colorectal Cancer. Anticancer Res. 2021, 41, 2349–2355. [Google Scholar] [CrossRef]
- Ohlsson, L.; Israelsson, A.; Öberg, Å.; Palmqvist, R.; Stenlund, H.; Hammarström, M.L.; Hammarström, S.; Lindmark, G. Lymph node CEA and MUC2 mRNA as useful predictors of outcome in colorectal cancer. Int. J. Cancer 2012, 130, 1833–1843. [Google Scholar] [CrossRef]
- Ohlsson, L.; Hammarström, M.-L.; Israelsson, A.; Näslund, L.; Öberg, Å.; Lindmark, G.; Hammarström, S. Biomarker selection for detection of occult tumour cells in lymph nodes of colorectal cancer patients using real-time quantitative RT-PCR. Br. J. Cancer 2006, 95, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.; AbdelMageed, M.; Olsson, L.; Israelsson, A.; Lindmark, G.; Hammarström, M.L.; Hammarström, S.; Sitohy, B. Utility of G protein-coupled receptor 35 expression for predicting outcome in colon cancer. Tumour Biol. 2019, 41, 1010428319858885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- AbdelMageed, M.; Ali, H.; Olsson, L.; Lindmark, G.; Hammarström, M.L.; Hammarström, S.; Sitohy, B. The chemokine CXCL16 is a new biomarker for lymph node analysis of colon cancer outcome. Int. J. Mol. Sci. 2019, 20, 5793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rashad, Y.; Olsson, L.; Israelsson, A.; Öberg, Å.; Lindmark, G.; Hammarström, M.L.; Hammarström, S.; Sitohy, B. Lymph node CXCL17 messenger RNA: A new prognostic biomarker for colon cancer. Tumour Biol. 2018, 40, 1010428318799251. [Google Scholar] [CrossRef] [Green Version]
- Ohlsson, L.; Hammarström, M.L.; Lindmark, G.; Hammarström, S.; Sitohy, B. Ectopic expression of the chemokine CXCL17 in colon cancer cells. Br. J. Cancer 2016, 114, 697–703. [Google Scholar] [CrossRef] [Green Version]
- AbdelMageed, M.; Ismail, H.T.H.; Olsson, L.; Lindmark, G.; Hammarström, M.L.; Hammarström, S.; Sitohy, B. Clinical Significance of Stem Cell Biomarkers EpCAM, LGR5 and LGR4 mRNA Levels in Lymph Nodes of Colon Cancer Patients. Int. J. Mol. Sci. 2021, 23, 403. [Google Scholar] [CrossRef]
- Olsson, L.; Lindmark, G.; Hammarström, M.L.; Hammarström, S.; Sitohy, B. Evaluating macrophage migration inhibitory factor 1 expression as a prognostic biomarker in colon cancer. Tumour Biol. 2020, 42, 1010428320924524. [Google Scholar] [CrossRef]
- Öberg, Å.; Lindmark, G.; Israelsson, A.; Hammarström, S.; Hammarström, M.-L. Detection of occult tumor cells in lymph nodes of colorectal cancer patients using real-time quantitative RT-PCR for CEA and CK20 mRNAs. Int. J. Cancer 2004, 111, 101–110. [Google Scholar] [CrossRef]
- Waldhauer, I.; Steinle, A. NK cells and cancer immunosurveillance. Oncogene 2008, 27, 5932–5943. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Ramirez, J.C.; Frampton, G.A.; Golden, L.E.; Quinn, M.A.; Pae, H.Y.; Horvat, D.; Liang, L.J.; DeMorrow, S. Anandamide exerts its antiproliferative actions on cholangiocarcinoma by activation of the GPR55 receptor. Lab. Investig. 2011, 91, 1007–1017. [Google Scholar] [CrossRef]
- Fahlgren, A.; Hammarström, S.; Danielsson, Å.; Hammarström, M.L. Increased expression of antimicrobial peptides and lysozyme in colonic epithelial cells of patients with ulcerative colitis. Clin. Exp. Immunol. 2003, 131, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.; Ohlsson, L.; Lindmark, G.; Hammarström, S.; Hammarström, M.-L.; Sitohy, B. The myeloid cell biomarker EMR1 is ectopically expressed in colon cancer. Tumor Biol. 2021, 43, 197–211. [Google Scholar] [CrossRef] [PubMed]


| CEA | CXCL17 | CXCL16 | GPR35 V2/3 | LGR5 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| r | p-Value | r | p-Value | r | p-Value | r | p-Value | r | p-Value | ||
| GPR55 | All CRC LNs | −0.08 | 0.123 | −0.04 | 0.421 | 0.33 | <0.0001 | 0.28 | <0.0001 | 0.33 | <0.0001 |
| TNM Stage I LNs | 0.21 | 0.075 | 0.43 | 0.0001 | 0.64 | <0.0001 | 0.58 | <0.0001 | 0.56 | <0.0001 | |
| TNM Stage II LNs | −0.10 | 0.155 | 0.004 | 0.958 | 0.47 | <0.0001 | 0.42 | <0.0001 | 0.48 | <0.0001 | |
| TNM Stage III LNs | −0.18 | 0.098 | −0.24 | 0.027 | 0.17 | 0.120 | 0.15 | 0.150 | 0.21 | 0.047 | |
| TNM Stage IV LNs | −0.46 | 0.009 | −0.38 | 0.036 | −0.50 | 0.005 | −0.39 | 0.028 | −0.31 | 0.092 | |
| Patient Group | Category | Number of Patients in Each Group Stratified by TNM Stage | Total | 5-Year Follow-Up after Surgery | 12-Year Follow-Up after Surgery | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Disease-Free Survival a | Risk for Recurrence b | Disease-Free Survival a | Risk for Recurrence b | |||||||||||||
| Stage I | Stage II | Stage III | Stage IV | Average (Months) | Difference (Months) | p-Value | Hazard Ratio (95% CI) | p-Value | Average (Months) | Difference (Months) | p-Value | Hazard Ratio (95% CI) | p-Value | |||
| GPR55 mRNA level | ||||||||||||||||
| All CC patients | GPR55(−) c | 7 | 10 | 12 | 4 | 33 | 46 | 7 | 0.12 | 0.6 (0.3–1.2) | 0.12 | 93 | 14 | 0.04 | 0.5 (0.3–1.0) | 0.05 |
| GPR55(+) | 16 | 42 | 25 | 5 | 88 | 53 | 107 | |||||||||
| CXCL16 e | GPR55(−) | 2 | 3 | 6 | 4 | 15 | 36 | 16 | 0.01 | 0.3 (0.1–0.8) | 0.02 | 60 | 40 | 0.04 | 0.4 (0.2–1.0) | 0.04 |
| GPR55(+) | 8 | 12 | 10 | 3 | 33 | 52 | 100 | |||||||||
| CXCL17f | GPR55(−) | 2 | 2 | 7 | 2 | 13 | 40 | 17 | 0.05 | 0.3 (0.1–1.1) | 0.07 | 64 | 44 | 0.05 | 0.3 (0.1–1.1) | 0.07 |
| GPR55(+) | 3 | 5 | 6 | 2 | 16 | 57 | 108 | |||||||||
| CEA g | GPR55(−) | 4 | 5 | 3 | 0 | 12 | 51 | 4 | 0.56 | 0.7 (0.2–2.6) | 0.56 | 86 | 26 | 0.05 | 0.3 (0.1–1.1) | 0.06 |
| GPR55(+) | 6 | 17 | 10 | 0 | 33 | 55 | 112 | |||||||||
| GPR55 mRNA/CEA mRNA ratio | ||||||||||||||||
| All CC patients | GPR55(−−) d | 1 | 2 | 9 | 7 | 19 | 40 | 14 | 0.0003 | 0.3 (0.1–0.6) | 0.001 | 80 | 33 | 0.003 | 0.4 (0.2–0.7) | 0.004 |
| GPR55(++) | 22 | 50 | 28 | 2 | 102 | 54 | 113 | |||||||||
| CXCL16 e | GPR55(−−) | 1 | 0 | 8 | 7 | 16 | 38 | 13 | 0.02 | 0.4 (0.1–0.9) | 0.02 | 60 | 40 | 0.04 | 0.4 (0.2–1.0) | 0.04 |
| GPR55(++) | 9 | 15 | 8 | 0 | 32 | 51 | 100 | |||||||||
| CXCL17 h | GPR55(−−) | 0 | 2 | 1 | 3 | 6 | 35 | 19 | 0.001 | 0.2 (0.1–0.6) | 0.004 | 73 | 40 | 0.04 | 0.3 (0.1–1.0) | 0.05 |
| GPR55(++) | 18 | 43 | 22 | 2 | 85 | 54 | 113 | |||||||||
| CEA i | GPR55(−−) | 1 | 2 | 9 | 7 | 19 | 40 | 13 | 0.002 | 0.3 (0.1–0.7) | 0.004 | 80 | 26 | 0.005 | 0.3 (0.1–0.8) | 0.008 |
| GPR55(++) | 12 | 28 | 15 | 2 | 57 | 53 | 106 | |||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ismail, H.T.H.; AbdelMageed, M.; Lindmark, G.; Hammarström, M.-L.; Hammarström, S.; Sitohy, B. Prognostic Significance of GPR55 mRNA Expression in Colon Cancer. Int. J. Mol. Sci. 2022, 23, 4556. https://doi.org/10.3390/ijms23094556
Ismail HTH, AbdelMageed M, Lindmark G, Hammarström M-L, Hammarström S, Sitohy B. Prognostic Significance of GPR55 mRNA Expression in Colon Cancer. International Journal of Molecular Sciences. 2022; 23(9):4556. https://doi.org/10.3390/ijms23094556
Chicago/Turabian StyleIsmail, Hager Tarek H., Manar AbdelMageed, Gudrun Lindmark, Marie-Louise Hammarström, Sten Hammarström, and Basel Sitohy. 2022. "Prognostic Significance of GPR55 mRNA Expression in Colon Cancer" International Journal of Molecular Sciences 23, no. 9: 4556. https://doi.org/10.3390/ijms23094556
APA StyleIsmail, H. T. H., AbdelMageed, M., Lindmark, G., Hammarström, M.-L., Hammarström, S., & Sitohy, B. (2022). Prognostic Significance of GPR55 mRNA Expression in Colon Cancer. International Journal of Molecular Sciences, 23(9), 4556. https://doi.org/10.3390/ijms23094556






