Global 5mC and 5hmC DNA Levels in Human Sperm Subpopulations with Differentially Protaminated Chromatin in Normo- and Oligoasthenozoospermic Males
Abstract
1. Introduction
2. Results
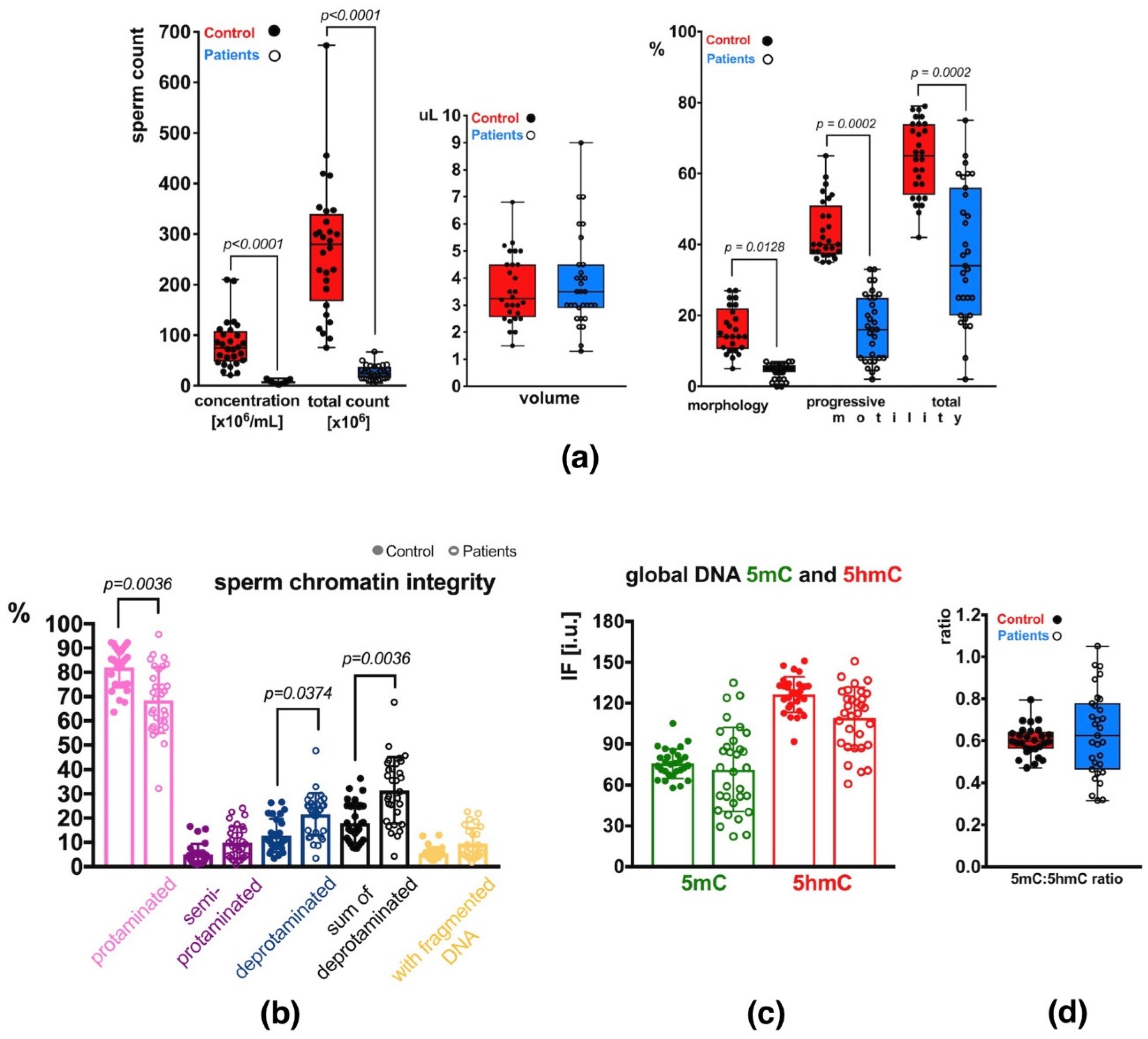
2.1. Semen Parameters
2.2. Sperm Chromatin Integrity
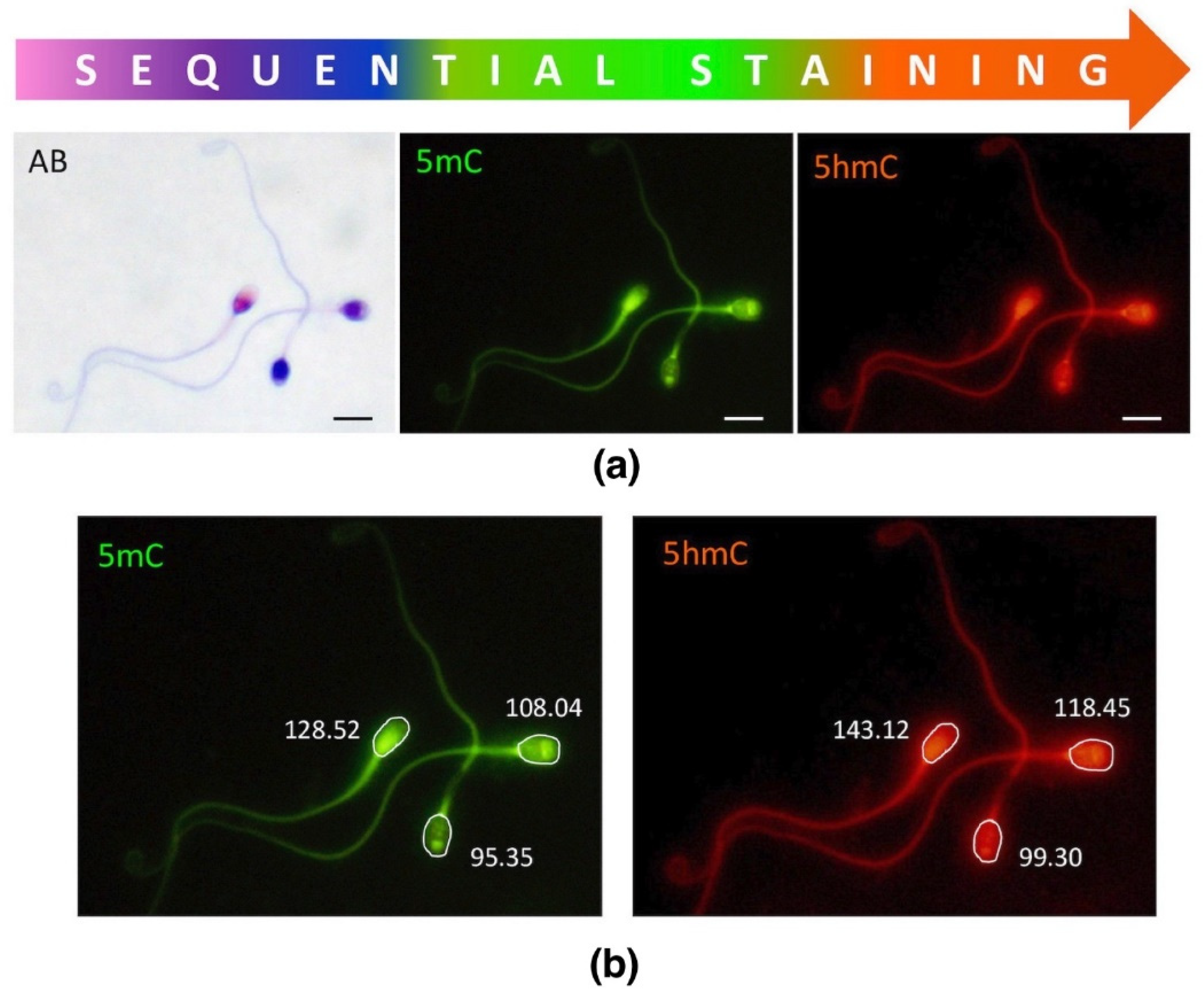
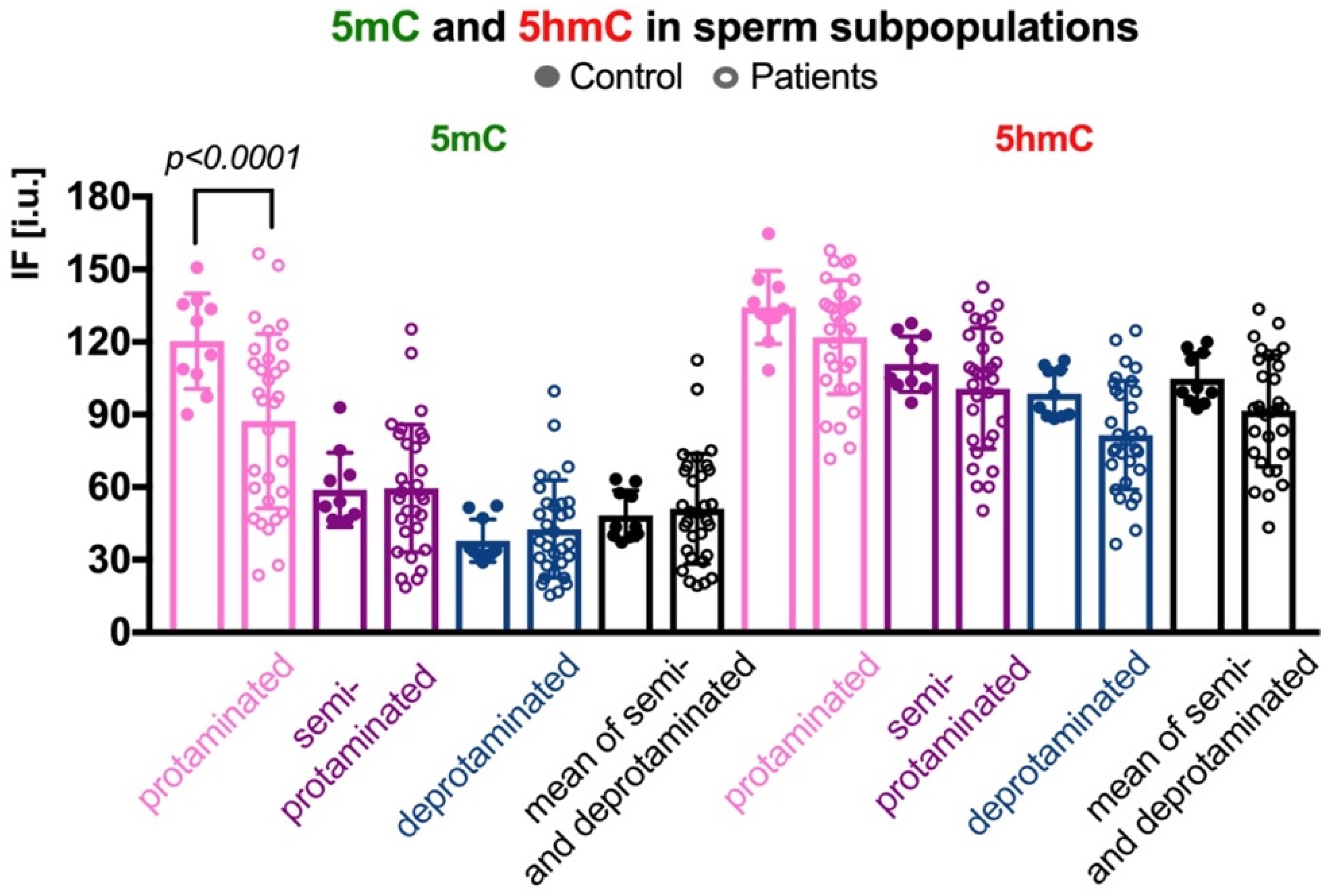
2.3. Global Methylation (5mC) and Hydroxymethylation (5hmC) of Sperm DNA
2.3.1. Unfractionated Total Sperm Population
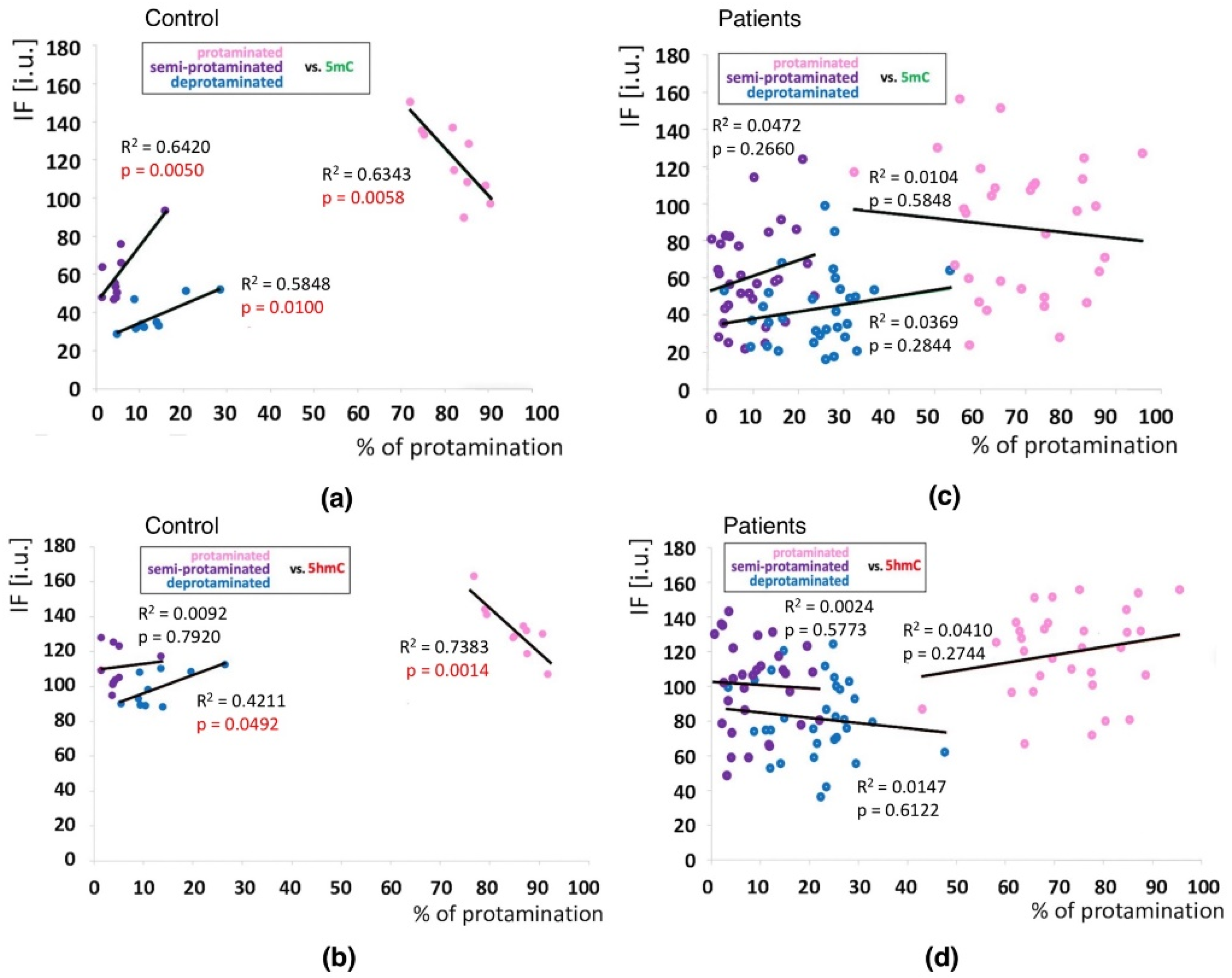
2.3.2. Sperm Populations According to Chromatin Protamination
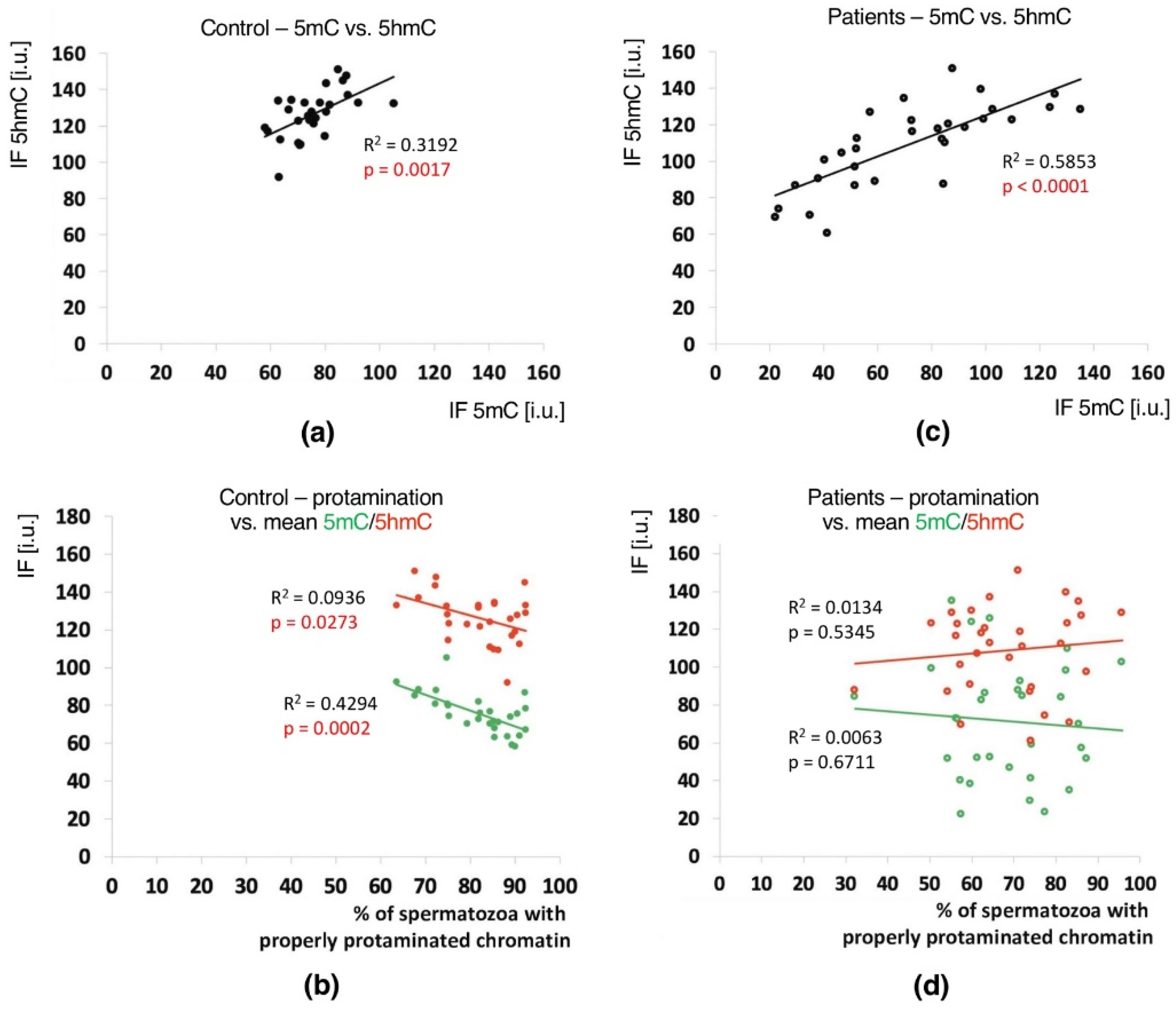
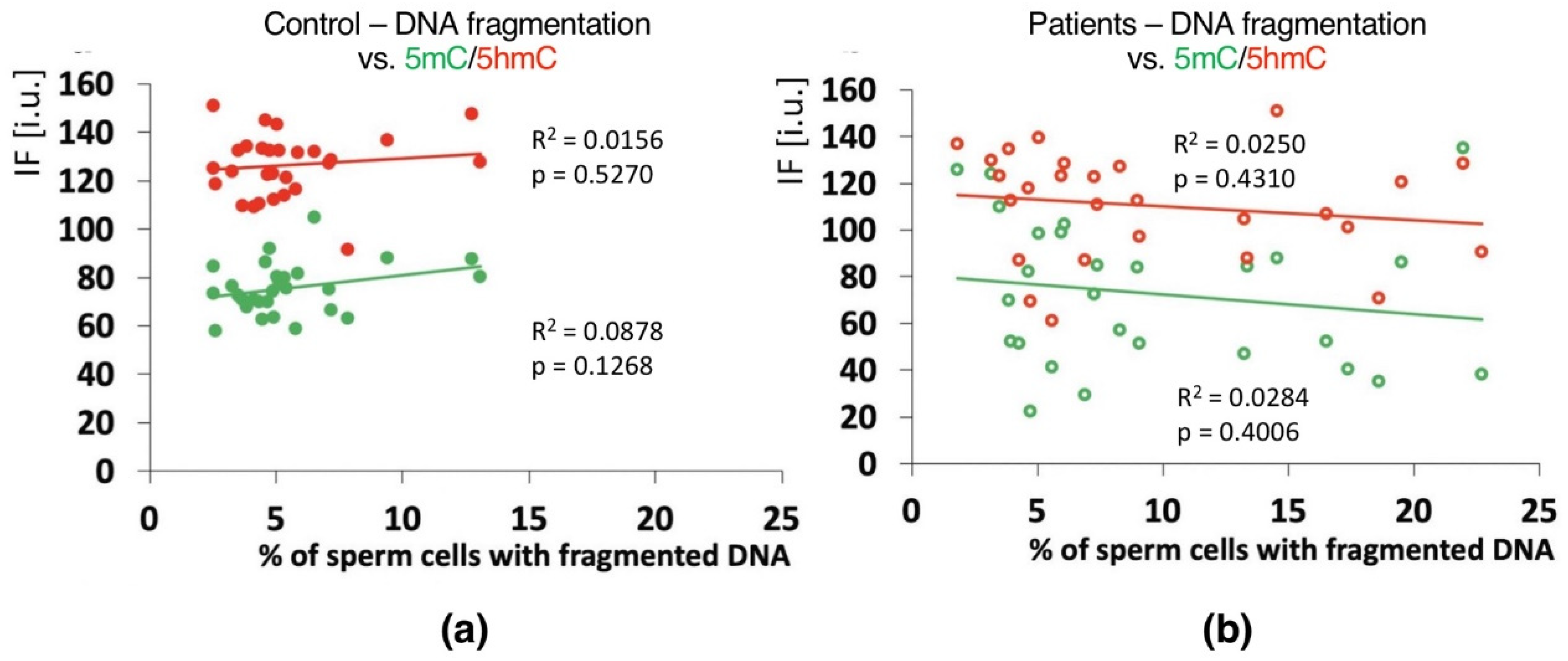
2.4. Correlations
3. Discussion
4. Materials and Methods
4.1. Ethics Approval and Consent to Participate
4.2. Participants
4.3. Sperm Chromatin Integrity
4.3.1. Sperm Chromatin Protamination Status
4.3.2. Sperm DNA Fragmentation
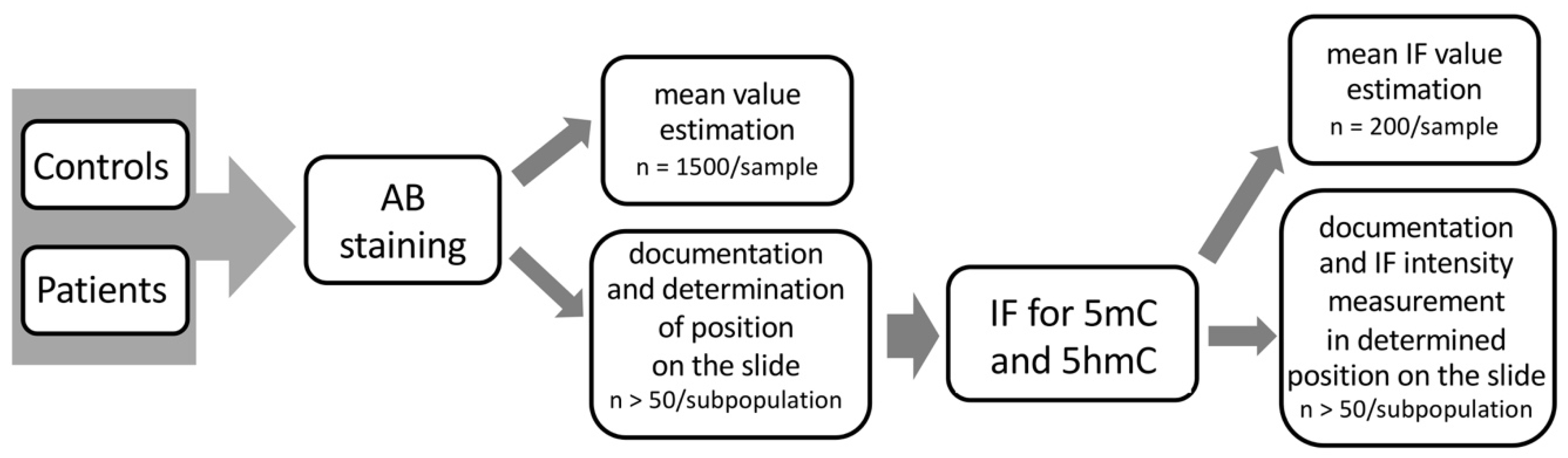
4.4. Immunofluorescence (IF)
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meyer, R.G.; Ketchum, C.C.; Meyer-Ficca, M.L. Heritable sperm chromatin epigenetics: A break to remember. Biol. Reprod. 2017, 97, 784–797. [Google Scholar] [CrossRef] [PubMed]
- Ben Maamar, M.; Sadler-Riegelman Beck, D.; Skinner, M.K. Epigenetic transgenerational inheritance of altered sperm histone retention sites. Sci Rep. 2018, 8, 5308. [Google Scholar] [CrossRef] [PubMed]
- Xavier, M.J.; Roman, S.D.; Aitken, R.J.; Nixon, B. Transgenerational inheritance: How impacts to the epigenetic and genetic information of parents affect offspring health. Hum. Reprod. Update 2019, 25, 519–541. [Google Scholar] [CrossRef] [PubMed]
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef]
- Bao, J.; Bedford, M.T. Epigenetic regulation of the histone–to–protamine transition during spermatogenesis. Reproduction 2016, 151, R55–R70. [Google Scholar] [CrossRef]
- Stewart, K.R.; Veselovska, L.; Kelsey, G. Establishment and functions of DNA methylation in the germline. Epigenomics 2016, 8, 1399–1413. [Google Scholar] [CrossRef]
- Luo, C.; Hajkova, P.; Ecker, J.R. Dynamic DNA methylation: In the right place at the right time. Science 2018, 361, 1336–1340. [Google Scholar] [CrossRef]
- Slieker, R.C.; Relton, C.L.; Gaunt, T.R.; Slagboom, P.E.; Heijmans, B.T. Age–related DNA methylation changes are tissue–specific with ELOVL2 promoter methylation as exception. Epigenet. Chromatin 2018, 11, 25. [Google Scholar] [CrossRef]
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 2013, 14, R115. [Google Scholar] [CrossRef]
- Barciszewska, A.M. Total DNA methylation as a biomarker of DNA damage and tumor malignancy in intracranial meningiomas. BMC Cancer 2020, 20, 509. [Google Scholar] [CrossRef]
- Monk, D.; Mackay, D.J.G.; Eggermann, T.; Maher, E.R.; Riccio, A. Genomic imprinting disorders: Lessons on how genome, epigenome and environment interact. Nat. Rev. Genet. 2019, 20, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; D’Alessio, A.C.; Taranova, O.V.; Hong, K.; Sowers, L.C.; Zhang, Y. Role of Tet proteins in 5mC to 5hmC conversion, ES–cell self–renewal and inner cell mass specification. Nature 2010, 466, 1129–1133. [Google Scholar] [CrossRef] [PubMed]
- Delatte, B.; Fuks, F. TET proteins: On the frenetic hunt for new cytosine modifications. Brief. Funct. Genom. 2013, 12, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Ecsedi, S.; Rodriguez-Aguilera, J.R.; Hernandez-Vargas, H. 5–hydroxymethylcytosine (5hmC), or how to identify your favorite cell. Epigenomes 2018, 2, 3. [Google Scholar] [CrossRef]
- Xu, T.; Gao, H. Hydroxymethylation and tumors: Can 5–hydroxymethylation be used as a marker for tumor diagnosis and treatment? Hum. Genom. 2020, 14, 15. [Google Scholar] [CrossRef]
- Guz, J.; Gackowski, D.; Foksinski, M.; Rozalski, R.; Olinski, R. Comparison of the absolute level of epigenetic marks 5-methylcytosine, 5-hydroxymethylcytosine, and 5-hydroxymethyluracil between human leukocytes and sperm. Biol. Reprod. 2014, 91, 55. [Google Scholar] [CrossRef]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5–methylcytosine to 5–hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef]
- Nabel, C.S.; Jia, H.; Ye, Y.; Shen, L.; Goldschmidt, H.L.; Stivers, J.T.; Znahg, Y.; Kohli, R.M. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat. Chem. Biol. 2012, 8, 751–758. [Google Scholar] [CrossRef]
- Szyf, M. The elusive role of 5’–hydroxymethylcytosine. Epigenomics 2016, 8, 1539–1551. [Google Scholar] [CrossRef]
- Allegrucci, C.; Thurston, A.; Lucas, E.; Young, L. Epigenetics and the germline. Reproduction 2005, 129, 137–149. [Google Scholar] [CrossRef]
- Hammoud, S.S.; Nix, D.A.; Zhang, H.; Purwar, J.; Carrell, D.T.; Cairns, B.R. Distinctive chromatin in human sperm packages genes for embryo development. Nature 2009, 460, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.-Q.; Lin, S.-L.; Zhao, Z.-H.; Sun, Q.-Y. Epigenetic dynamics and interplay during spermatogenesis and embryogenesis: Implications for male fertility and offspring health. Oncotarget 2017, 8, 53804–53818. [Google Scholar] [CrossRef] [PubMed]
- Mayer, W.; Niveleau, A.; Walter, J.; Fundele, R.; Haaf, T. Embryogenesis: Demethylation of the zygotic paternal genome. Nature. 2000, 403, 501–502. [Google Scholar] [CrossRef] [PubMed]
- Puri, D.; Dhawan, J.; Mishra, R.K. The paternal hidden agenda: Epigenetic inheritance through sperm chromatin. Epigenetics 2010, 5, 386–391. [Google Scholar] [CrossRef]
- Aston, K.I.; Punj, V.; Liu, L.; Carrell, D.T. Genome–wide sperm deoxyribonucleic acid methylation is altered in some men with abnormal chromatin packaging or poor in vitro fertilization embryogenesis. Fertil. Steril. 2012, 97, 285–292. [Google Scholar] [CrossRef]
- Carrell, D.T. Epigenetics of the male gamete. Fertil. Steril. 2012, 97, 267–274. [Google Scholar] [CrossRef]
- Castillo, J.; Jodar, M.; Oliva, R. The contribution of human sperm proteins to the development and epigenome of the preimplantation embryo. Hum. Reprod. Update 2018, 24, 535–555. [Google Scholar] [CrossRef]
- Mudrak, O.; Tomilin, N.; Zalensky, A. Chromosome architecture in the decondensing human sperm nucleus. J. Cell. Sci. 2005, 118, 4541–4550. [Google Scholar] [CrossRef]
- Vavouri, T.; Lehner, B. Chromatin organization in sperm may be the major functional consequence of base composition variation in the human genome. PLoS Genet. 2011, 7, e1002036. [Google Scholar] [CrossRef]
- De Vries, M.; Ramos, L.; Housein, Z.; De Boer, P. Chromatin remodelling initiation during human spermiogenesis. Biol. Open 2012, 1, 446–457. [Google Scholar] [CrossRef]
- Venkatesh, S.; Workman, J.L. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell. Biol. 2015, 16, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Champroux, A.; Cocquet, J.; Henry-Berger, J.; Drevet, J.R.; Kocer, A.A. Decade of Exploring the Mammalian Sperm Epigenome: Paternal Epigenetic and Transgenerational Inheritance. Front. Cell. Dev. Biol. 2018, 6, 50. [Google Scholar] [CrossRef]
- Wang, T.; Gao, H.; Li, W.; Liu, C. Essential role of histone replacement and modifications in male fertility. Front. Genet. 2019, 10, 962. [Google Scholar] [CrossRef] [PubMed]
- Denomme, M.M.; McCallie, B.R.; Parks, J.C.; Schoolcraft, W.B.; Katz-Jaffe, M.G. Alterations in the sperm histone–retained epigenome are associated with unexplained male factor infertility and poor blastocyst development in donor oocyte IVF cycles. Hum. Reprod. 2017, 32, 2443–2455. [Google Scholar] [CrossRef] [PubMed]
- Brunner, A.M.; Nanni, P.; Mansuy, I.M. Epigenetic marking of sperm by post–translational modification of histones and protamines. Epigenet. Chromatin 2014, 7, 2. [Google Scholar] [CrossRef]
- Sillaste, G.; Kaplinski, L.; Meier, R.; Jaakma, U.; Eriste, E. A novel hypothesis for histone–to–protamine transition in Bos taurus spermatozoa. Reproduction 2017, 153, 241–251. [Google Scholar] [CrossRef]
- Hammoud, S.S.; Purwar, J.; Pflueger, C.; Cairns, B.R.; Carrell, D.T. Alterations in sperm DNA methylation patterns at imprinted loci in two classes of infertility. Fertil. Steril. 2010, 94, 1728–1733. [Google Scholar] [CrossRef]
- Nanassy, L.; Carrell, D.T. Analysis of the methylation pattern of six gene promoters in sperm of men with abnormal protamination. Asian J. Androl. 2011, 13, 342–346. [Google Scholar] [CrossRef]
- Hamad, M.F. Quantification of histones and protamines mRNA tran–scripts in sperms of infertile couples and their impact on sperm’s quality and chromatin integrity. Reprod. Biol. 2019, 19, 6–13. [Google Scholar] [CrossRef]
- Rajabi, H.; Mohseni-Kouchesfehani, H.; Mohammadi-Sangcheshmen, A.; Farifteh-Nobijari, F.; Salehi, M. Pronuclear epigenetic modification of protamine deficient human sperm following injection into mouse oocytes. Syst. Biol. Reprod. Med. 2016, 62, 125–132. [Google Scholar] [CrossRef]
- Menezo, Y.J.R.; Silvestris, E.; Dale, B.; Elder, K. Oxidative stress and alterations in DNA methylation: Two sides of the same coin in reproduction. Reprod. BioMed. Online 2016, 33, 668–683. [Google Scholar] [CrossRef]
- Olszewska, M.; Barciszewska, M.Z.; Fraczek, M.; Huleyuk, N.; Chernykh, V.B.; Zastavna, D.; Barciszewski, J.; Kurpisz, M. Global methylation status of sperm DNA in carriers of chromosome structural aberrations. Asian J. Androl. 2017, 19, 117–124. [Google Scholar] [CrossRef]
- Doshi, T.; Mehta, S.S.; Dighe, V.; Balasinor, N.; Vanage, G. Hypermethylation of estrogen receptor promoter region in adult testis of rats exposed neonatally to bisphenol A. Toxicology 2011, 289, 74–82. [Google Scholar] [CrossRef]
- Benchaib, M.; Ajina, M.; Lornage, J.; Niveleau, A.; Durand, P.; Guerin, J.F. Quantitation by image analysis of global DNA methylation in human spermatozoa and its prognostic value in in vitro fertilization: A preliminary study. Fertil. Steril. 2003, 80, 947–953. [Google Scholar] [CrossRef]
- Benchaib, M.; Braun, V.; Ressnikof, D.; Lornage, J.; Durand, P.; Niveleau, A.; Guerin, J.F. Influence of global sperm DNA methylation on IVF results. Hum. Reprod. 2005, 20, 768–773. [Google Scholar] [CrossRef]
- El Hajj, N.; Haaf, T. Epigenetic disturbances in in vitro cultured gametes and embryos: Implications for human assisted reproduction. Fertil. Steril. 2013, 3, 632–641. [Google Scholar] [CrossRef]
- Feinberg, J.I.; Bakulski, K.M.; Jaffe, A.E.; Tryggvadottir, R.; Brown, S.C.; Goldman, L.R.; Croen, L.A.; Hertz-Picciotto, I.; Newschaffer, C.J.; Fallin, M.D.; et al. Paternal sperm DNA methylation associated with early signs of autism risk in an autism–enriched cohort. Int. J. Epidemiol. 2015, 44, 1199–1210. [Google Scholar] [CrossRef]
- Stuppia, L.; Franzago, M.; Ballerini, P.; Gatta, V.; Antonucci, I. Epigenetics and male reproduction: The consequences of paternal lifestyle on fertility, embryo development, and children lifetime health. Clin. Epigenet. 2015, 7, 120. [Google Scholar] [CrossRef]
- White, C.R.; Denomme, M.M.; Tekpetey, F.R.; Feyles, V.; Power, S.G.A.; Mann, M.R.W. High frequency of imprinted methylation errors in human preimplantation embryos. Sci. Rep. 2015, 5, 17311. [Google Scholar] [CrossRef]
- Miller, D.; Brinkworth, M.; Iles, D. Paternal DNA packaging in spermatozoa: More than the sum of its parts? DNA, histones, protamines and epigenetics. Reproduction 2010, 139, 287–301. [Google Scholar] [CrossRef]
- Zini, A. Are sperm chromatin and DNA defects relevant in the clinic? Syst. Biol. Reprod. Med. 2011, 57, 78–85. [Google Scholar] [CrossRef]
- Aberg, K.A.; McClay, J.L.; Nerella, S.; Clark, S.; Kumar, G.; Chen, W.; Khachane, A.N.; Xie, L.; Hudson, A.; Gao, G.; et al. Methylome-wide association study of schizophrenia: Identifying blood biomarker signatures of environmental insults. JAMA Psychiatry 2014, 71, 255–264. [Google Scholar] [CrossRef]
- Besaratinia, A.; Tommasi, S. Epigenetics of human melanoma: Promises and challenges. J. Mol. Cell. Biol. 2014, 6, 356–367. [Google Scholar] [CrossRef][Green Version]
- Bennett, D.A.; Yu, L.; Yang, J.; Srivastava, G.P.; Aubin, C.; De Jager, P.L. Epigenomics of Alzheimer’s disease. Transl. Res. 2015, 165, 200–220. [Google Scholar] [CrossRef]
- Küçükali, C.I.; Kürtüncü, M.; Çoban, A.; Çebi, M.; Tüzün, E. Epigenetics of multiple sclerosis: An updated review. Neuromolecular Med. 2015, 17, 83–96. [Google Scholar] [CrossRef]
- Wei, Y.; Schatten, H.; Sun, Q.-Y. Environmental epigenetic inheritance through gametes and implications for human reproduction. Hum. Reprod. Update 2015, 21, 194–208. [Google Scholar] [CrossRef]
- Ioannou, D.; Miller, D.; Griffin, D.K.; Tempest, H. Impact of sperm DNA chromatin in the clinic. J. Assist. Reprod. Genet. 2016, 33, 157–166. [Google Scholar] [CrossRef]
- Schutte, B.; El Hajj, N.; Kuhtz, J.; Nanda, I.; Gromoll, J.; Hahn, T.; Dittrich, M.; Schorsch, M.; Müller, T.; Haaf, T. Broad DNA methylation changes of spermatogenesis, inflammation and subgroup of sperm samples for assisted reproduction. Andrology 2013, 1, 822–829. [Google Scholar] [CrossRef]
- Camprubi, C.; Salas-Huetos, A.; Aiese-Cigliano, R.; Godo, A.; Pons, M.-C.; Castellano, G.; Grossmann, M.; Sanseverino, W.; Martin-Subero, J.I.; Garrido, N.; et al. Spermatozoa from infertile patients exhibit differences of DNA methylation associated with spermatogenesis–related processes: An array–based analysis. Reprod. Biomed. Online 2016, 33, 709–719. [Google Scholar] [CrossRef]
- De Geyter, C.; Calhaz-Jorge, C.; Kupka, M.S.; Wyns, C.; Mocanu, E.; Motrenko, T.; Scaravelli, G.; Smeenk, J.; Vidakovic, S.; Goossens, V.; et al. The European IVF–monitoring Consortium (EIM) for the European Society of Human Reproduction and Embryology (ESHRE) (2018) ART in Europe, 2014: Results generated from European registries by ESHRE. Hum. Reprod. 2018, 33, 1586–1601. [Google Scholar] [CrossRef]
- Wyns, C.; Bergh, C.; Calhaz-Jorge, C.; De Geyter, C.; Kupka, M.S.; Motrenko, T.; Rugescu, I.; Smeenk, J.; Tandler-Schneider, A.; Vidakovic, S.; et al. The European IVF–monitoring Consortium (EIM) for the European Society of Human Reproduction and Embryology (ESHRE) (2020) ART in Europe, 2016: Results generated from European registries by ESHRE. Hum. Reprod. Open 2020, 3, hoaa032. [Google Scholar] [CrossRef]
- Kobayashi, H.; Hiura, H.; John, R.M.; Sato, A.; Otsu, E.; Kobayashi, N.; Suzuki, R.; Suzuki, F.; Hayashi, C.; Utsunomiya, T.; et al. DNA methylation errors at imprinted loci after assisted conception originate in the parental sperm. Eur. J. Hum. Genet. 2009, 17, 1582–1591. [Google Scholar] [CrossRef]
- Depa-Martynow, M.; Kempisty, B.; Jagodzinski, P.P.; Pawelczyk, L.; Jedrzejczak, P. Impact of protamine transcripts and their proteins on the quality and fertilization ability of sperm and the development of preimplantation embryos. Reprod. Biol. 2012, 12, 57–72. [Google Scholar] [CrossRef]
- Jenkins, T.G.; Aston, K.I.; Cairns, B.R.; Carrell, D.T. Paternal aging and associated intraindividual alterations of global sperm 5-methylcytosine and 5-hydroxymethylcytosine levels. Fertil. Steril. 2013, 100, 945–951. [Google Scholar] [CrossRef]
- Jenkins, T.G.; Aston, K.I.; Trost, C.; Farley, J.; Hotaling, J.M.; Carrell, D.T. Intra–sample heterogeneity of sperm DNA methylation. Mol. Hum. Reprod. 2015, 21, 313–319. [Google Scholar] [CrossRef]
- Jenkins, T.G.; Aston, K.I.; Hotaling, J.M.; Shamsi, M.B.; Simon, L.; Carrell, D.T. Teratozoospermia and asthenozoospermia are associated with specific epigenetic signatures. Andrology. 2016, 4, 843–849. [Google Scholar] [CrossRef]
- Jenkins, T.G.; Aston, K.I.; Meyer, T.D.; Hotaling, J.M.; Shamsi, M.B.; Johnstone, E.B.; Cox, K.J.; Stanford, J.B.; Porucznik, C.; Carrell, D.T. Decreased fecundity and sperm DNA methylation patterns. Fertil. Steril. 2016, 105, 51–57.e1–3. [Google Scholar] [CrossRef]
- Jenkins, T.G.; Aston, K.I.; Cairns, B.; Smith, A.; Carrell, D.T. Paternal germ line aging: DNA methylation age prediction from human sperm. BMC Genom. 2018, 19, 763. [Google Scholar] [CrossRef]
- Botezatu, A.; Socolov, R.; Socolov, D.; Iancu, I.V.; Anton, G. Methylation pattern of methylene tetrahydrofolate reductase and small nuclear ribonucleoprotein polypeptide N promoters in oligoasthenospermia: A case–control study. Reprod. Biomed. Online 2014, 28, 225–231. [Google Scholar] [CrossRef]
- Montjean, D.; Zini, A.; Ravel, C.; Belloc, S.; Dalleac, A.; Copin, H.; Boyer, P.; McElreavey, K.; Benkhalifa, M. Sperm global DNA methylation level: Association with semen parameters and genome integrity. Andrology 2015, 3, 235–240. [Google Scholar] [CrossRef]
- Laqqan, M.; Solomayer, E.-F.; Hammadeh, M. Aberrations in sperm DNA methylation patterns are associated with abnormalities in semen parameters of subfertile males. Reprod. Biol. 2017, 17, 246–251. [Google Scholar] [CrossRef]
- Laqqan, M.; Tierling, S.; Alkhaled, Y.; LoPorto, C.; Hammadeh, M.E. Alterations in sperm DNA methylation patterns of oligospermic males. Reprod. Biol. 2017, 17, 396–400. [Google Scholar] [CrossRef]
- Krausz, C.; Riera-Escamilla, A. Genetics of male infertility. Nat. Rev. Urol. 2018, 15, 369–384. [Google Scholar] [CrossRef]
- Montjean, D.; Ravel, C.; Benkhalifa, M.; Cohen-Bacrie, P.; Berthaut, I.; Bashamboo, A.; McElreavey, K. Methylation changes in mature sperm deoxyribonucleic acid from oligozoospermic men: Assessment of genetic variants and assisted reproductive technology outcome. Fertil. Steril. 2013, 100, 1241–1247. [Google Scholar] [CrossRef]
- Wu, W.; Shen, O.; Qin, Y.; Niu, X.; Lu, C.; Xia, Y.; Song, L.; Wang, S.; Wang, X. Idiopathic male infertility is strongly associated with aberrant promoter methylation of methylenetetrahydrofolate reductase (MTHFR). PLoS ONE. 2010, 5, e13884. [Google Scholar] [CrossRef]
- Kitamura, A.; Miyauchi, N.; Hamada, H.; Hiura, H.; Chiba, H.; Okae, H.; Sato, A.; John, R.M.; Arima, T. Epigenetic alterations in sperm associated with male infertility. Congenit. Anom. 2015, 55, 133–144. [Google Scholar] [CrossRef]
- Sujit, K.M.; Sarkar, S.; Singh, V.; Pandey, R.; Agrawal, N.K.; Trivedi, S.; Singh, K.; Gupta, G.; Rajender, S. Genome–wide differential methylation analyses identifies methylation signatures of male infertility. Hum. Reprod. 2018, 33, 2256–2267. [Google Scholar] [CrossRef]
- Luján, S.; Caroppo, E.; Niederberger, C.; Arce, J.-C.; Sadler-Riggleman, I.; Beck, D.; Nilsson, E.; Skinner, M.K. Sperm DNA Methylation Epimutation Biomarkers for Male Infertility and FSH Therapeutic Responsiveness. Sci. Rep. 2019, 9, 16786. [Google Scholar] [CrossRef]
- Wang, X.; Suo, Y.; Yin, R.; Shen, H.; Wang, H. Ultra-performance liquid chromatography/tandem mass spectrometry for accurate quantification of global DNA methylation in human sperms. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2011, 879, 1647–1652. [Google Scholar] [CrossRef]
- Barzideh, J.; Scott, R.J.; Aitken, R.J. Analysis of the global methylation status of human spermatozoa and its association with the tendency of these cells to enter apoptosis. Andrologia 2012, 45, 424–429. [Google Scholar] [CrossRef]
- Milekic, M.H.; Xin, Y.; O’Donnell, A.; Kumar, K.K.; Bradley-Moore, M.; Malaspina, D.; Moore, H.; Brunner, D.; Ge, Y.; Edwards, J.; et al. Age-related sperm DNA methylation changes are transmitted to offspring and associated with abnormal behavior and dysregulated gene expression. Mol. Psychiatry 2015, 20, 995–1001. [Google Scholar] [CrossRef]
- Jenkins, T.G.; Aston, K.I.; James, E.R.; Carrell, D.T. Sperm epigenetics in the study of male infertility, offspring health, and potential clinical applications. Syst. Biol. Reprod. Med. 2017, 63, 69–76. [Google Scholar] [CrossRef]
- Soubry, A.; Guo, L.; Huang, Z.; Hoyo, C.; Romanus, S.; Price, T.; Murphy, S.K. Obesity–related DNA methylation at imprinted genes in human sperm: Results from the TIEGER study. Clin. Epigenet. 2016, 8, 51. [Google Scholar] [CrossRef]
- Skinner, M.K.; Ben Maamar, M.; Sadler-Riggleman, I.; Beck, D.; Nilsson, E.; McBirney, M.; Klukovich, R.; Xie, Y.; Tang, C.; Yan, W. Alterations in sperm DNA methylation, non–coding RNA and histone retention associate with DDT–induced epigenetic transgenerational inheritance of disease. Epigenet. Chromatin 2018, 11, 8. [Google Scholar] [CrossRef]
- Potabattula, R.; Dittrich, M.; Schorsch, M.; Hahn, T.; Haaf, T.; El Hajj, N. Male obesity effects on sperm and next–generation cord blood DNA methylation. PLoS ONE 2019, 14, e0218615. [Google Scholar] [CrossRef]
- Zhang, W.; Li, M.; Sun, F.; Xu, X.; Zhang, Z.; Liu, J.; Sun, X.; Zhang, A.; Shen, Y.; Xu, J.; et al. Association of Sperm Methylation at LINE–1, Four Candidate Genes, and Nicotine/Alcohol Exposure With the Risk of Infertility. Front. Genet. 2019, 10, 1001. [Google Scholar] [CrossRef]
- Aston, K.I.; Uren, P.J.; Jenkins, T.G.; Horsager, A.; Cairns, B.R.; Smith, A.D.; Carrell, D.T. Aberrant sperm DNA methylation predicts male fertility status and embryo quality. Fertil. Steril. 2015, 104, 1388–1397.e5. [Google Scholar] [CrossRef]
- Urdinguio, R.G.; Bayon, G.F.; Dmitrijeva, M.; Torano, E.G.; Bravo, C.; Fraga, M.F.; Bassas, L.; Larriba, S.; Fernandez, A.F. Aberrant DNA methylation patterns of spermatozoa in men with unexplained infertility. Hum. Reprod. 2015, 30, 1014–1028. [Google Scholar] [CrossRef]
- Rogenhofer, N.; Dansranjavin, T.; Schorsch, M.; Spiess, A.; Wang, H.; von Schönfeldt, W.; Capallo-Obermann, H.; Baukloh, V.; Yang, H.; Paradowska, A.; et al. The sperm protamine mRNA ratio as a clinical parameter to estimate the fertilizing potential of men taking part in an ART programme. Hum. Reprod. 2013, 28, 969–978. [Google Scholar] [CrossRef]
- Mengual, L.; Oriola, J.; Ascaso, C.; Ballescà, J.L.; Oliva, R. An increased CAG repeat length in the androgen receptor gene in azoospermic ICSI candidates. J. Androl. 2003, 24, 279–284. [Google Scholar] [CrossRef]
- Aoki, V.W.; Emery, B.R.; Carrell, D.T. Global sperm deoxyribonucleic acid methylation is unaffected in protamine–deficient infertile males. Fertil. Steril. 2006, 86, 1541–1543. [Google Scholar] [CrossRef]
- Bahreinian, M.; Tavalaee, M.; Abbasi, H.; Kiani-Esfahani, A.; Shiravi, A.H.; Nasr-Esfahani, M.H. DNA hypomethylation predisposes sperm to DNA damage in individuals with varicocele. Syst. Biol. Reprod. Med. 2015, 61, 179–186. [Google Scholar] [CrossRef]
- Rahiminia, T.; Farashahi Yazd, E.; Ghasemi-Esmailabad, S.; Talebi, A.R. Relation between sperm protamine transcripts with global sperm DNA methylation and sperm DNA methyltransferases mRNA in men with severe sperm abnormalities. Hum. Fertil. Online 2021, 24, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Ribas–Maynou, J.; Abad, C.; Garcia-Segura, S.; Oliver-Bonet, M.; Prada, E.; Amengual, M.J.; Navarro, J.; Benet, J. Sperm chromatin condensation and single– and double–stranded DNA damage as important parameters to define male factor related recurrent miscarriage. Mol. Reprod. Dev. 2020, 87, 1126–1132. [Google Scholar] [CrossRef]
- Amor, H.; Shelko, N.; Hamad, M.F.; Zeyad, A.; Hammadeh, M.E. An additional marker for sperm DNA quality evaluation in spermatozoa of male partners of couples undergoing assisted reproduction technique (IVF/ICSI): Protamine ratio. Andrologia 2019, 51, e13400. [Google Scholar] [CrossRef] [PubMed]
- Berg, E.; Houska, P.; Nesheim, N.; Schuppe, H.-C.; Pilatz, A.; Fijak, M.; Manthey, M.; Steger, K.; Wagenlehner, F.; Schagdarsurengin, U. Chronic prostatitis/chronić pelvic pain syndrome leads to impaired semen parameters, increased sperm DNA fragmentation and unfavorable changes of sperm protamine mRNA ratio. Int. J. Mol. Sci. 2021, 22, 7854. [Google Scholar] [CrossRef]
- Marques, C.J.; Costa, P.; Vaz, B.; Carvalho, F.; Fernandes, S.; Barros, A.; Sousa, M. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol. Hum. Reprod. 2008, 14, 67–73. [Google Scholar] [CrossRef]
- Molaro, A.; Hodges, E.; Fang, F.; Song, Q.; McCombie, R.W.; Hannon, G.J.; Smith, A.D. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell 2011, 146, 1029–1041. [Google Scholar] [CrossRef]
- Darbandi, M.; Darbandi, S.; Agarwal, A.; Baskaran, S.; Dutta, S.; Sengupta, P.; Khorshid, H.R.K.; Esteves, S.; Gilany, K.; Hedayati, M.; et al. Reactive oxygen species–induced alterations in H19–IGF2 methylation patterns, seminal plasma metabolites, and semen quality. J. Assist. Reprod. Genet. 2019, 36, 241–253. [Google Scholar] [CrossRef]
- Valinluck, V.; Tsai, H.-H.; Rogstad, D.K.; Burdzy, A.; Bird, A.; Sowers, L.C. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2). Nucleic Acids Res. 2004, 32, 4100–4108. [Google Scholar] [CrossRef]
- Tavalaee, M.; Razavi, S.; Nasr-Esfahani, M.H. Influence of sperm chromatin anomalies on assisted reproductive technology outcome. Fertil. Steril. 2009, 91, 1119–1126. [Google Scholar] [CrossRef] [PubMed]
- Tunc, O.; Tremellen, K. Oxidative DNA damage impairs global sperm DNA methylation in infertile men. J. Assist. Reprod. Genet. 2009, 26, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Gharagozloo, P.; Aitken, R.J. The role of sperm oxidative stress in male infertility and the significance of oral antioxidant therapy. Hum. Reprod. 2001, 7, 1628–1640. [Google Scholar] [CrossRef]
- Brugnon, F.; Van Assche, E.; Verheyen, G.; Sion, B.; Boucher, D.; Pouly, J.L.; Janny, L.; Devroey, P.; Liebaers, I.; Van Steirteghem, A. Study of two markers of apoptosis and meiotic segregation in ejaculated sperm of chromosomal translocation carrier patients. Hum. Reprod. 2006, 21, 685–693. [Google Scholar] [CrossRef][Green Version]
- Perrin, A.; Caer, E.; Oliver-Bonet, M.; Navarro, J.; Benet, J.; Amice, V.; De Braekeleer, M.; Morel, F. DNA fragmentation and meiotic segregation in sperm of carriers of a chromosomal structural abnormality. Fertil. Steril. 2009, 92, 583–589. [Google Scholar] [CrossRef]
- Nicopoullos, J.; Vicens-Morton, A.; Lewis, S.E.M.; Lee, K.; Larsen, P.; Ramsay, J.; Yap, T.; Minhas, S. Novel use of COMET parameters of sperm DNA damage may increase its utility to diagnose male infertility and predict live births following both IVF and ICSI. Hum. Reprod. 2019, 34, 1915–1923. [Google Scholar] [CrossRef]
- Kobayashi, H.; Sato, A.; Otsu, E.; Hiura, H.; Tomatsu, C.; Utsunomiya, T.; Sasaki, H.; Yaegashi, N.; Arima, T. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum. Mol. Genet. 2007, 16, 2542–2551. [Google Scholar] [CrossRef]
- Kobayashi, N.; Miyauchi, N.; Tatsua, N.; Kitamura, A.; Okae, H.; Hiura, H.; Sato, A.; Utsunomiya, T.; Yaegashi, N.; Nakai, K.; et al. Factors associated with aberrant imprint methylation and oligozoospermia. Sci. Rep. 2017, 7, 42336. [Google Scholar] [CrossRef]
- Yu, B.; Zhou, H.; Liu, M.; Zheng, T.; Jiang, L.; Zhao, M.; Xu, X.; Huang, Z. Epigenetic alterations in density selected human spermatozoa for assisted reproduction. PLoS ONE. 2015, 10, e0145585. [Google Scholar] [CrossRef]
- Cassuto, N.G.; Montjean, D.; Siffroi, J.-P.; Bouret, D.; Marzouk, F.; Copin, H.; Benkhalifa, M. Different levels of DNA methylation detected in human sperms after morphological selection using high magnification microscopy. Biomed. Res. Int. 2016, 2016, 6372171. [Google Scholar] [CrossRef]
- Du, Y.; Li, M.; Chen, J.; Duan, Y.; Wang, X.; Qiu, Y.; Cai, Z.; Gui, Y.; Jiang, H. Promoter targeted bisulfite sequencing reveals DNA methylation profiles associated with low sperm motility in asthenozoospermia. Hum. Reprod. 2016, 31, 24–33. [Google Scholar] [CrossRef]
- Louie, K.; Minor, A.; Ng, R.; Poon, K.; Chow, V.; Ma, S. Evaluation of DNA methylation at imprinted DMRs in the spermatozoa of oligozoospermic men in association with MTHFR C677T genotype. Andrology 2016, 4, 825–831. [Google Scholar] [CrossRef]
- Xu, A.; Hua, Y.; Zhang, J.; Chen, W.; Zhao, K.; Xi, W.; Wang, H.; Fang, J.; Su, S.; Tang, M.; et al. Abnormal hypermethylation of the VDAC2 promoter is a potential cause of idiopathic asthenospermia in men. Sci. Rep. 2016, 6, 37836. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, A.; Zhang, Z.; Wang, P.; Qian, Y.; He, L.; Shi, H.; Xing, Q.; Du, J. DNA methylation levels of imprinted and nonimprinted genes DMRs associated with defective human spermatozoa. Andrologia 2016, 48, 1027–1035. [Google Scholar] [CrossRef]
- Santi, D.; De Vincentis, S.; Magnani, E.; Spaggiari, G. Impairment of sperm DNA methylation in male infertility: A meta–analytic study. Andrology 2017, 5, 695–703. [Google Scholar] [CrossRef]
- Tang, Q.; Pan, F.; Yang, J.; Fu, Z.; Lu, Y.; Wu, X.; Han, X.; Chen, M.; Lu, C.; Xia, Y.; et al. Idiopathic male infertility is strongly associated with aberrant DNA methylation of imprinted loci in sperm: A case–control study. Clin. Epigenet. 2018, 10, 134. [Google Scholar] [CrossRef]
- Efimova, O.A.; Pendina, A.A.; Tikhonov, A.V.; Parfenyev, S.E.; Mekina, I.D.; Komarova, E.M.; Mazilina, M.A.; Daev, E.V.; Chiryaeva, O.G.; Galembo, I.A.; et al. Genome–wide 5–hydroxymethylcytosine patterns in human spermatogenesis are associated with semen quality. Oncotarget 2017, 8, 88294–88307. [Google Scholar] [CrossRef]
- Pacheco, S.E.; Houseman, E.A.; Christensen, B.C.; Marsit, C.J.; Kelsey, K.T.; Sigman, M.; Boekelheide, K. Integrative DNA methylation and gene expression analyses identify DNA packaging and epigenetic regulatory genes associated with low motility sperm. PLoS ONE. 2011, 6, e20280. [Google Scholar] [CrossRef]
- Kläver, R.; Tüttelmann, F.; Bleiziffer, A.; Haaf, T.; Kliesch, S.; Gromoll, J. DNA methylation in spermatozoa as a prospective marker in andrology. Andrology 2013, 1, 731–740. [Google Scholar] [CrossRef]
- Gunes, S.; Agarwal, A.; Henkel, R.; Mahmutoglu, A.M.; Sharma, R.; Esteves, S.C.; Aljowair, A.; Emirzeoglu, D.; Alkhani, A.; Pelegrini, L.; et al. Association between promoter methylation of MLH1 and MSH2 and reactive oxygen species in oligozoospermic men—A pilot study. Andrologia 2018, 50, e12903. [Google Scholar] [CrossRef]
- Nanassy, L.; Carrell, D.T. Abnormal methylation of the promoter of CREM is broadly associated with male factor infertility and poor sperm quality but is improved in sperm selected by density gradient centrifugation. Fertil. Steril. 2011, 95, 2310–2314. [Google Scholar] [CrossRef] [PubMed]
- Laurentino, S.; Beygo, J.; Nordhoff, V.; Kliesch, S.; Wistuba, J.; Borgmann, J.; Buiting, K.; Horsthemke, B.; Gromoll, J. Epigenetic germline mosaicism in infertile men. Hum. Mol. Genet. 2015, 24, 1295–1304. [Google Scholar] [CrossRef] [PubMed]
- Dere, E.; Huse, S.; Hwang, K.; Sigman, M.; Boekelheide, K. Intra– and inter–individual differences in human sperm DNA methylation. Andrology 2016, 4, 832–842. [Google Scholar] [CrossRef] [PubMed]
- Leitao, E.; Di Persio, S.; Laurentino, S.; Wöste, M.; Dugas, M.; Kliesch, S.; Neuhaus, N.; Horsthemke, B. The sperm epigenome does not display recurrent epimutations in patients with severely impaired spermatogenesis. Clin. Epigenet. 2020, 12, 61. [Google Scholar] [CrossRef]
- Stimpfel, M.; Vrtacnik-Bokal, E. Minor DNA methylation changes are observed in spermatozoa prepared using different protocols. Andrology 2020, 8, 1312–1323. [Google Scholar] [CrossRef]
- World Health Organization. WHO Laboratory Manual for the Examination and Processing of Human Semen, 5th ed.; World Health Organization: Geneva, Switzerland, 2010. [Google Scholar]
- Olszewska, M.; Fraczek, M.; Huleyuk, N.; Czernikiewicz, A.; Wiland, E.; Boksa, M.; Zastavna, D.; Panasiuk, B.; Midro, A.T.; Kurpisz, M. Chromatin structure analyzis of spermatozoa from reciprocal chromosome translocation carriers (RCT) with known meiotic segregation patterns. Reprod. Biol. 2013, 13, 209–220. [Google Scholar] [CrossRef]
- Olszewska, M.; Huleyuk, N.; Fraczek, M.; Zastavna, D.; Wiland, E.; Kurpisz, M. Sperm FISH and chromatin integrity in spermatozoa from t(6;10;11) carrier. Reproduction 2014, 147, 659–670. [Google Scholar] [CrossRef]
- Wiland, E.; Fraczek, M.; Olszewska, M.; Kurpisz, M. Topology of chromosome centromeres in human sperm nuclei with high levels of DNA damage. Sci. Rep. 2016, 6, 31614. [Google Scholar] [CrossRef]
- Selvam, M.K.P.; Agarwal, A. A systematic review on sperm DNA fragmentation in male factor infertility: Laboratory assessment. Arab. J. Urol. 2018, 16, 65–76. [Google Scholar] [CrossRef]
- Ribas–Maynou, J.; Garcia-Peiro, A.; Fernandez-Encinas, A.; Abad, C.; Amengual, M.J.; Prada, E.; Navarro, J.; Benet, J. Comperhensive analysis of sperm DNA fragmentation by five different assays: TUNEL assay, SCSA, SCD test and alkaline and neutral Comet assay. Andrology 2013, 1, 715–722. [Google Scholar] [CrossRef]
- Kim, S.K.; Jee, B.C.; Kim, S.H. Histone methylation and acetylation in ejaculated human sperm: Effects of swim-up and smoking. Fertil. Steril. 2015, 103, 1425–1431. [Google Scholar] [CrossRef] [PubMed]
| Control Group: Mean C ± SD | Patients’ Group: Mean P ± SD | |||
|---|---|---|---|---|
| SPERM CHROMATIN INTEGRITY | sperm chromatin protamination [%; aniline blue assay, AB] | protaminated [pink] | 82.02 ± 8.31 | 68.56 * ± 13.59 |
| semiprotaminated [purple] | 5.17 ± 4.21 | 9.86 ± 6.68 | ||
| deprotaminated [navy blue] | 12.82 ± 6.86 | 21.59 * ± 8.82 | ||
| sum of semi- and deprotaminated | 17.98 ± 8.31 | 31.44 * ± 13.59 | ||
| spermatozoa with fragmented DNA [%; TUNEL assay] | 5.52 ± 2.62 | 9.55 ± 6.29 | ||
| EPI-MARKS | in unfractionated total sperm population | mean 5mC [IF i.u.] | 75.61 ± 10.66 | 71.32 ± 30.82 |
| mean 5hmC [IF i.u.] | 126.33 ± 13.17 | 109.03 ± 22.74 | ||
| 5mC/5hmC ratio | 0.60 ± 0.07 | 0.64 ± 0.20 | ||
| Subpopulation | Control Group: Mean C ± SD | Patients’ Group: Mean P ± SD | |
|---|---|---|---|
| 5mC [IF i.u.] | protaminated [pink] | 120.28 ± 19.65 | 87.29 * ± 36.01 |
| semiprotaminated [purple] | 58.92 ± 15.41 | 59.54 ± 26.34 | |
| deprotaminated [navy blue] | 37.90 ± 8.84 | 42.73 ± 20.11 | |
| mean of semi- and deprotaminated | 48.41 ± 10.23 | 51.13 ± 22.72 | |
| 5hmC [IF i.u.] | protaminated [pink] | 134.26 ± 15.12 | 121.20 ± 23.42 |
| semiprotaminated [purple] | 110.85 ± 11.48 | 100.76 ± 24.95 | |
| deprotaminated [navy blue] | 98.71 ± 9.97 | 81.53 ± 22.39 | |
| mean of semi- and deprotaminated | 104.78 ± 10.51 | 91.15 ± 23.03 | |
| 5mC/5hmC ratio | protaminated [pink] | 0.90 ± 0.12 | 0.70 ** ± 0.23 |
| semiprotaminated [purple] | 0.53 ± 0.12 | 0.58 ± 0.20 | |
| deprotaminated [navy blue] | 0.38 ± 0.07 | 0.52 * ± 0.19 | |
| mean of semi- and deprotaminated | 0.46 ± 0.07 | 0.55 ± 0.19 |
| Control Group (C) | Sperm Chromatin Integrity [%] | Unfractionated Total Sperm Population | Properly Protaminated Subpopulation | Semen Parameters | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients’ Group (P) | Protaminated | Fragmented DNA | Glo-bal 5mC | Glo-bal 5hmC | 5mC/5hmC Ratio | Mean 5mC | Mean 5hmC | 5mC/5hmC Ratio | Concen-tration [×106/mL] | Volume [mL] | Total Count [×106] | Morphology [%] | Motility [%] | |||
| Progres-sive | Total | |||||||||||||||
| Sperm Chromatin Integrity [%] | Protaminated | ns | ** | * | * | ** | ** | ns | ns | ns | * | ns | ns | ns | ||
| Fragmented DNA | ns | ns | ns | ns | ns | ns | ns | ns | ns | ns | ns | ns | ns | |||
| Unfractionated Total Sperm Population | Global 5mC | ns | ns | ** | *** | ns | ns | ns | ns | ns | #ns | ns | ns | ns | ||
| Global 5hmC | ns | ns | *** | ns | ** | ns | * | ns | ns | ns | ns | ns | ns | |||
| 5mC/5hmC Ratio | ns | ns | *** | ** | ns | ns | ns | ns | ns | ns | ns | ns | ns | |||
| Properly Protaminated Subpopulation | Mean 5mC | ns | ns | *** | *** | *** | #ns | * | ns | ns | ns | ns | ns | #ns | ||
| Mean 5hmC | ns | ns | *** | *** | ** | *** | ns | #ns | ns | ns | ns | ns | ns | |||
| 5mC/5hmC Ratio | ns | ns | *** | ** | *** | *** | * | ns | ns | ns | ns | ns | #ns | |||
| Semen Parameters | Concentration [×106/mL] | ns | ns | ns | ns | ns | ns | ns | ns | * | *** | ns | * | * | ||
| Volume [mL] | ns | ns | ns | ns | ns | ns | ns | ns | ns | ns | ns | ns | ns | |||
| Total Count [×106] | ns | ns | ns | ns | ns | ns | ns | ns | *** | ** | ns | ns | ns | |||
| Morphology [%] | ns | ns | * | ns | ** | ** | ns | ** | ns | ns | #ns | * | ** | |||
| Motility [%] | Progressive | ns | ns | ns | ns | ns | ns | ns | ns | ns | ns | ns | ns | ** | ||
| Total | ns | ns | ns | ns | ns | ns | *** | ns | ns | ns | ns | ns | *** | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olszewska, M.; Kordyl, O.; Kamieniczna, M.; Fraczek, M.; Jędrzejczak, P.; Kurpisz, M. Global 5mC and 5hmC DNA Levels in Human Sperm Subpopulations with Differentially Protaminated Chromatin in Normo- and Oligoasthenozoospermic Males. Int. J. Mol. Sci. 2022, 23, 4516. https://doi.org/10.3390/ijms23094516
Olszewska M, Kordyl O, Kamieniczna M, Fraczek M, Jędrzejczak P, Kurpisz M. Global 5mC and 5hmC DNA Levels in Human Sperm Subpopulations with Differentially Protaminated Chromatin in Normo- and Oligoasthenozoospermic Males. International Journal of Molecular Sciences. 2022; 23(9):4516. https://doi.org/10.3390/ijms23094516
Chicago/Turabian StyleOlszewska, Marta, Oliwia Kordyl, Marzena Kamieniczna, Monika Fraczek, Piotr Jędrzejczak, and Maciej Kurpisz. 2022. "Global 5mC and 5hmC DNA Levels in Human Sperm Subpopulations with Differentially Protaminated Chromatin in Normo- and Oligoasthenozoospermic Males" International Journal of Molecular Sciences 23, no. 9: 4516. https://doi.org/10.3390/ijms23094516
APA StyleOlszewska, M., Kordyl, O., Kamieniczna, M., Fraczek, M., Jędrzejczak, P., & Kurpisz, M. (2022). Global 5mC and 5hmC DNA Levels in Human Sperm Subpopulations with Differentially Protaminated Chromatin in Normo- and Oligoasthenozoospermic Males. International Journal of Molecular Sciences, 23(9), 4516. https://doi.org/10.3390/ijms23094516








